Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(23):7214-7222. doi:10.7150/jca.63517 This issue Cite
Research Paper
Decipher the Helicobacter pylori Protein Targeting in the Nucleus of Host Cell and their Implications in Gallbladder Cancer: An insilico approach
1. Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Tumor Hospital of Zhengzhou University, Zhengzhou City, Henan Province, 450008, China.
2. King Abdullah Institute for Nanotechnology, King Saud University, Riyadh 11451, Saudi Arabia.
3. Department of Biology, College of Sciences, Princess Nourah bint Abdulrahman University, Riyadh 11617, Saudi Arabia.
4. Department of Biology, College of Science, Imam Mohammad Ibn Saud Islamic University (IMSIU), Riyadh 11623, Saudi Arabia.
5. Department of Health Sciences, Novel Global Community Educational Foundation, Australia.
6. Department of Bioscience, Shri Ram Group of College (SRGC), Muzaffarnagar, UP, India.
7. Department of Pharmaceutics, College of Pharmacy, PO Box 2457, King Saud University, Riyadh 11451, Saudi Arabia.
Received 2021-6-4; Accepted 2021-10-3; Published 2021-10-25
Abstract
Gallbladder cancer (GBC) is one of the leading causes of cancer-related mortality worldwide. Researchers have investigated that specific strains of bacteria are connected with growth of different types of cancers in human. Some reports show possible implication of Helicobacter pylori (H. pylori) in the etiology of gallbladder cancer (GBC). Their enigmatic mechanisms, nevertheless, are not still well clear. We sought to predict whether various proteins of H. pylori targeted to nucleus of host cells and their implication in growth of gallbladder cancer. GBC is one of the leading causes of cancer mortality worldwide. We applied bioinformatics approach to analyze the H. pylori proteins targeting into the nucleus of host cells using different bioinformatics predictors including nuclear localization signal (NLS) mapper Balanced Subcellular Localization (BaCelLo) and Hum-mPLoc 2.0. Various nuclear targeting proteins may have a potential role in GBC etiology during intracellular infection. We identified 46 H. pylori proteins targeted into nucleus of host cell through bioinformatics tools. These H. pylori nucleus-targeting proteins might alter the normal function of host cells by disturbing the different pathways including replication, transcription, translation etc. Various nucleus-targeted proteins can affect the normal growth and development of infected cells. We propose that H. pylori proteins targeting into the nucleus of host cells regulate GBC growth using different strategies. These integrative bioinformatics research demonstrated several H. pylori proteins that may serve as possible targets or biomarkers for early cure and treatment or diagnosis GBC.
Keywords: Helicobacter pylori, Protein targeting, Host cell, Nucleus, Health informatics, Gallbladder cancer
Introduction
Cancer is the second leading cause of death in the United States and a predominant public health problem worldwide [1]. Gallbladder cancer (GBC) is the fifth most common gastrointestinal cancer worldwide with poor prognosis [2-4]. The number of new cases of GBC and other biliary cancers in the United States was estimated to be approximately 11,740, with 3,830 deaths reported in 2016 [1]. GBC is most frequently associated with the biliary tract. GBC shows the highest incidence in the sixth and seventh decades of life, and females are affected two to six times more often than males [2, 5, 6]. Although GBC is more common in Korea, Japan, Northern India, and Eastern European countries, elevated incidence rates have been observed in Latin America [7]. The infections of different types of bacteria are associated with the progress and development of many diseases including typhoid, diarrhea and different types of cancer [8-10]. Various factors are involved in the process of carcinogenesis for the growth of different types of cancer such as exposure to specific chemicals, obesity, diet, reproductive factors, hepato-biliary anomalies, cholelithiasis (particularly mixed gallstone or gallstone disease), and poor prognosis, unsatisfactory treatment [4], and late diagnosis of chronic gallbladder infections [11]. Similarly different factors have been associated with the growth and progression of GBC. The percentage of patients suffering from GBC after cholecystectomy for assumed gallbladder stone disease is 0.5-1.5% [12]. In addition, genetic disorders such as Peutz-Jegher syndrome, anomalous pancreaticobiliary ductal union, and multiple familial polyposis/Gardener syndrome are associated with GBC [13-15]. The relationship between life style, genetic predisposition, and previous infection in GBC is not well understood [7]. The existence of H. pylori and H. bilis, both in the bile and gallbladder, was confirmed in more than 75% of patients with GBC and more than 50% of patients with chronic cholecystitis that underwent surgery [16-18]. H. pylori is a gram-negative, micro-aerophilic, spiral-shaped, flagellated, and slow-growing bacterium and probably the cause of the most common chronic bacterial infections in humans, present in almost half of the world's population [19, 20]. KHP30 phage observed to be associated as an episome with NY43 strain of H. pylori [21, 22].
Recent reports have indicated the presence of H. pylori in the gallbladders and bile of approximately 75% of patients with GBC and about 50% of patients with chronic cholecystitis [18]. Although studies have revealed some possible mechanisms involved in biliary carcinogenesis, most key events and specific connections to H. pylori infection in this multifaceted cascade that directs the transformation of epithelial cells in the gallbladder remain unknown and require additional investigation. The aim of the present work was to determine H. pylori proteins that are localized into the host cell nucleus and their potential associations with GBC. In this study, we focused on the association between chronic H. pylori infection and GBC development.
Materials and methods
Retrieve the H. pylori proteome
We performed various specified searches to retrieve the whole proteome of H. pylori. Eventually we were focused to the UniProt (Universal Protein Resource) database to predict the nucleus-targeting proteins of H. pylori in the host cell [23]. This UniProt database developed through the collection of PIR protein database, SWISS-PROT, and TrEMBL [23-25] contains immense information regarding the H. pylori proteome. The proteomes of various strains of H. pylori such as strains ATCC 700392/26695 and ATCC 27545, are available in these databases [21, 26].
Selection of a predictive computational tool
The whole proteome of H. pylori strain ATCC 700392/26695 was selected for the prediction of nucleus-targeting proteins in human gallbladder cells. We were used different tools including ExPASy Compute pI/Mw tool, cNLS mapper, Balanced Subcellular Localization (BaCelLo) and Hum-mPLoc 2.0 bioinformatics predictor.
Prediction of pI values and MWs using the ExPASy Compute pI/Mw tool
The ExPASy Compute pI/Mw tool was used to predict the theoretical isoelectric point (pI) and molecular weight (MW) of the query sequence of a particular protein [27]. The tool was utilized to access the extensive annotations available in the SWISS-PROT database [24].
Prediction of NLS in the H. pylori proteome using cNLS mapper
We were used cNLS predictor to analyze the possible monopartite and bipartite NLSs in whole protein sequences of H. pylori proteome [28]. NLS prediction may be used to predict the nucleus-targeting ability of specific proteins [28]. The cNLS predictor shows NLS values in the form of an NLS cut-off, and protein sequences with cut-off values of 10 to 8, 7 to 8, 5 to 3, and 1 to 2 were identified as absolutely targeting the nucleus, partly targeting the nucleus, targeting both the cytoplasm and nucleus, and targeting the cytoplasm, respectively. Moreover, protein sequences with cut-off values between two ranges were rounded to the closest whole integer.
Prediction of protein targeting using the BaCelLo predictor
H. pylori proteins targeting the nucleus of the host cell were predicted using BaCelLo [29]. This predictor may be used to identify proteins in organisms of three different kingdoms (Fungi, Plants, and Animals). In the current study, we analyzed proteins of the organisms from the animal kingdom. The BaCelLo predictor is a computational tool based on diverse support vector machines (SVMs) structured in a decision tree [29].
Selection of BaCeILo-predicted proteins using the Hum-mPLoc 2.0 predictor
H. pylori proteins targeting the nucleus and other compartments in humans were predicted by utilizing the Hum-mPLoc 2.0 subcellular localization predictor [30]. This predictor is based on a top-down approach to increase the power to predict human proteins targeting subcellular components, including the nucleus. Hum-mPLoc 2.0 predicted 14 different classes of subcellular localization, including the nucleus, mitochondrion, cytoplasm, centriole, endoplasmic reticulum, Golgi apparatus, and lysosome, etc.
Results
Search for the H. pylori proteome
UniProt is a comprehensive database that includes the whole H. pylori proteome. We select the ATCC 700392/26695 strain of H. pylori because it had the highest number of proteins (1,552) identified among the available proteomes [26].
Selection of computational tools for the prediction study
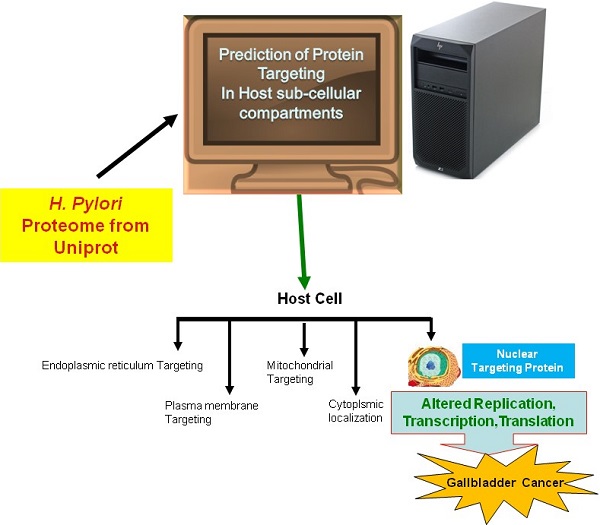
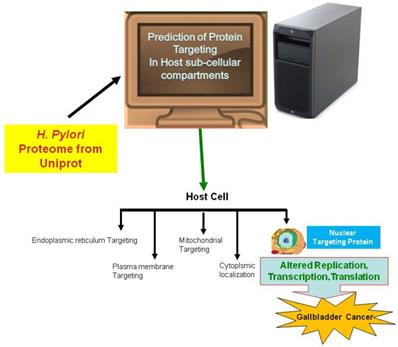
In the current study, we employed the ExPASy Compute pI/Mw tool to predict the pI and MW of proteins, cNLS mapper to determine NLSs, BaCelLo to identify proteins targeting different components of host cells, and Hum-mPLoc 2.0 to predict proteins targeting the nucleus of the host cell because of the relative specificities of their predictive approaches (Fig. 1).
Prediction of pI values and MWs using the ExPASy Compute pI/Mw tool
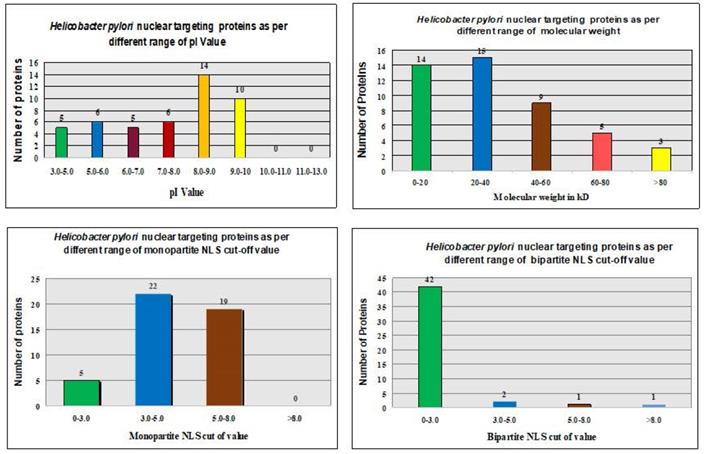
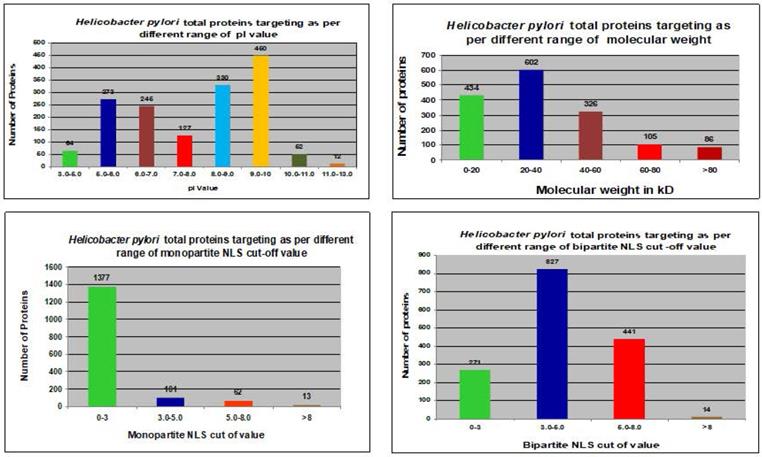
The ExPASy Compute pI/Mw tool calculated theoretical pI values and MWs of proteins in the H. pylori proteome (Fig. 2 and Table 1).
The pI values showed no consistent pattern of proteins targeting in the nucleus of the host cell [31, 32]. However, the maximum number of nucleus-targeting proteins (14 proteins) was observed in the pI range of 8.0-9.0. Increase in the MWs, consistently decreased the frequency of nuclear targeting proteins, except in the 0-20 kDa range (Fig. 2 and Table 1). The least proteins targeting observed in the nucleus of the host cell with MW > 80 kDa (Fig. 2 and Table 1).
The image shows the method use for the prediction of nuclear targeting proteins using in silico approach.
In silico analysis of H. pylori proteins that target the nucleus of host cells and their relationship to different parameters.
In silico analysis of total H. pylori proteins in host cells and their relationship to different parameters.
In silico analysis of Helicobacter pylori proteins targeted to the host cell nuclei their relation to all proteins with similar molecular weight
| S. no. | Molecular weight (kW) | Number of proteins targeting nucleus | Total number of proteins | Percentage |
|---|---|---|---|---|
| 1 | 0-20 | 14 | 434 | 3.22 |
| 2 | 20-40 | 15 | 602 | 2.49 |
| 3 | 40-60 | 9 | 326 | 2.76 |
| 4 | 60-80 | 5 | 105 | 4.76 |
| 5 | > 80 | 3 | 86 | 3.48 |
Prediction of NLSs in the H. pylori proteome using the cNLS mapper
We utilized the cNLS mapper to analyze NLSs in whole protein sequences of the H. pylori proteome. Both monopartite and bipartite NLSs in the H. pylori proteome were determined (Fig. 3). Proteins with NLS cutoff values of 3.0-5.0 were reported to mostly target the nucleus of the host cell with monopartite NLSs (Table 3). Proteins with NLS cutoff values of 0-3.0 were mostly found to target the nucleus of the host cell with bipartite NLSs (Table 3).
Prediction of protein targeting using the BaCelLo predictor
A total of 85 (out of 1,552) proteins in the H. pylori proteome were predicted to target the nucleus of the host cell using the BaCeILo predictor [29]. The details of H. pylori proteins that target the host cell nucleus based on various parameters are shown in Table 4.
Selection of BaCeILo-predicted proteins using the Hum-mPLoc 2.0 predictor
Only 46 proteins were consistently shown to target the nucleus of the host cell by the software of Hum-mPlooc 2.0 [30]. Fig. 2 and Fig. 3 illustrate the patterns of H. pylori proteins targeting host cell nucleus according to different parameters. Moreover, the details of the 46 proteins along with their functions are shown in Table 4.
Discussion
Various studies have revealed the different possible factors involved in the development of cancer, including genetic factors, gender, age, diet, consumption of tobacco, inflammation, and infections by various pathogens. Infection is considered a leading factor involved in the development of about 16% of cancers [33]. It has confirmed that various specific bacterial strains have the ability to alter numerous pathways and molecular events in the host cell for their own survival and involved in the growth and development of different types of cancer [10, 32, 34]. In a report Arthur et. al (2012) demonstrated the involvement of E. coli NC101 strain in the progression of invasive carcinoma in azoxymethane (AOM)-treated Il10(-/-) mice. We have illustrated the involvement of mycoplasma hominis and Chlamydia pneumoniae protein targeting and their implication in the progression of prostate cancer and lungs cancer in recently published study [31, 32, 34]. It was only in the early 1990s that the role of H. pylori as a causative agent of cancer was highlighted [35]. The molecular mechanisms underlying gallbladder carcinogenesis remain unclear even today. We have proposed that various nucleus-targeting proteins of H. pylori alter the normal function of host cells. pI values failed to explain the pattern for nuclear targeting (Table 2). The association between H. pylori proteins that targeted the host cell nucleus and various parameters is shown in Fig. 2. The process of targeting the host nucleus is a key event that involves the regulation of the host cell. This is generally analyzed through specific motifs in protein sequences called NLSs. The NLS predictor allows prediction of the possible activity of an NLS in the amino acid sequences of different proteins. Various predictors may analyze the specific motifs in the amino acid sequences. The NLS mapper identified six classes of NLSs such that the nuclear import proteins are transported through the α/β pathways of importin. Therefore, we utilized the NLS mapper in our study to predict NLS activity in both monopartite and bipartite NLSs (enriched basic amino acid stretches) [36]. The NLS predictor identified potential localization sites of the proteins, including the nucleus, partially in the nucleus, the cytoplasm, and equally in both the cytoplasm and nucleus of the host cell.
In silico analysis of H. pylori proteins targeted to the host cell nucleus and their relation to all proteins with similar pI value
| S. no. | Isoelectric point | Number of proteins targeting nuclei | Total number of proteins | Percentage |
|---|---|---|---|---|
| 1 | 3.0-5.0 | 5 | 64 | 7.81 |
| 2 | 5.0-6.0 | 6 | 273 | 2.19 |
| 3 | 6.0-7.0 | 5 | 245 | 2.04 |
| 4 | 7.0-8.0 | 6 | 127 | 4.72 |
| 5 | 8.0-9.0 | 14 | 330 | 4.24 |
| 6 | 9.0-10.0 | 10 | 450 | 2.22 |
| 7 | 10.0-11.0 | 0 | 52 | 0 |
| 8 | 11.0-13.0 | 0 | 12 | 0 |
In silico analysis of H. pylori proteins targeted to the host cell nucleus and their relation to all proteins with similar monopartite and bipartite nuclear localization signals (NLS)
| NLS | NLS cut-off | Number of proteins targeting nucleus | Total number of proteins | Percentage |
|---|---|---|---|---|
| Monopartite | 0-3.0 | 5 | 1377 | 0.36 |
| Monopartite | 3.0-5.0 | 22 | 101 | 21.78 |
| Monopartite | 5.0-8.0 | 19 | 62 | 30.64 |
| Monopartite | > 8.0 | 0 | 13 | 0 |
| Bipartite | 0-3.0 | 42 | 271 | 15.49 |
| Bipartite | 3.0-5.0 | 2 | 827 | 0.24 |
| Bipartite | 5.0-8.0 | 1 | 441 | 0.22 |
| Bipartite | > 8.0 | 1 | 14 | 7.14 |
Descriptions of Helicobacter pylori proteins targeted into the nucleus of host cells as predicted using various tools
| Accession number | Protein name | Function in bacteria | Evidence of protein | Isoelectric point | Molecular weight in daltons | NLS Mapper Monopartite Bipartite | BaCeILo | Hum-mPLoc 2.0 | |
|---|---|---|---|---|---|---|---|---|---|
| P55973 | Translation initiation factor IF-3 | Translation initiation factor activity | Protein inferred from homology | 9.57 | 23342 | 0 | 4.9 | Nucleus | Nucleus |
| P56131 | tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase (EC 2.8.4.3) (Dimethylallyl)adenosine tRNA methylthiotransferase MiaB) (tRNA-i(6)A37 methylthiotransferase) | 4 iron, 4 sulfur cluster binding, metal ion binding, transferase activity | Protein inferred from homology | 8.75 | 49423 | 0 | 5.5 | Nucleus | Nucleus |
| O25029 | DEAD-box ATP-dependent RNA helicase RhpA (EC 3.6.4.13) | ATP binding, helicase activity, RNA binding | Experimental evidence at the protein level | 8.78 | 55806 | 2 | 6.3 | Nucleus | Nucleus |
| P56398 | Ribosome-recycling factor (RRF) (Ribosome-releasing factor) | Translation | Protein inferred from homology | 7.79 | 20915 | 0 | 3.7 | Nucleus | Nucleus |
| O26061 | Flavin-dependent thymidylate synthase (FDTS) (EC 2.1.1.148) (FAD-dependent thymidylate synthase) (Thymidylate synthase ThyX) (TS) (TSase) | Flavin adenine dinucleotide binding, thymidylate synthase (FAD) activity | Experimental evidence at the protein level | 6.76 | 24071 | 0 | 3.2 | Nucleus | Nucleus |
| P55991 | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I) (Omega-protein) (Relaxing enzyme) (Swivelase) (Untwisting enzyme) | DNA binding, DNA topoisomerase type I activity, metal ion binding | Experimental evidence at the protein level | 9.04 | 83196 | 2 | 4.9 | Nucleus | Nucleus |
| P55986 | Uncharacterized RNA pseudouridine synthase HP_1459 (EC 5.4.99.-) (RNA pseudouridylate synthase) (RNA-uridine isomerase) | Pseudouridine synthase activity, RNA binding | Protein inferred from homology | 9.9 | 30228 | 3 | 6.8 | Nucleus | Nucleus |
| O25506 | DNA-binding protein HU | DNA binding | Protein inferred from homology | 9.05 | 10384 | 0 | 5.8 | Nucleus | Nucleus |
| O25242 | DNA polymerase III subunit beta (EC 2.7.7.7) | 3'-5' exonuclease activity, DNA binding, DNA-directed DNA polymerase activity | Experimental evidence at the protein level | 5.51 | 42185 | 0 | 4.2 | Nucleus | Nucleus |
| P57798 | Putative Fe (2+) transport protein A | Ion transport, iron, ion homeostasis | Protein inferred from homology | 8.62 | 8719 | 0 | 5.8 | Nucleus | Nucleus |
| O25929 | Flagellar assembly factor FliW 2 | Bacterial-type flagellum assembly, regulation of translation | Experimental evidence at the protein level | 5.92 | 14862 | 0 | 2.8 | Nucleus | Nucleus |
| P55976 | Transcription termination/antitermination protein NusG | DNA-templated transcription, termination, regulation of DNA-templated transcription, elongation, transcription antitermination | Protein inferred from homology | 6.98 | 20261 | 0 | 2.9 | Nucleus | Nucleus |
| P55993 | RNA polymerase sigma factor RpoD (Sigma-70) | DNA binding, DNA binding transcription factor activity, sigma factor activity | Experimental evidence at the protein level | 7.94 | 77737 | 4 | 5.2 | Nucleus | Nucleus |
| P56123 | Ribonuclease R (RNase R) (EC 3.1.13.1) (VacB protein homolog) | Exoribonuclease II activity, RNA binding | Protein inferred from homology | 9.17 | 74163 | 0 | 4.4 | Nucleus | Nucleus |
| P66621 | 30S ribosomal protein S8 | rRNA binding, structural constituent of ribosome | Protein inferred from homology | 9.78 | 15184 | 0 | 4.3 | Nucleus | Nucleus |
| O25475 | Protein translocase subunit SecA | ATP binding, metal ion binding | Protein inferred from homology | 5.62 | 99084 | 0 | 5.6 | Nucleus | Nucleus |
| Q09064 | Urease accessory protein UreE | Nickel cation binding | Experimental evidence at the protein level | 8.58 | 19382 | 0 | 5.4 | Nucleus | Nucleus |
| O25448 | Flagellar protein FliS | Bacterial-type flagellum assembly | Experimental evidence at the protein level | 5.03 | 14543 | 0 | 0 | Nucleus | Nucleus |
| O25684 | Response regulator | DNA binding | Experimental evidence at the protein level | 5.22 | 25468 | 0 | 3.1 | Nucleus | Nucleus |
| O25998 | Secreted protein involved in flagellar motility | Unknown | Experimental evidence at the protein level | 6.54 | 20480 | 3 | 3.3 | Nucleus | Nucleus |
| O25262 | Cag pathogenicity island protein (Cag7) | Unknown | Protein predicted | 5.61 | 219401 | 2.5 | 6.8 | Nucleus | Nucleus |
| O25085 | Uncharacterized protein | Unknown | Protein predicted | 6.74 | 15304 | 0 | 5.4 | Nucleus | Nucleus |
| O24903 | Uncharacterized protein | Unknown | Protein predicted | 5.24 | 57408 | 8 | 7.5 | Nucleus | Nucleus |
| O25555 | Uncharacterized protein | Unknown | Protein predicted | 8.8 | 14714 | 0 | 5 | Nucleus | Nucleus |
| O25800 | Uncharacterized protein | Unknown | Protein predicted | 9.45 | 30632 | 0 | 4.4 | Nucleus | Nucleus |
| O25576 | Uncharacterized protein | Metal ion binding | Protein predicted | 10.13 | 16137 | 0 | 3.3 | Nucleus | Nucleus |
| O25834 | Uncharacterized protein | Unknown | Protein predicted | 5.57 | 22149 | 5 | 5.7 | Nucleus | Nucleus |
| O24939 | Uncharacterized protein | Unknown | Protein predicted | 9.54 | 45264 | 2 | 4.3 | Nucleus | Nucleus |
| O24937 | Uncharacterized protein | Unknown | Protein predicted | 9.46 | 45136 | 0 | 3.9 | Nucleus | Nucleus |
| O25553 | Uncharacterized protein | Unknown | Protein predicted | 9.79 | 11356 | 0 | 6.6 | Nucleus | Nucleus |
| O25652 | Conjugal transfer protein (TraG) | Unidirectional conjugation | Protein predicted | 8.89 | 20442 | 0 | 4 | Nucleus | Nucleus |
| O25650 | Uncharacterized protein | Unknown | Protein predicted | 8.76 | 32600 | 0 | 6.5 | Nucleus | Nucleus |
| O25105 | Uncharacterized protein | Unknown | Protein predicted | 9.81 | 21978 | 0 | 4.5 | Nucleus | Nucleus |
| O25188 | DNA topoisomerase (EC 5.99.1.2) | DNA binding, DNA topoisomerase type I activity | Protein inferred from homology | 8.47 | 77677 | 0 | 5.8 | Nucleus | Nucleus |
| O25799 | Uncharacterized protein | Unknown | Protein predicted | 9.56 | 43994 | 0 | 3.5 | Nucleus | Nucleus |
| O25195 | Uncharacterized protein | Unknown | Protein predicted | 5.36 | 41437 | 0 | 5.1 | Nucleus | Nucleus |
| O24905 | Uncharacterized protein | Unknown | Protein predicted | 4.73 | 14086 | 0 | 2.7 | Nucleus | Nucleus |
| O25102 | Uncharacterized protein | Unknown | Protein predicted | 6.56 | 6989 | 0 | 2.1 | Nucleus | Nucleus |
| O25304 | Uncharacterized protein | Transport | Protein predicted | 9.46 | 37382 | 0 | 4.3 | Nucleus | Nucleus |
| O25419 | DNA polymerase III gamma and tau subunits (DnaX) | ATP binding, DNA-directed DNA polymerase activity | Protein predicted | 5.87 | 66245 | 0 | 4.8 | Nucleus | Nucleus |
| O25379 | Uncharacterized protein | ATP binding, DNA binding, hydrolase activity | Protein predicted | 8.05 | 68760 | 10.5 | 6.4 | Nucleus | Nucleus |
| O25131 | Uncharacterized protein | Unknown | Protein predicted | 7.85 | 23211 | 0 | 6.1 | Nucleus | Nucleus |
| O25010 | Uncharacterized protein | Regulation of transcription, DNA-templated | Protein predicted | 9.52 | 8599 | 0 | 4.2 | Nucleus | Nucleus |
| O25373 | Uncharacterized protein | Unknown | Protein predicted | 8.93 | 47633 | 0 | 4.7 | Nucleus | Nucleus |
| O25265 | Cag pathogenicity island protein (Cag10) | Unknown | Protein predicted | 9.54 | 29095 | 0 | 5.9 | Nucleus | Nucleus |
| O34550 | IS200 insertion sequence from SARA17 | DNA binding, transposase activity | Protein predicted | 8.44 | 15965 | 0 | 4.6 | Nucleus | Nucleus |
In addition, BaCeILo and Hum-mPLoc 2.0 were employed in the current study to analyze H. pylori protein targeting to different host cell compartments. The results of BaCeILo and Hum-mPLoc 2.0 revealed the variation in protein targeting to the nucleus because of the utilization of different datasets during prediction. Such differences in the results from various predictors are acceptable. The Hum-mPLoc predictor analyzes the targeting of proteins to different compartments of cells using sequential evolution and domain information. The predictor computes 14 subcellular compartments such as the nucleus, mitochondrion, cytoplasm, endoplasmic reticulum, lysosome, Golgi apparatus, plasma membrane, and peroxisome. Proteins with MW < 40 kDa may be transported to the nucleus through passive transport mechanisms [28]. In the present study, we predicted various H. pylori proteins with MW < 40 kDa that affected the normal pathways of cells and may be involved in the progression of GBC. Furthermore, the nucleus-targeting proteins in humans determined by Hum-mPLoc 2.0 were compared with those determined by BaCelLo to more accurately define the subcellular localization of H. pylori proteins. The Hum-mPLoc 2.0 predictor confirmed only 46 H. pylori proteins that were targeted to the nuclei of host cells.
We focused on evaluating the involvement of the nucleus-targeting proteins of H. pylori in the progression and development of GBC. H. pylori-derived effector proteins may alter the host cell internal environment through the induction of immunosuppression, suppression of tumor suppressor genes, activation of chronic inflammation, and transformation of normal cells [37].
H. pylori proteins that target the nuclei of host cells and their implications in GBC
From the whole H. pylori proteome with 1,552 proteins, only 46 proteins were predicted to be targeted to the host cell nucleus during intracellular infection. This specific targeting may alter the homeostasis of normal cells. The results of the current study should be validated through experimental research in wet laboratories prior to drawing any final conclusions. The corresponding results may be used to develop therapies to manage and cure cancer.
Replication, DNA binding, and DNA repair in the development of GBC
Various factors such as genomic instability determine cancer susceptibility. However, the molecular mechanisms that lead to the development of cancer are incompletely understood. A report showed that H. pylori infection suppressed the expression of p53 protein [38]. A prominent hypothesis is that alterations in replication or the establishment of error-prone DNA synthesis phenotypes originating in genomic instability may serve as a source of cancer [39]. Progression of cancer is affected by different DNA-binding proteins such as the methyl CpG-binding protein, which detects the methylation of DNA and its components. Together these proteins play an important role in the development of cancer [40]. Furthermore, the tightly controlled DNA replication is essential for the multiplication of normal cells, and mutations in proteins involved in DNA replication have been associated with the development of different types of cancers [41, 42].
Diverse DNA-binding proteins have been predicted to target the nuclei of host cells, including DNA topoisomerase 1 (accession no. P55991), DNA polymerase III subunit beta (accession no. O25242), ribonuclease R (RNase R) (accession no. P56123), DNA topoisomerase (accession no. O25188), DNA polymerase III gamma and tau subunits (accession no. O25419), and the IS200 insertion sequence from SARA17 (accession no. O34550). These nucleus-targeting proteins and other uncharacterized proteins may affect the replication process in the nucleus of the host cell. The functions and other details of these proteins are shown in Table 4. Bacterial insertion sequences IS200 and IS607 encode a transposase (TnpA) and one protein with unknown function (TnpB) that is believed to act as a methyltransferase [43]. The levels of methyltransferase are increased in some cancer cell lines and cancer tissues, wherein these enzymes may be involved in the hypermethylation of the promoter CpG-rich regions of the tumor suppressor genes [44].
Transcription and translation regulatory proteins in the development of GBC
The progression from normal to cancerous cells is associated with alterations in protein-protein interactions, either in the transcription or translation regulatory proteins. The dysregulation in the expression of various genes may lead to the suppression of different anti-oncogenes and activation of proto-oncogenes during bacterial infection [45]. Conserved structural similarities in different subunits of RNA polymerase as well as antigenicity are specific features of eukaryotes. The current study showed that H. pylori RNA polymerase sigma factor RpoD (accession no. P55993), response regulator (accession no. O25684), and transcription termination/antitermination protein NusG (accession no. P55976) target the host cell nucleus and may alter the normal pathways in the host cell. The unfolded response regulator has been reported as a new predictive biomarker for the identification of cancers [46]. Nevertheless, the possible involvement of such proteins in the dysregulation of normal pathways must be experimentally demonstrated before making final conclusions.
Various H. pylori translation regulatory proteins similarly target the host cell nucleus, including translation initiation factor IF-3 (accession no. P55973), ribosome-recycling factor (RRF) (accession no. P56398), and 30S ribosomal protein S8 (accession no. P66621). These proteins also disturb the normal functioning of protein synthesis by altering gene expression. Alterations in gene expression may lead to the progression of GBC.
Uncharacterized proteins in the development of GBC
Various uncharacterized H. pylori proteins were predicted to target the nucleus of the host cell, including Cag pathogenicity island protein (Cag7) (accession no. O25262), Cag pathogenicity island protein (Cag10) (accession no. O25265), and another uncharacterized protein (accession no. O25010). These proteins may also act as factors that promote carcinogenesis in the gallbladder. For instance, CagA may interact with a tumor suppressor protein (RUNX3) that is commonly inactivated in gastric carcinomas [47].
Conclusions
The current work examines the mechanisms underlying the progression of GBC during H. pylori infection and the possible implications of the nucleus-targeting proteins in the development of GBC. The novel findings of this study may suggest new approaches to manage and cure GBC.
Abbreviations
H. pylori: Helicobacter pylori; GBC: Gallbladder cancer; NLS: Nuclear localization signal; BaCelLo: Balanced Subcellular Localization; Hum-mPLoc 2.0.: Human protein subcellular localization; UniProt: Universal Protein Resource; pI: Isoelectric point; MW: Molecular weight.
Acknowledgements
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Availability of data and materials
Data of whole proteome of H. pylori with Proteome ID UP000000429 have download from UniProt database (https://www.uniprot.org/uniprot/?query=taxonomy:85962) in current study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167-76
3. Mondal SK, Bhattacharjee D, Mandal PK, Biswas S. Histopathological study of gallbladder carcinoma and its mimics with role of carcinoembryonic antigen immunomarker in resolving diagnostic difficulties. Indian J Med Paediatr Oncol. 2017;38:411-15
4. Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II. et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349-64
5. Rakic M, Patrlj L, Kopljar M, Klicek R, Kolovrat M, Loncar B. et al. Gallbladder cancer. Hepatobiliary Surg Nutr. 2014;3:221-6
6. Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23:3978-98
7. Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer. 2006;118:1591-602
8. Crump JA. Progress in Typhoid Fever Epidemiology. Clin Infect Dis. 2019;68:S4-S9
9. Li J, Zakariah M, Malik A, Ola MS, Syed R, Chaudhary AA. et al. Analysis of Salmonella typhimurium Protein-Targeting in the Nucleus of Host Cells and the Implications in Colon Cancer: An in-silico Approach. Infect Drug Resist. 2020;13:2433-42
10. Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120-3
11. Wernberg JA, Lucarelli DD. Gallbladder cancer. Surg Clin North Am. 2014;94:343-60
12. Han HS, Cho JY, Yoon YS, Ahn KS, Kim H. Preoperative inflammation is a prognostic factor for gallbladder carcinoma. Br J Surg. 2011;98:111-6
13. Wada K, Tanaka M, Yamaguchi K. Carcinoma and polyps of the gallbladder associated with Peutz-Jeghers syndrome. Dig Dis Sci. 1987;32:943-6
14. Chang HJ, Kim SW, Kim YT, Kim WH. Loss of heterozygosity in dysplasia and carcinoma of the gallbladder. Mod Pathol. 1999;12:763-9
15. Walsh N, Qizilbash A, Banerjee R, Waugh GA. Biliary neoplasia in Gardner's syndrome. Arch Pathol Lab Med. 1987;111:76-7
16. Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J. et al. Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology. 2014;60:1963-71
17. de Martel C, Plummer M, Parsonnet J, van Doorn LJ, Franceschi S. Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br J Cancer. 2009;100:194-9
18. Hassan EH, Gerges SS, El-Atrebi KA, El-Bassyouni HT. The role of H. pylori infection in gall bladder cancer: clinicopathological study. Tumour Biol. 2015;36:7093-8
19. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449-90
20. Camilo V, Sugiyama T, Touati E. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2017 22 Suppl 1
21. Uchiyama J, Takemura-Uchiyama I, Kato S, Takeuchi H, Sakaguchi Y, Ujihara T. et al. Screening of KHP30-like prophages among Japanese Helicobacter pylori strains, and genetic analysis of a defective KHP30-like prophage sequence integrated in the genome of the H. pylori strain NY40. FEMS Microbiol Lett. 2016;363:1-9
22. Uchiyama J, Takeuchi H, Kato S, Gamoh K, Takemura-Uchiyama I, Ujihara T. et al. Characterization of Helicobacter pylori bacteriophage KHP30. Appl Environ Microbiol. 2013;79:3176-84
23. Consortium U. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014;42:D191-8
24. Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365-70
25. Consortium U. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43-7
26. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-47
27. Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531-52
28. Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171-6
29. Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics. 2006;22:e408-16
30. Shen HB, Chou KC. A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0. Anal Biochem. 2009;394:269-74
31. Khan S, Zakariah M, Rolfo C, Robrecht L, Palaniappan S. Prediction of mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget. 2017;8:30830-43
32. Khan S, Imran A, Khan AA, Abul Kalam M, Alshamsan A. Systems Biology Approaches for the Prediction of Possible Role of Chlamydia pneumoniae Proteins in the Etiology of Lung Cancer. PLoS One. 2016;11:e0148530
33. Gisserot O, Romeo E, Boudin L, Tsitsi Nding Tsogou P, Abed S, Blade JS. et al. [Can we prevent or cure infection-related cancers?]. Rev Med Interne. 2014;35:259-63
34. Khan S, Zakariah M, Palaniappan S. Computational prediction of Mycoplasma hominis proteins targeting in nucleus of host cell and their implication in prostate cancer etiology. Tumour Biol. 2016;37:10805-13
35. Mishra RR, Tewari M, Shukla HS. Helicobacter pylori and pathogenesis of gallbladder cancer. J Gastroenterol Hepatol. 2011;26:260-6
36. Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101-5
37. Mishra RR, Tewari M, Shukla HS. Association of Helicobacter pylori infection with inflammatory cytokine expression in patients with gallbladder cancer. Indian J Gastroenterol. 2013;32:232-5
38. Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J. et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333-43
39. Arana ME, Kunkel TA. Mutator phenotypes due to DNA replication infidelity. Semin Cancer Biol. 2010;20:304-11
40. Parry L, Clarke AR. The Roles of the Methyl-CpG Binding Proteins in Cancer. Genes Cancer. 2011;2:618-30
41. Suzuki M, Takahashi T. Aberrant DNA replication in cancer. Mutat Res. 2013;743-744:111-7
42. Hoffmann JS, Cazaux C. Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Semin Cancer Biol. 2010;20:312-9
43. Bao W, Jurka J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob DNA. 2013;4:12
44. Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80
45. Chen DF, Hu L, Yi P, Liu WW, Fang DC, Cao H. Helicobacter pylori damages human gallbladder epithelial cells in vitro. World J Gastroenterol. 2008;14:6924-8
46. Thornton M, Aslam MA, Tweedle EM, Ang C, Campbell F, Jackson R. et al. The unfolded protein response regulator GRP78 is a novel predictive biomarker in colorectal cancer. Int J Cancer. 2013;133:1408-18
47. Tsang YH, Lamb A, Romero-Gallo J, Huang B, Ito K, Peek RM Jr. et al. Helicobacter pylori CagA targets gastric tumor suppressor RUNX3 for proteasome-mediated degradation. Oncogene. 2010;29:5643-50
Author contact
![]() Corresponding authors: Dr. Shahanavaj Khan, E-mail: sdkhanedu.sa; Tel.: +91-921-999-3262; Department of Biosciences, Shri Ram Group of College (SRGC), Muzaffarnagar 251001, India; Department of Pharmaceutics, College of Pharmacy, P.O. Box 2457, King Saud University, Riyadh 11451, Saudi Arabia; Novel Global Community Educational Foundation, Australia. Dr, Ashwag Shami, E-mail: ayshamiedu.sa; Department of Biology, College of Sciences, Princess Nourah bint Abdulrahman University, Riyadh 11617, Saudi Arabia.
Corresponding authors: Dr. Shahanavaj Khan, E-mail: sdkhanedu.sa; Tel.: +91-921-999-3262; Department of Biosciences, Shri Ram Group of College (SRGC), Muzaffarnagar 251001, India; Department of Pharmaceutics, College of Pharmacy, P.O. Box 2457, King Saud University, Riyadh 11451, Saudi Arabia; Novel Global Community Educational Foundation, Australia. Dr, Ashwag Shami, E-mail: ayshamiedu.sa; Department of Biology, College of Sciences, Princess Nourah bint Abdulrahman University, Riyadh 11617, Saudi Arabia.

 Global reach, higher impact
Global reach, higher impact