3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(23):7201-7213. doi:10.7150/jca.58397 This issue Cite
Research Paper
INPP4B exerts a dual role in gastric cancer progression and prognosis
1. Department of General Surgery, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, People's Republic of China.
2. Department of Endoscopy Center, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, People's Republic of China.
3. Department of General Surgery, the Armed Police Corps Hospital of Anhui, Hefei 230041, People's Republic of China.
Received 2021-1-19; Accepted 2021-9-29; Published 2021-10-22
Abstract
Inositol polyphosphate 4-phosphatase type II (INPP4B) negatively regulates PI3K-Akt signalling and plays diverse roles in different types of cancer, but its role in gastric cancer (GC) is still unknown. Our study aimed to investigate the function and clinical relevance of INPP4B in GC. INPP4B expression was detected in GC tissues and nontumour tissues. The effect of INPP4B on the phenotypic changes of AGS and BGC-823 cells was investigated in vitro. The activation of serum and glucocorticoid-regulated kinase 3 (SGK3) and AKT were used to evaluate the specific mechanistic function of INPP4B in GC cells. The messenger RNA (mRNA) and protein expression levels of INPP4B were decreased in GC tissues compared with nontumour tissues. INPP4B expression was associated with tumour-node-metastasis (TNM) stage and histopathological differentiation. In addition, high INPP4B expression in GC patients with large tumour size/low-undifferentiated/TNM's III-IV stage was correlated with a poor prognosis but it was correlated with a better prognosis in patients with small tumour size/high-moderate differentiated/TNM's I-II stage patients. In addition, INPP4B knockdown inhibited proliferation, clonal formation and migration and promoted cell apoptosis in vitro, while INPP4B overexpression led to the opposite effects. Mechanistically, we found that INPP4B overexpression enhanced the phosphorylation of SGK3 (p-SGK3) in AGS cells, whereas INPP4B knockdown enhanced the p-Akt level in BGC823 cells. These findings suggested that the expression of INPP4B in GC is lower than that in normal tissues. Based on stratification survival analysis and in vitro cell experiments, INPP4B may play dual roles as an oncogene and tumour suppressor gene in different tissue grades and clinical stages.
Keywords: gastric cancer, INPP4B, tumour suppressor gene, oncogene, prognostic biomarker
Introduction
Gastric cancer (GC) is the fourth most common cancer and the second most lethal malignancy worldwide, resulting in 951,600 novel cases and 723,100 deaths in 2012 worldwide, especially in East Asian countries [1]. Due to the lack of effective early biomarkers, most GC patients are already in advanced stages with a poor prognosis when diagnosed. Therefore, it is imperative to identify additional molecular prognostic biomarkers for GC.
The phosphoinositide 3-kinase (PI3K)-Akt pathway is one of the most common dysregulated cancer-associated signalling pathways and it regulates many cellular functions, such as cell proliferation, growth, survival, migration and metabolism; targeting enzymes in this network has become a major goal of cancer drug development [2]. Hyperactivation of PI3K-Akt signalling frequently occurs in human cancers including GC, and it leads to an accumulation of phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] or phosphatidyl inositol (3,4)-bisphosphate [PI(3,4)P2], thus increasing the recruitment and activation of protein kinase Akt to the cytoplasmic membrane. It is the key mediator of PI3K signalling to enhance carcinogenesis [3]. However, the signalling lipids PI(3,4,5)P3 and PI(3,4)P2 are degraded by PTEN (phosphatase and tensin homolog) and INPP4B (inositol polyphosphate 4-phosphatase type II) [4, 5].
The tumour suppressor gene (TSG) PTEN is frequently deleted or inactivated in human cancers, including GC. It encodes a lipid phosphatase that dephosphorylates the 3-position phosphate from the inositol ring of PtdIns(3,4,5)P3 to form PtdIns(4,5)P2 to antagonize PI3K-Akt signalling [6]. INPP4B, another lipid phosphatase, dephosphorylates the 4-position phosphate from the inositol ring of PtdIns(3,4)P2 to form PtdIns(3)P. As both PI(3,4)P2 and PI(3,4,5)P3 are required for Akt recruitment to the plasma membrane and maximal Akt activation, INPP4B was predicted to act as a tumour suppressor by inhibiting PI3K-Akt signalling [7-9]. Indeed, increasing amounts of evidence have identified INPP4B as a potential TSG. LOH (loss of heterozygosity) at 4q31.21, the chromosomal region containing INPP4B, is reported to occur in basal-like breast tumours (55.6%), ovarian cancers (39.8%) and melanomas (21.6%) [10]. In addition, INPP4B protein expression is reduced in several cancers, and its low expression is associated with a poor prognosis [11-14]. In agreement with these findings, knockdown of INPP4B enhanced Akt activation, anchorage-independent growth, and proliferation in melanoma, breast cancer, and prostate cancer cell lines [11, 13, 15]. INPP4B, like PTEN, is anticipated to act as a tumour suppressor by antagonizing PI3K-Akt signalling. However, unlike PTEN, the underlying molecular mechanisms by which INPP4B exerts its tumour suppressive function are poorly understood.
More recently, an increasing number of studies have shown that INPP4B may be upregulated frequently and may play an oncogenic role in several types of cancers [16-18]. Even for the same tumour, different researchers have different views on the expression and role of INPP4B [18, 19]. In short, the role of INPP4B in oncogenesis and development remains controversial. The contradictory role of INPP4B in different tumours or the same tumour may reflect its extremely complex function in tumours. To date, however, the biological function of INPP4B in GC remains unexplored. To further understand the potential value of INPP4B in GC, detection of its expression level, evaluation of its clinical prognostic significance, and conducting in vitro functional studies are very useful.
In our study, we examined the expression of INPP4B in GC using qPCR, western blotting, and immunohistochemistry. We analysed correlations of INPP4B expression with clinicopathological factors by chi-square tests. Furthermore, the prognostic roles of INPP4B in GC were analysed using Cox regression and Kaplan-Meier analysis. Moreover, we examined the biological function of INPP4B in GC cell proliferation, apoptosis, colony formation and migration in vitro.
Materials and Methods
Ethics approval and consent to participate
This study obtained approval from the Institute Research Ethics Committee of the First Affiliated Hospital of Anhui Medical University, and written informed consent was obtained from all the patients enrolled in this study.
Patients and tissue specimens
In our study, to construct a tissue microarray (TMA), a total of 178 GC and 41 randomly selected normal gastric tissues were gathered from the First Affiliated Hospital of Anhui Medical University of Anhui (Hefei, China) from December 2006 to September 2008. All patients were followed up to December 2014 according to the following rules: regular follow-up every 3 months in the first 2 years and every 6 months in the following years.
Immunohistochemical staining and all of the patients' pathological features were confirmed by two experienced pathologists. Tumour node metastasis (TNM) staging was classified based on the 2010 7th edition of the American Joint Committee on Cancer (AJCC) TNM classification criteria. The detailed clinicopathologic parameters are described in Table 1.
Additionally, thirty-seven matched fresh GC tissues and adjacent nontumour tissue samples were collected and immediately frozen in liquid nitrogen and stored at -80°C from May to August 2019 until use for qPCR and western blotting analysis to explore the differential expression of INPP4B mRNA and protein between the GC and adjacent nontumour tissues. All diagnoses were histopathologically confirmed. The inclusion criteria were as follows: 1) histologically confirmed gastric cancer; 2) no preoperative anticancer treatment was given; 3) radical resection of primary lesion; 4) complete clinical information; 5) no other organ or system severe disorders or malignancies. Patients who did not meet the above criteria were excluded.
RNA preparation, reverse transcription and real-time qPCR
Total RNA was isolated from fresh tissues using TRIzol Reagent (Invitrogen), and reverse transcription (RT) was performed to obtain first-strand cDNA using ReverTra Ace qPCR RT Master Mix (Toyobo) according to the protocol supplied by the manufacturer. The PCR primers were as follows: INPP4B, forward primer 5′-ACGCAGGAAAGTCAGGCTAA-3′, reverse primer 5′-TGCCAGGTAACACCATTTCTT-3′; GAPDH was applied as the internal control: forward primer 5′-ATCAAGAAGGTGGTGAAGCAGG-3′, reverse primer 5′-CGTCAAAGGTGGAGGAGTGG-3′. qPCR was carried out using an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems, USA) in the presence of SYBR-Green mix (Toyobo, Japan). Amplification was carried out under the following conditions: denaturation program (95°C for 5 min), followed by an amplification and quantification program for 40 cycles (95°C for 15 s and 60°C for 45 s). Each sample was tested in triplicate. The relative expression of mRNA was calculated using the 2-ΔΔCT method.
Protein extraction and western blotting
Total proteins were extracted from fresh tissues and cell lines with RIPA lysis buffer containing a cocktail of protease inhibitors (Beyotime Institute of Biotechnology). The protein samples were then separated by 10% SDS-PAGE and electrotransferred to nitrocellulose membranes (Millipore). After blocking with 5% nonfat milk dilution with TBST for 1 h at room temperature, the membrane was incubated with primary antibodies against INPP4B, p-SGK3T320, SGK3, p-AKTT308, and AKT at 4°C overnight. After washing with TBST, the membranes were incubated with the secondary antibody (Cell Signaling Technology) at a dilution of 1:3000 at room temperature for 60 min. The membranes were then washed with TBST, and the bound antibodies were detected with an enhanced chemiluminescence system. Anti-GAPDH antibody (1:3000; Vazyme) was used as a loading control.
Immunohistochemistry (IHC)
We provided the tissue samples, and tissue microarray (TMA) manufacturing was done by Shanghai Biochip Company, Ltd., Shanghai, China. Briefly, the immunohistochemistry (IHC) process consisted of the following steps: baking, deparaffinization, antigen retrieval, quenching the endogenous peroxidase activity, blocking nonspecific staining, and incubation with the primary antibody (INPP4B, 1:50, Abcam) and the secondary antibody. The immunoreactivity score (IRS) was defined by “cell staining percentage×staining intensity”. The specimens were defined as negative (IRS =0~2) and positive (IRS=3~9). The detailed immunohistochemical process and staining evaluation were performed as previously described [20].
Cell culture and lentivirus infection
All cell lines used in this study were obtained from GeneChem (Shanghai, China). All cell lines were cultured in RPMI 1640 supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml) and maintained at 37°C in a humidified incubator with 5% CO2. To establish the INPP4B overexpression AGS cells, AGS cells were infected with INPP4B overexpression lentivirus (GV492-INPP4B, OE) and the negative control (GV492, NC, purchased from GeneChem, Shanghai, China). To interfere with INPP4B expression and to establish INPP4B knockdown BGC-823 cells, the shRNA target sequence for INPP4B was 5'-CCATCTGAGTATCCCATCTAT-3′. BGC-823 cells were infected with INPP4B shRNA lentivirus (GV248-shINPP4B, KD) and a negative control (GV248, NC, purchased from GeneChem, Shanghai, China). q-PCR and immunoblotting assays were used to estimate the efficiency of the overexpression and knockdown of the INPP4B gene.
Cell proliferation assay and clonogenic assay
Cell proliferation assays and clonogenic assays were used to evaluate the effect of INPP4B on the cell growth and proliferation capabilities of GC cells. These assays were performed as previously described [21]. For the cell proliferation assay, different lentivirus (OE, KD and NC)-infected cell lines (AGS and BGC-823) were seeded into 96-well plates (approximately 2000 cells/well) in sextuplicate and the cell proliferation assay was performed by MTT (Genview, JT343) according to the manufacturer's instructions. Each experiment was performed at least three times. For the colony formation assay, different lentivirus-infected cell lines were seeded into 6-well plates (approximately 800 cells) in triplicate. These cells were cultured for 14 days, and the medium was replaced every 3 days. Then, the cells were fixed with 4% polyoxymethylene and stained with crystal violet, and the colony numbers (more than 50 cells/colony) were counted using an inverted microscope. The experiments were performed independently in triplicate.
Apoptosis assay
For apoptosis analysis, different lentivirus-infected cell lines were used with an Annexin V-APC apoptosis detection kit (eBioscience, 88-8007) following the manufacturer's instructions. The apoptosis rate was detected by flow cytometry (BD, FACSCalibur). The detailed apoptotic analysis process was performed as previously described [22].
Scratch wound-healing assay and Transwell assay
Scratch wound-healing assays and Transwell assays were used to evaluate the cell migration ability and were performed as previously described [14, 23]. Briefly, for the scratch wound-healing assay, different lentivirus-infected cell lines were seeded in 96-well plates at approximately 5×105 cells/well. The next day, a scratch tester was used to cross the centre of the cell monolayer. The images were collected by a Celigo instrument, and the migration area of the cells was analysed by its software. For the Transwell assay, different lentivirus-infected cell lines (approximately 1×105 cells/well) in 200 μl serum-free medium were added to the upper chamber of 24-well plates with 8-μm pores, and then 600 μl RPMI-1640 containing 30% FBS was added to the lower chamber. After 24 h of incubation, the noninvading cells were removed by cotton swabs, and the invading cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet solution. Several random fields of low magnification (×100) and high magnification (×200) were photographed under a microscope. The migratory cell counts were performed using 200× magnification.
Statistical analysis
All statistical analyses were carried out with SPSS 16.0 software (SPSS Inc., Chicago, IL) and GraphPad Prism 5. Data are expressed as the mean ± SEM. Student's t-test and one-way analysis of variance (ANOVA) were used to evaluate the statistical significance of differences between two groups and multiple groups. The relationships between INPP4B expression and the clinicopathologic characteristics were analysed by chi-square tests. Overall survival (OS) was assessed by the Kaplan-Meier method, and statistical significance was analysed using the log-rank test. Cox regression analysis was used to estimate the independent risk factors for OS in GC patients after surgery. P<0.05 indicates statistical significance.
Results
Expression of INPP4B mRNA and protein in GC tissues
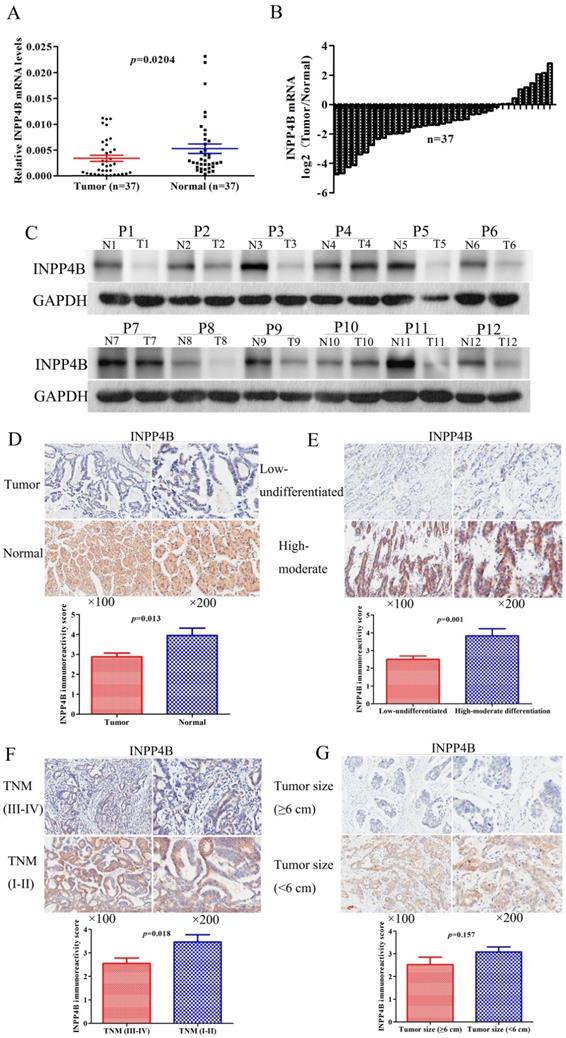
The expression level of INPP4B in tumours is controversial [24, 25]. The INPP4B expression pattern in GC is still unknown. qPCR and western blotting were used to investigate INPP4B mRNA and protein expression levels in paired fresh frozen tumour-normal tissue specimens. We found that the mean expression level of INPP4B mRNA in GC tissues was significantly lower than that in matched controls (P=0.0204, Figure 1A); moreover, 75.68% (28/37) of the tumour tissues had a lower level of INPP4B mRNA than the matched controls (Figure 1B). In addition, the expression of INPP4B protein in tumour tissues was decreased compared with that in normal controls (Figure 1C). Taken together, these results suggest that the mRNA and protein expression of INPP4B in GC tissues is lower than that in normal controls.
To determine the clinical significance of INPP4B in GC, we analysed the expression of INPP4B in the TMA by immunohistochemistry. We found that the positive immunohistochemical staining was mainly localized in the cytoplasm and that it exhibited a significant difference: the positive rate was significantly lower in the GC tissues than in normal controls [72/178 (40.45%) vs. 31/41 (75.61%), P < 0.001]. Representative images of INPP4B immunohistochemical staining in GC tissue and normal controls are shown in Figure 1D. In summary, our results showed that INPP4B is expressed at low levels in GC and it is mainly located in the cytoplasm.
Associations of INPP4B expression with clinicopathological parameters
To further elucidate the roles of INPP4B in the pathogenesis of GC, we analysed the relationships between the expression of INPP4B and clinicopathological parameters. The tumour TNM stage was classified as early stage (I-II) and advanced stage (III-IV). The histopathological differentiation was divided into high-moderate and low-undifferentiated. The lesional lymph node stages were classified as lymph node-negative (No) and lymph node-positive (Yes). As shown in Table 1, INPP4B expression was significantly correlated with TNM tumour stage (I-II vs. III-IV, P=0.033) and histopathological differentiation (high-moderate vs. low-undifferentiated, P=0.019) and weakly related to tumour size (<6 cm vs. ≥6 cm, P=0.08) but not related to any other clinical parameters. The IRS in the tumour tissues was decreased compared with that in the normal controls (Figure 1D). Compared to high-moderate differentiation, TNM early stage (I-II) and small tumour size (<6 cm), the IRS of INPP4B was significantly decreased in low-undifferentiated and TNM advanced stage (III-IV, Figure 1E, 1F), while the IRS of INPP4B was slightly decreased in large tumour size (≥6 cm, Figure 1G), not reaching statistical significance. Collectively, our results indicate that the expression level of INPP4B may be related to tumour size, histopathological differentiation and TNM tumour stage.
Survival analysis
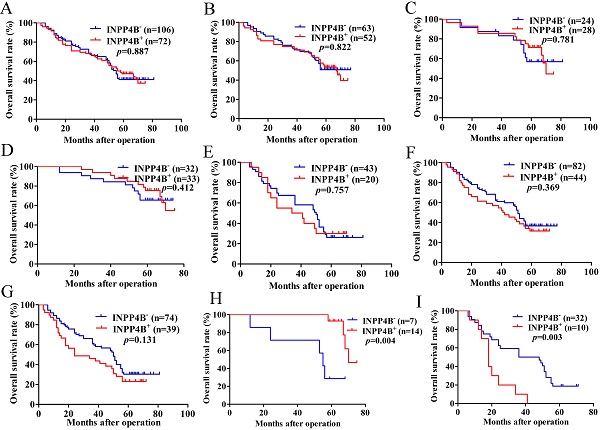
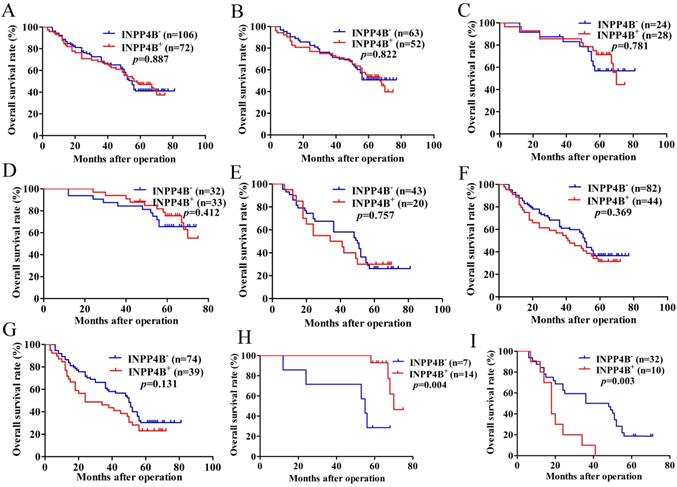
Based on the above results, we surmised that INPP4B might be related to the GC prognosis. Kaplan-Meier analysis and log-rank tests were used to evaluate the prognostic value of INPP4B in GC patients. However, when we did not stratify the relationship between INPP4B and GC prognosis, we found that INPP4B expression was not associated with overall survival (OS). There was no significant difference in OS between GC patients with INPP4B- expression (median 55 months) and those with INPP4B+ (median 56 months, P=0.887, Figure 2A). Because TNM stage contains information on the depth of invasion and lymph node metastasis, we did not add these two factors in the stratification analysis of tumour size, differentiation and TNM stage. When we stratified the relationship between INPP4B and the prognosis of GC in terms of tumour size, differentiation, and TNM staging, we obtained the following interesting findings.
In the small tumour size (<6 cm) group, high-moderate differentiation group and TNM early stage (I-II) group, INPP4B+ GC patients seems to have a longer OS than INPP4B- GC patients (Figure 2B, 2C, 2D, respectively). However, in the large tumour size (≥6 cm) group, low-undifferentiated group and TNM advanced stage group (III-IV), GC patients with INPP4B+ seemed to have a shorter OS than patients with INPP4B- (Figure 2E, 2F, 2G, respectively). These results did not reach statistical significance, which may be due to the small number of cases. It can be inferred from the existing results that INPP4B may act as a TSG in the well differentiated tissue grade and early clinical stage, and as an oncogene in the worse differentiated tissue grade and advanced clinical stage. To eliminate the interaction between these factors, we further stratified the relationship between these factors and the clinical prognosis. We found that small tumour size (<6 cm)/high-moderate differentiation/TNM early stage (I-II) patients with INPP4B+ had a more favourable prognosis than patients with INPP4B- (Figure 2H), whereas large tumour size (≥ 6cm)/low-undifferentiated/TNM advanced stage (III-IV) patients with INPP4B+ had a worse prognosis than patients with INPP4B- (Figure 2I). Taken together, these results indicate that INPP4B plays different roles in GC progression in different tissue grades and clinical stages.
Finally, we further assessed the predictive values of INPP4B in GC prognosis by Cox regression analysis. When we did not stratify the relationship between INPP4B and GC prognosis, multivariate analyses only revealed that the depth of invasion, lymph node metastasis and differentiation were independent predictor factors of GC patients (Table 2). When we stratified the relationship between INPP4B and GC prognosis, univariate analysis revealed that INPP4B was a significant predictor of a good outcome for GC patients in the small tumour size (<6 cm)/high-moderate differentiation/TNM early stage (I-II) group (Table 3), but it was a significant predictor of a poor outcome in the large tumour size (≥ 6cm)/low-undifferentiated/TNM advanced stage (III-IV) group (Table 4). Taken together, our results indicate that INPP4B plays a contradictory predictive role in GC after stratification by tissue grade and clinical stage.
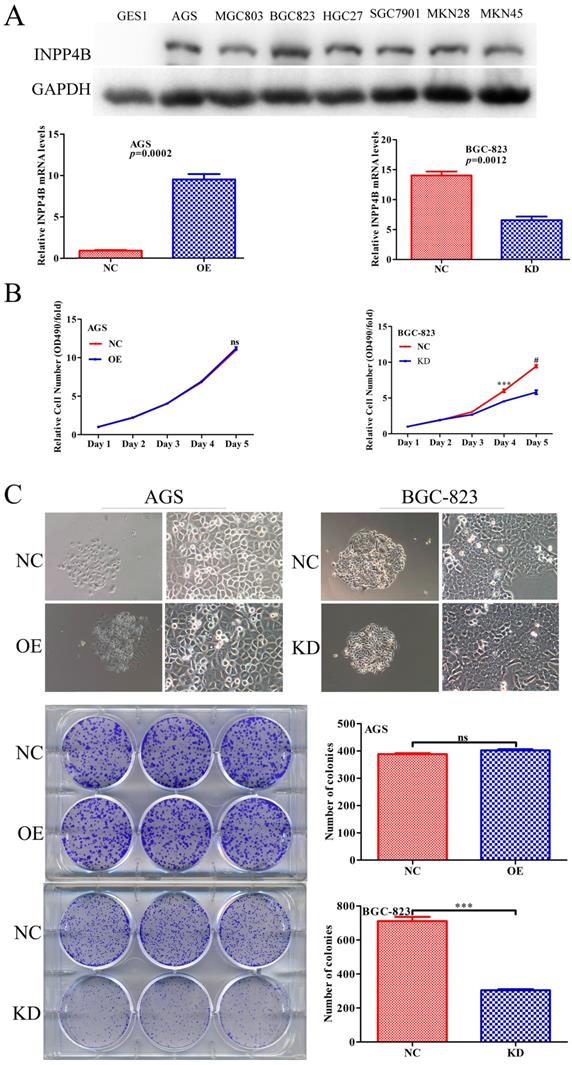
INPP4B regulates GC cells proliferation in vitro
Given that INPP4B has dual functions in GC clinical prognosis, we inferred that INPP4B might have dual functions in vitro. We chose AGS cells for overexpression and BGC-823 cells for knockdown because INPP4B was highly expressed in BGC-823 cells, while AGS cells and other GC cell lines were relatively low expressed (Figure 3A). To verify our hypothesis, we established AGS and BGC-823 cells stably overexpressing, silenced and negative control INPP4B by infection with different lentiviruses. Then, we performed MTT and colonigenic assays to examine the effects of INPP4B on the growth and proliferation of AGS and BGC-823 cells. As a result, INPP4B knockdown significantly inhibited BGC-823 cell proliferation and colony formation, while INPP4B overexpression weakly promoted AGS cell proliferation and colony formation (Figure 3B and 3C), which did not reach a statistically significant level. In summary, our findings suggest that the inhibitory effect of INPP4B knockdown on GC cell proliferation and colony formation is stronger than the promoting effect of INPP4B overexpression.
Relationship between INPP4B expression and clinicopathological variables (n=178).
| Clinicopathological variables | Total | INPP4B expression | χ2 | P value | |
|---|---|---|---|---|---|
| positive | negative | ||||
| Gender | 0.236 | 0.627 | |||
| Male | 132 | 52 | 80 | ||
| Female | 46 | 20 | 26 | ||
| Age (y) | 0.298 | 0.585 | |||
| <61 | 86 | 33 | 53 | ||
| ≥61 | 92 | 39 | 53 | ||
| Tumour size (cm) | 3.066 | 0.08 | |||
| <6 | 115 | 52 | 63 | ||
| ≥6 | 63 | 20 | 43 | ||
| Differentiation | 5.473 | 0.019 | |||
| High/moderate | 52 | 28 | 24 | ||
| Low/undifferentiated | 126 | 44 | 82 | ||
| Depth of invasion | 0.125 | 0.724 | |||
| T1/T2 | 30 | 13 | 17 | ||
| T3/T4 | 148 | 59 | 89 | ||
| Lymph node metastasis | 2.18 | 0.14 | |||
| Yes | 127 | 47 | 80 | ||
| No | 51 | 25 | 26 | ||
| TNM | 4.527 | 0.033 | |||
| I/II | 65 | 33 | 32 | ||
| III/IV | 113 | 39 | 74 | ||
| Location | 0.587 | 0.746 | |||
| Upper | 86 | 37 | 49 | ||
| Middle | 39 | 14 | 25 | ||
| Lower | 53 | 21 | 32 | ||
Note: TNM, tumour-node-metastasis; P values were detected by the Pearson Chi-square test; P<0.05 was defined statistically significant and was given in bold.
Low expression of INPP4B mRNA and protein in GC samples. (A) Scatter plots of the relative expression of INPP4B mRNA in tumour tissues and normal controls. (B) Bar plots of INPP4B mRNA expression in GC tissues compared with normal controls. (C) The protein expression level of INPP4B was analysed by western blot assay. Representative protein expression level of INPP4B in 12 pairs of tumour samples (T) and corresponding normal controls (N). GAPDH was used as an endogenous control. Immunohistochemistry analysis of INPP4B protein expression in GC and random normal controls. (D) Representative images and immunoreactivity scores of INPP4B in tumour tissues (T) and normal tissues (N). (E) Representative images and immunoreactivity scores of INPP4B in low-undifferentiated and high-moderate differentiation. (F) Representative images and immunoreactivity scores of INPP4B in TNM early stage (I-II) and TNM advanced stage (III-IV); (G) Representative images and immunoreactivity scores of INPP4B in small tumour size (< 6 cm) and large tumour size (≥ 6 cm).
Kaplan-Meier survival analysis with the log-rank test for the OS of 178 GC patients. (A) The OS of GC patients with INPP4B- and INPP4B+. (B) The OS of GC patients with small tumour sizes (< 6 cm) who were INPP4B- and INPP4B+. (C) The OS of GC patients with high-moderate differentiation with INPP4B- and INPP4B+. (D) The OS of GC patients in TNM early stage (I-II) with INPP4B- and INPP4B+. (E) The OS of GC patients with a large tumour size (≥6 cm) who were INPP4B- and INPP4B+. (F) The OS of GC patients in low undifferentiated grade with INPP4B- and INPP4B+. (G) The OS of GC patients in TNM advanced stage (III-IV) with INPP4B- and INPP4B+. (H) The OS of GC patients in small tumour size (< 6cm)/high-moderate differentiation/TNM early stage (I-II) with INPP4B- and INPP4B+. (I) The OS of GC patients in large tumour size (≥ 6cm)/low-undifferentiated/TNM advanced stage (III-IV) with INPP4B- and INPP4B+.
Univariate and multivariate analysis of the correlation between clinicopathological parameters and prognostic significance of GC patients (n=178).
| Variables | Univariate analysis | p value | Multivariate analysis | P value |
|---|---|---|---|---|
| HR(95%CI) | HR(95%CI) | |||
| Gender (male vs. female) | 0.778(0.489-1.238) | 0.289 | NA | |
| Age (y) (<61 vs. ≥61) | 1.053(0.715-1.550) | 0.794 | NA | |
| Tumour diameter (cm) (<6 vs. ≥6) | 0.545(0.368-0.807) | 0.002 | 0.793(0.531-1.184) | 0.257 |
| Differentiation (high/moderate vs. low/undifferentiated) | 2.207(1.364-3.570) | 0.001 | 2.147(1.311-3.514) | 0.002 |
| Location (upper vs middle vs. lower) | 0.916(0.732-1.145) | 0.439 | NA | |
| Depth of invision (T1/TI vs. T3/T4) | 6.083(2.473-14.965) | <0.0001 | 4.027(1.507-10.763) | 0.005 |
| Lymph node metastasis (yes vs. no) | 3.880(2.203-6.833) | <0.0001 | 2.718(1.313-5.623) | 0.007 |
| TNM stages (I/II vs. III/IV) | 3.390(2.109-5.449) | <0.0001 | 1.134(0.585-2.197) | 0.71 |
| INPP4B (positive vs. negative) | 0.972(0.655-1.443) | 0.888 | NA |
Note: Variables with P values more then 0.05 in the univariate models were not adapted (NA) in the multivariate analysis. P<0.05 was defined statistically significant and was given in bold. CI: confidence interval. HR: Hazard ratio.
Univariate analysis of GC patients with small tumour size (<6cm)/high-moderate differentiation/TNM early stage (I-II) (n=21).
| Variables | Univariate analysis | P value |
|---|---|---|
| HR(95%CI) | ||
| Gender (male vs. female) | 5.223(0.984-27.734) | 0.052 |
| Age (y) (<61 vs. ≥61) | 0.670(0.179-2.508) | 0.552 |
| Location (upper vs middle vs. lower) | 0.674(0.226-2.011) | 0.479 |
| Depth of invision (T1/TI vs. T3/T4) | 5.333(0.660-43.085) | 0.116 |
| Lymph node metastasis (yes vs. no) | 2.864(0.710-11.555) | 0.139 |
| INPP4B (positive vs. negative) | 0.154(0.036-0.662) | 0.012 |
Note: P<0.05 was defined statistically significant and was given in bold. CI: confidence interval. HR: Hazard ratio.
Univariate analysis of GC patients with large tumour size (≥6cm)/low-undifferentiated/ TNM advanced stage (III-IV) (n=42).
| Variables | Univariate analysis | P value |
|---|---|---|
| HR(95%CI) | ||
| Gender (male vs. female) | 0.729(0.317-1.675) | 0.457 |
| Age (y) (<61 vs. ≥61) | 1.058(0.549-2.038) | 0.867 |
| Location (upper vs middle vs. lower) | 0.948(0.642-1.402) | 0.79 |
| Depth of invision (T1/TI vs. T3/T4) | 0.921(0.124-6.817) | 0.936 |
| Lymph node metastasis (yes vs. no) | 3.146(0.430-23.048) | 0.259 |
| INPP4B (positive vs. negative) | 3.219(1.417-7.312) | 0.005 |
Note: P<0.05 was defined statistically significant and was given in bold. CI: confidence interval. HR: Hazard ratio.
INPP4B controls GC cell growth in vitro. (A) The protein and mRNA expression of INPP4B in GC cell lines before and after infection with different lentiviruses. (B) Proliferation curve for AGS cells and BGC-823 cells with INPP4B overexpression (OE), knockdown (KD) and negative control (NC). (C) Colony formation of AGS cells and BGC-823 cells with INPP4B overexpression (OE), knockdown (KD) and negative control (NC). Each experiment was repeated three times. Values are expressed as the means ± SEM. ***, P<0.001; #, P<0.0001; ns, not significant.
INPP4B controls GC apoptosis in vitro. (A) Overexpression of INPP4B significantly inhibits AGS apoptosis. (B) Knockdown of INPP4B significantly induces BGC-823 apoptosis. ***, P<0.001.
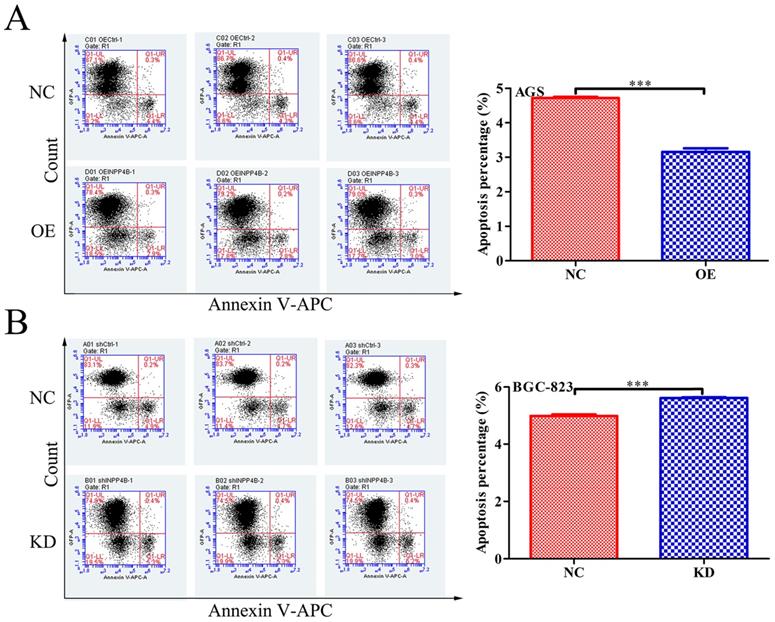
INPP4B inhibits GC cells apoptosis in vitro
To further verify our hypothesis, we next investigated the effect of INPP4B on apoptosis in GC cells using flow cytometry. Our date showed that INPP4B overexpression significantly reduced the apoptosis rate of AGS cells (Figure 4A), and INPP4B knockdown significantly increased the apoptosis rate of BGC-823 cells (Figure 4B). These date indicate that INPP4B inhibits the apoptosis of GC cells.
INPP4B promotes GC cells migration in vitro
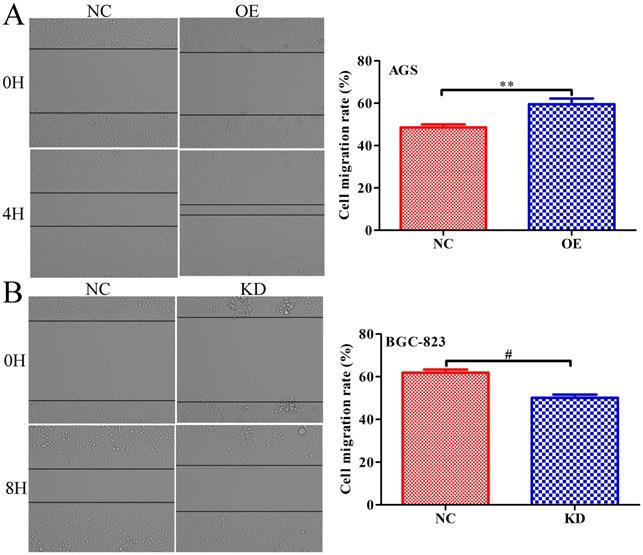
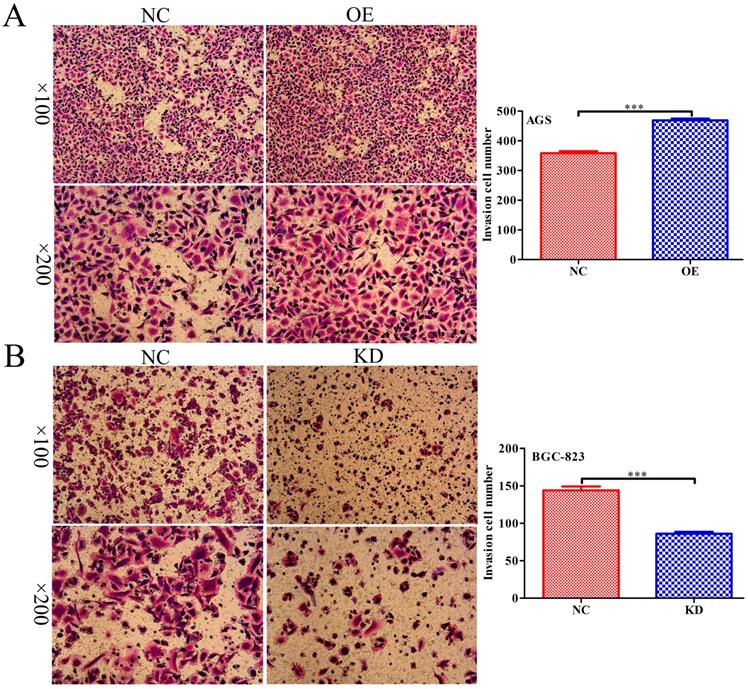
To further explore whether INPP4B affects the migration of GC cells, scratch wound healing and Transwell assays were performed. Scratch wound-healing assays and Transwell assays revealed that INPP4B overexpression notably increased AGS cell migration (Figure 5A and Figure 6A), while INPP4B knockdown significantly reduced BGC-823 cell migration (Figure 5B and Figure 6B). Taken together, our findings indicate that INPP4B promotes GC cell migration in vitro.
INPP4B mediates SGK3 and Akt activation in GC cells
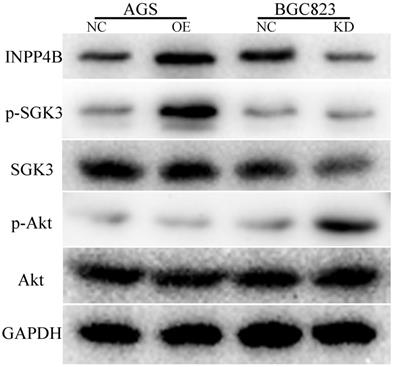
To explore the possible mechanism by which INPP4B promotes GC cell progression, we used WB to detect the changes in some important downstream proteins of INPP4B, such as p-SGK3(Thr320), SGK3, p-Akt(Ser473) and AKT, in AGS-overexpressing and BGC-823-knockdown cell lines. Our results showed that INPP4B overexpression enhanced the phosphorylation of SGK3 (p-SGK3), but did not affect the phosphorylation of Akt (p-Akt) in AGS cells; whereas INPP4B knockdown enhanced the p-Akt level but did not enhance the p-SGK3 level in BGC823 cells (Figure 7). Taken together, these results suggest that INPP4B may affect the biological functions of AGS and BGC823 cells through different signalling pathways.
Discussion
GC is one of the major threats to human health and it has a poor prognosis. Despite great advances in diagnosis and treatment methods for GC, the long-term survival of patients remains unsatisfactory. Currently, the prognostic system routinely employed for tumour management is primarily dependent on the American Joint Committee on Cancer (AJCC) TNM staging system [24]. However, we cannot predict the clinical outcome of patients after surgery depending on clinical parameters alone, because the biological aggressiveness of each individual disease is characterized by its potential for metastasis and its resistance to anticancer therapy. Therefore, identifying significant molecular biological prognostic factors may help achieve a more accurate prediction of the clinical outcome and may also reveal novel predictive factors and therapeutic targets [25].
The PI3K-Akt signalling pathway was confirmed to be a typical pathway that triggers a cascade of responses, including cell growth, metastasis, EMT, angiogenesis, and the development of chemoresistance in a wide range of tumour types [26-28]. Deregulation of the PI3K-Akt signalling pathway is associated with numerous human cancers, including GC [29-31]. INPP4B negatively regulates the PI3K-Akt signalling pathway, which is expressed at low levels and has a tumour-suppressive role in several types of human malignancies [13-15]. However, unexpected findings from recent reports indicated that INPP4B is highly expressed in some malignancies and plays a role as an oncogene [16-18]. Evidence accumulated from basic and clinical studies suggests that INPP4B may play a very controversial role in cancer progression.
There was only one previous report on INPP4B in GC. Choi et al. reported that they found a frameshift mutation of the INPP4B gene in GC, which created a premature stop codon, leading to functional inactivation of the protein [32]. However, the expression level and clinical significance of INPP4B in GC are unknown. In our study, we analysed the expression of INPP4B in GC and its prognostic implications. The mRNA expression level of INPP4B was consistent with the protein expression level, and the expression level was low in GC tissues compared with normal tissues. These results indicate that the INPP4B protein level is low in GC tumour tissues, which may be at least partially caused by decreased transcription of the INPP4B gene.
INPP4B controls GC cell migration as demonstrated by a scratch wound-healing assay. (A). Overexpression of INPP4B significantly promotes AGS cell migration. (B). Knockdown of INPP4B significantly inhibits BGC-823 cell migration. **, P<0.01; #, P<0.0001.
INPP4B regulates GC cell migration as demonstrated by a Transwell assay. INPP4B overexpression enhances AGS cell migration. (B). INPP4B knockdown inhibits BGC-823 cell migration. ***, P<0.001.
INPP4B affects the biological functions of AGS and BGC823 cells through different signalling pathways. INPP4B overexpression enhanced the phosphorylation of SGK3 (p-SGK3) in AGS cells; whereas INPP4B knockdown enhanced the p-Akt level in BGC823 cells.
As shown in Table 1 and Figure 1, we found an interesting phenomenon in which the expression of INPP4B in small tumour size (< 6cm), high-moderate histopathological differentiation and TNM early stage (I-II) was increased compared with that in large tumour size (≥ 6cm), low-undifferentiated histopathological differentiation and TNM advanced stage (III-IV). However, survival analysis and Cox proportional hazards model analysis found that when we did not stratify the relationship between INPP4B and GC prognosis, INPP4B expression was independent of OS. When we stratified the relationship between INPP4B and the prognosis of GC in terms of tumour size, differentiation, and TNM staging, we found that GC patients with high expression of INPP4B had a better prognosis in the well differentiated tissue grade and early clinical stage but had a poor prognosis in the worse tissue grade and advanced clinical stage, which indicated a contradictory role. These findings demonstrated the dual function of INPP4B in different tissue grades and clinical stages.
Previous studies on primary non-metastatic and metastatic colorectal cancer stem-like cells (CR-CSLCs) have found that INPP4B is expressed at low levels in non-metastatic CR-CSLCs and at high levels in metastatic CR-CSLCs, plays a tumour suppressor role in non-metastatic CR-CSLCs cells and plays an oncogenic role in metastatic CR-CSLCs, depending on different molecular mechanisms [4]. This has only been demonstrated at the cellular level in vitro so far, but we found for the first time that INPP4B plays a dual role in different tissue grades and clinical stages of the same type of tumour. We cannot explain this phenomenon fully, but it is likely that INPP4B regulates different molecular signalling pathways in different tissue grades and clinical stages of GC. These results suggested that INPP4B was likely to play important roles in the progression of GC. However, these results need to be validated by further study of a larger cohort of GC patients. Next, we further elucidated whether INPP4B has dual functions in GC cells in vitro.
Previous studies on the role of INPP4B in tumour cells have shown that INPP4B plays different roles in different tumour cells. Knockdown of INPP4B in breast cancer cells results in enhanced Akt activation, cell proliferation, anchorage-independent growth and motility [10, 15]. Overexpression of INPP4B in prostate cancer cells results in suppressed migration, invasion and angiogenesis [33]. Zhang et al. found that IRF2 serves as an oncogenic protein in human AML by promoting the expression of INPP4B to promote the growth of AML cells [34]. Guo et al. found that silencing INPP4B blocks the activation of Akt and serum- and glucocorticoid-regulated kinase 3 (SGK3), inhibits colon cancer cell proliferation and delays colon cancer xenograft growth [18]. These results indicate that INPP4B functions as an oncogene or TSG in different types of cancer cells.
When we tested the basal expression of INPP4B in GC cell lines by western blot, it was found that INPP4B was expressed in all gastric cancer cells but not in normal (GES1) cells, which was consistent with the role of INPP4B as an oncogene in the phenotype experiment of gastric cancer cells. We chose two GC cell lines (AGS and BGC-823) for the in vitro functional assays. Overexpression of INPP4B in AGS cells significantly reduced the cell apoptosis rate, increased the cell migration capability, and weakly promoted cell proliferation and colony formation; knockdown of INPP4B in BGC-823 cells significantly increased the apoptosis rate, decreased the cell migration capability, and decreased proliferation and colony formation. All of the above phenotypic experiments support the hypothesis that INPP4B plays an oncogenic in gastric cancer cells.
However, we also used WB to explore the possible mechanism by which INPP4B promotes GC cell progression. We found that INPP4B may affect the biological functions of AGS and BGC823 cells through different signalling pathways. What makes us wonder is that INPP4B knockdown in BGC823 cells can lead to the activation of p-Akt, a typical role of tumour suppressors, but the phenotypic experiment suggested it functioned like an oncogene, which seems to be contradictory. This may be because INPP4B in BGC823 cells can affect other unknown signalling pathways in addition to p-Akt activation. According to these results, INPP4B is more likely to act as an oncogene in GC cells. However, the mechanism by which INPP4B acts as an oncogene in GC cells is not fully understood. Therefore, more biological experiments and mechanistic studies should be carried out to verify this finding using more cell lines.
In conclusion, our study has demonstrated for the first time that INPP4B is expressed at low levels in Chinese GC tissues and it plays a dual role in the prognosis of GC patients. Our results show that GC patients with high expression of INPP4B have a better prognosis in the well-differentiated tissue grade and early clinical stage group, but patients in the worse-differentiated tissue grade and advanced clinical stage group have poor prognosis, indicating that INPP4B plays a contradictory role. Additionally, INPP4B may act as an oncoprotein in GC cells. Using in vitro analyses, we found that knockdown of INPP4B in BGC-823 cells could increase the apoptosis rate, decrease cell migration capability, and reduce proliferation and colony formation, while overexpression of INPP4B in AGS cells had the opposite effect, suggesting that INPP4B is an oncogene in GC cells. However, due to the paradoxical roles of INPP4B in the prognosis of GC patients and other cancers, further biological experiments and mechanistic studies of INPP4B in GC are necessary. Moreover, understanding the molecular mechanisms underlying the role of INPP4B in the development of GC will provide a novel treatment approach for GC.
Abbreviations
INPP4B: inositol polyphosphate 4-phosphatase type II; GC: gastric cancer; SGK3: serum and glucocorticoid-regulated kinase 3; TNM: tumour-node-metastasis; PTEN: phosphatase and tensin homolog; TSG: tumour suppressor gene; LOH: loss of heterozygosity; TMA: tissue microarray; AJCC: American Joint Committee on Cancer; IHC: immunohistochemistry; IRS: immunoreactivity score; OS: Overall survival.
Acknowledgements
We thank Qiang Yan for his technical support. We thank Shangxin Zhang for the helpful suggestions on this manuscript. We further thank all volunteers who participated in this study.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81874063) and the Provincial Natural Science Foundation of Anhui (No. 1808085MH238, No. 1908085MH261 and No.2008085QH408).
Author Contributions
Youliang Wu and Yida Lu: Experimentation and writing of the manuscript. Xiaodong Wang and Huizhen Wang: Clinical data collection and analysis. Mingliang Wang, Yexiang You and Xiaoli Su: Assisted with the cell functional experiments and the clinical data compilation. Dengqun Sun and Yanjun Sun: Assemblage of the tissue microarray. Yongxiang Li: Concept and design, supervision and writing of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140-56
3. Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10:342-52
4. Yang L, Ding C, Tang W. et al. INPP4B exerts a dual function in the stemness of colorectal cancer stem-like cells through regulating Sox2 and Nanog expression. Carcinogenesis. 2020;41(1):78-90
5. Malek M, Kielkowska A, Chessa T. et al. PTEN Regulates PI(3,4)P2 Signaling Downstream of Class I PI3K. Mol Cell. 2017;68( 3):566-580
6. Zhao C, Tao T, Yang L. et al. Loss of PDZK1 expression activates PI3K/AKT signaling via PTEN phosphorylation in gastric cancer. Cancer Lett. 2019;453:107-121
7. Rodgers SJ, Ferguson DT, Mitchell CA. et al. Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases. Biosci Rep. 2017;37(1):BSR20160432
8. Mangialardi EM, Chen K, Salmon B. et al. Investigating the duality of Inpp4b function in the cellular transformation of mouse fibroblasts. Oncotarget. 2019;10(59):6378-6390
9. Dzneladze I, Woolley JF, Rossell C. et al. SubID, a non-median dichotomization tool for heterogeneous populations, reveals the pan-cancer significance of INPP4B and its regulation by EVI1 in AML. PLoS One. 2018;13(2):e0191510
10. Gewinner C, Wang ZC, Richardson A. et al. Evidence that Inositol Polyphosphate 4-Phosphatase Type II Is a Tumor Suppressor that Inhibits PI3K Signaling. Cancer Cell. 2009;16(2):115-25
11. Perez-Lorenzo R, Gill KZ, Shen CH. et al. A tumor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. The Journal of investigative dermatology. 2014;134:1359-68
12. Yuen JW, Chung GT, Lun SW. et al. Epigenetic inactivation of inositol polyphosphate 4-phosphatase B (INPP4B), a regulator of PI3K/AKT signaling pathway in EBV-associated nasopharyngeal carcinoma. PloS one. 2014;9:e105163
13. Hodgson MC, Shao LJ, Frolov A. et al. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer research. 2011;71:572-82
14. Tang W, Yang L, Yang T. et al. INPP4B inhibits cell proliferation, invasion and chemoresistance in human hepatocellular carcinoma. Onco Targets Ther. 2019;12:3491-3507
15. Fedele CG, Ooms LM, Ho M. et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci USA. 2010;107:22231e6
16. Gasser JA, Inuzuka H, Lau AW. et al. SGK3 mediates INPP4B- dependent PI3K signaling in breast cancer. Mol Cell. 2014;56(4):595-607
17. Dzneladze I, He R, Woolley JF. et al. INPP4B overexpression is associated with poor clinical outcome and therapy resistance in acute myeloid leukemia. Leukemia. 2015;29(7):1485-95
18. Guo ST, Chi MN, Yang RH. et al. INPP4B is an oncogenic regulator in human colon cancer. Oncogene. 2016;35(23):3049-61
19. Sung JY, Na K, Kim HS. Down-regulation of Inositol Polyphosphate 4-Phosphatase Type II Expression in Colorectal Carcinoma. Anticancer Res. 2017;37(10):5525-5531
20. Wu Y, Yun D, Zhao Y. et al. Down regulation of RNA binding motif, single-stranded interacting protein 3, along with up regulation of nuclear HIF1A correlates with poor prognosis in patients with gastric cancer. Oncotarget. 2017;8(1):1262-1277
21. Zhai S, Liu Y, Lu X. et al. INPP4B As A Prognostic And Diagnostic Marker Regulates Cell Growth Of Pancreatic Cancer Via Activating A KT. Onco Targets Ther. 2019;12:8287-8299
22. Shen Y, Wang X, Xu J. et al. SerpinE2, a poor biomarker of endometrial cancer, promotes the proliferation and mobility of EC cells. Cancer Biomark. 2017;19:271-278
23. Chen Y, Sun Z, Qi M. et al. INPP4B restrains cell proliferation and metastasis via regulation of the PI3K/AKT/SGK pathway. J Cell Mol Med. 2018;22(5):2935-2943
24. Amin MB, Greene FL, Edge SB. et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67(2):93-99
25. Oldenhuis CN, Oosting SF, Gietema JA. et al. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008;44(7):946-53
26. Kim D, Cheng GZ, Lindsley CW. et al. Targeting the phosphatidylinositol-3 kinase/Akt pathway for the treatment of cancer. Curr Opin Investig Drugs. 2005;6:1250-8
27. Liu X, Li Z, Song Y. et al. AURKA induces EMT by regulating histone modification through Wnt/beta-catenin and PI3K/Akt signaling pathway in gastric cancer. Oncotarget. 2016;7:33152-64
28. Liang H. Advanced glycation end products induce proliferation, invasion and epithelial-mesenchymal transition of human SW480 colon cancer cells through the PI3K/AKT signaling pathway. Oncol Lett. 2020;19(4):3215-3222
29. Ahmad A, Biersack B, Li Y. et al. Deregulation of PI3K/Akt/mTOR signaling pathways by isoflavones and its implication in cancer treatment. Anticancer Agents Med Chem. 2013;13(7):1014-24
30. Tapia O, Riquelme I, Leal P. et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465(1):25-33
31. Riquelme I, Tapia O, Espinoza JA. et al. The Gene Expression Status of the PI3K/AKT/mTOR Pathway in Gastric Cancer Tissues and Cell Lines. Pathol Oncol Res. 2016;22(4):797-805
32. Choi EJ, Kim MS, Yoo NJ. et al. Inactivating Frameshift Mutation of INPP4B Encoding a PI3K PathwayPhosphatase in Gastric and Colorectal Cancers. Pathol Oncol Res. 2016;22(3):653-4
33. Chen H, Li H, Chen Q. INPP4B overexpression suppresses migration, invasion and angiogenesis of human prostate cancer cells. Clin Exp Pharmacol Physiol. 2017;44(6):700-708
34. Zhang F, Zhu J, Li J. et al. IRF2-INPP4B axis participates in the development of acute myeloid leukemia by regulating cell growth and survival. Gene. 2017;627:9-14
Author contact
![]() Corresponding author: Yongxiang Li, Department of General Surgery, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, People's Republic of China. Tel: +86 13615601088; Fax: +86 0551-62923887; E-mail: liyongxiangedu.cn.
Corresponding author: Yongxiang Li, Department of General Surgery, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, People's Republic of China. Tel: +86 13615601088; Fax: +86 0551-62923887; E-mail: liyongxiangedu.cn.

 Global reach, higher impact
Global reach, higher impact