3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):62-75. doi:10.7150/jca.66016 This issue Cite
Research Paper
Ago-RIP Sequencing Identifies New MicroRNA-449a-5p Target Genes Increasing Sorafenib Efficacy in Hepatocellular Carcinoma
1. Department of Human Genetics, Hannover Medical School, Hannover, Germany.
2. Research Core Unit Genomics, Hannover Medical School, Hannover, Germany.
3. Hannover Unified Biobank (HUB), Hannover Medical School, Hannover, Germany.
Received 2021-8-12; Accepted 2021-10-23; Published 2022-1-1
Abstract
BACKGROUND: Patients with hepatocellular carcinoma (HCC) have very limited treatment options. For the last fourteen years, the multi-tyrosine kinase inhibitor sorafenib has been used as standard-of-care therapeutic agent in advanced HCC. Unfortunately, drug resistance develops in many cases. Therefore, we aimed to find a way to mitigate drug resistance and to improve the sorafenib efficacy in HCC cells. MicroRNAs play a significant role in targeting genes involved in tumor control suggesting microRNA/sorafenib combination therapy as a promising treatment option in advanced HCC.
METHODS: MiR-449a-5p target genes were identified by Ago-RIP sequencing and validated by luciferase reporter assays and expression analyses. Target gene expression and survival data were analyzed in public HCC datasets. Tumor-relevant functional effects of miR-449a-5p and its target genes as well as their impact on the effects of sorafenib were analyzed using in vitro assays. An indirect transwell co-culture system was used to survey anti-angiogenic effects of miR-449a-5p.
RESULTS: PEA15, PPP1CA and TUFT1 were identified as direct target genes of miR-449a-5p. Overexpression of these genes correlated with a poor outcome of HCC patients. Transfection with miR-449a-5p and repression of miR-449a-5p target genes inhibited cell proliferation and angiogenesis, induced apoptosis and reduced AKT and ERK signaling in HLE and Huh7 cells. Importantly, miR-449a-5p potentiated the efficacy of sorafenib in HCC cells via downregulation of PEA15, PPP1CA and TUFT1.
CONCLUSIONS: This study provides detailed insights into the targetome and regulatory network of miR-449a-5p. Our results demonstrate for the first time that targeting PEA15, PPP1CA and TUFT1 via miR-449a overexpression could have significant implications in counteracting sorafenib resistance suggesting miR-449a-5p as a promising candidate for a microRNA/sorafenib combination therapy.
Keywords: Liver cancer, microRNA combination therapy, drug resistance, microRNA target genes, Ago-RIP sequencing, multi-tyrosine kinase inhibitor
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related death worldwide, estimated to have been responsible for 781,631 deaths in 2018 [1]. While patients with early HCC benefit from potentially curative treatment options such as surgical resection or liver transplantation, patients with advanced HCC have a poor prognosis and very limited treatment options [2, 3]. However, more than 80% of HCC patients are diagnosed at an advanced stage and are treated with the multi-tyrosine kinase inhibitor sorafenib as the first-line therapy [3, 4]. Unfortunately, drug resistance in cancer therapy develops very often and is one of the main obstacles to overcome in curing this malignant disease [5, 6]. According to statistical reports, more than 90% of deaths of tumor patients are associated with tumor drug resistance [7, 8]. Therefore, a major goal of HCC research is to identify ways to mitigate drug resistance and to develop new strategies to improve the efficacy of therapeutic agents.
In recent years, it has become evident that the expression level of microRNAs (miRNAs) is globally reduced in cancer [9, 10]. MicroRNAs are small non-coding RNAs involved in the posttranscriptional regulation of gene expression. By binding to complementary sites in the 3' untranslated regions (3'UTRs) of mRNAs they promote mRNA degradation or translational repression [11]. MicroRNAs regulate hundreds of target genes and are powerful regulators of cell proliferation, apoptosis, cell migration and angiogenesis [12, 13]. We have previously reported that miRNA expression profiles of HCC cell lines are regulated by epigenetic mechanisms and have demonstrated the tumor suppressive potential of miR-449a-5p [10, 14]. Beyond this, significantly altered miRNA expression has been observed in a variety of drug-resistant HCC cells compared to drug-sensitive cells, suggesting that miRNAs may promote personalized HCC therapy [6,15]. Consequently, microRNAs are a promising option in cancer therapy to increase drug efficacy and to improve patient outcome.
Sorafenib was the only available standard-of-care for advanced HCC for a decade [4]. Currently, six systemic therapies have been FDA approved based on phase III trials (sorafenib, atezolizumab plus bevacizumab, lenvatinib, regorafenib, cabozantinib and ramucirumab) [4]. Among them the application of multi-tyrosine kinase inhibitor sorafenib is still an effective first-line therapy exhibiting anti-angiogenic and antiproliferative effects [5, 16]. Sorafenib suppresses tumor cell proliferation by inhibiting serine/threonine kinase Raf-1 in the RAF/MEK/ERK signaling pathway. In addition, sorafenib targets vascular endothelial growth factor receptor 2 (VEGFR) tyrosine kinase and other proteins to reduce tumor angiogenesis [17]. However, only approximately 30% of patients can benefit from sorafenib and most of them acquire drug resistance within 6 months [5].
In the publication at hand, we focus on the characterization of the miR-449a-5p targetome and analyze the impact of miR-449a-5p and its target genes PEA15, PPP1CA and TUFT1 on the efficacy of sorafenib in hepatocellular carcinoma.
Materials and Methods
Detailed information is provided in the supplementary data.
Ago2 RNA immunoprecipitation (Ago2-IP)
Ago2-IP was performed to identify direct miR-449a-5p target genes. 1 x 107 HLE cells were transfected with miR-449a-5p or microRNA negative control. Lysed cell extracts were immunoprecipitated with Ago2 antibody (Chromotek, Planegg-Martinsried, Germany) or IgG isotype control (Chromotek) using Dynabeads Protein G (Invitrogen, Carlsbad, USA) and is described in detail in the supplementary data.
Library generation and sequencing
Sequencing libraries were generated with the NEBNext Single Cell/ Low Input RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, USA). Enrichment and size distribution of the libraries were quality-assessed by Agilent Bioanalyzer 2100 on a DNA high sensitivity chip (Agilent, Santa Clara, USA). Three biological replicates were analyzed. Single-read sequencing was performed on an Illumina NextSeq 550 sequencer. Details are provided in the supplementary data.
Statistical modeling and analysis of Ago-RIP sequencing (Ago-RIP-Seq)
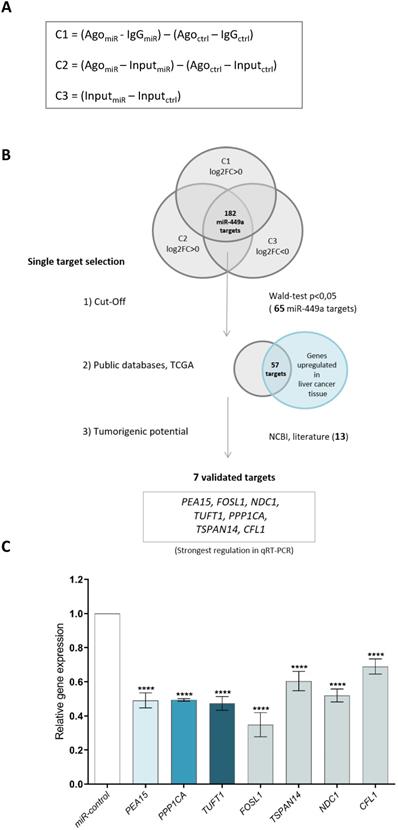
The identification of direct miR-449a-5p targets was achieved by using the concept of linear contrasts [18]. Following miR-449a-5p overexpression, the fraction of miR-449a-5p and its target genes is increased in the Ago complex. To reduce the noise due to unspecific RNA binding to the Protein G beads in the Ago-RIP-Seq experiment, the Ago-IP fractions were adjusted to IgG-IP [Contrast 1, C1] before comparing the Ago-IP fractions of miR-449a-5p with the Ago-IP of miR-control as follows:
[Contrast 1] = (AgomiR-449a - IgGmiR-449a) - (AgomiR-Ctrl - IgGmiR-Ctrl)
Since miR-449a-5p overexpression leads to widespread secondary changes within the gene expression profiles that might impact the profiles of immunoprecipitation [19], it was necessary to adjust the levels of RNAs detected in the Ago-IP fractions to their expression levels measured in the total lysates (=Input) [Contrast 2, C2] as follows:
[Contrast 2] = (AgomiR-449a - InputmiR-449a) - (AgomiR-Ctrl - InputmiR-Ctrl)
As microRNA targets are decreased after miR-449a-5p expression, the comparison [Contrast 3, C3] for identifying whole transcriptional changes was defined as follows:
[Contrast 3] = (InputmiR-449a - InputmiR-Ctrl)
The contrasts C1, C2 and C3 define the type of comparison between factor levels. Wald-tests were applied for testing against zero.
Analysis of mRNA, miRNA and protein expression
Total RNA was isolated using the Direct-zol RNA MiniPrep Kit (Zymo Research). 250 ng total RNA was transcribed into complementary DNA (cDNA) with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, USA). Relative mRNA expression was measured in triplicate by quantitative real-time PCR using TaqMan Gene Expression Assays (Life Technologies) with TBP as reference gene. All TaqMan Assays used in this study are listed in the supplementary data.
For protein analysis, whole cell lysates were prepared with RIPA buffer and protein concentration was measured by Bradford protein assay. Equal amounts of protein were separated in a sodium dodecyl sulfate-polyacrylamide gel and protein levels were analyzed using a standard western blot protocol. Antibodies against PEA15, PPP1CA, TUFT1, (phosphor‑) AKT, (phospho-) ERK, α-actinin, β-actin and GAPDH were used (antibody dilutions are provided in the supplementary data).
Luciferase reporter assay
To validate that PEA15, PPP1CA and TUFT1 are direct target genes of miR-449a-5p, luciferase reporter assays were performed. For this, luciferase reporter vectors were constructed containing the 3'UTR of PEA15, PPP1CA or TUFT1 either with intact miR-449a-5p binding sites or with mutated miR-449a-5p binding sites. Luciferase reporter assays were performed as described in the supplementary data.
Assays to determine proliferation, apoptosis and angiogenesis
Cell viability and apoptosis were measured in triplicate every 24 h at four different times using WST-1 Proliferation Reagent (Roche) and the Caspase3/7 Glo Assay (Promega, Madison, USA), respectively. Angiogenesis was determined by performing a tube formation assay in an indirect co-culture system of HLE or Huh7 and HUVEC cells. Tube formation assays were performed as described in the supplementary methods.
Analysis of public data sets
To analyze the expression of PEA15, PPP1CA and TUFT1 in non-tumorous liver tissue and HCC tissue, expression levels of the NCBI GEO data sets GSE14520 and GSE22058 were downloaded (https://www.ncbi.nlm.nih.gov/geo/, 07.01.2021). In addition, clinical data of the TCGA-LIHC cohort were downloaded from the Cell Index Database CELLX [20]. For survival analysis of the TCGA-LIHC cohort, survival data together with expression levels of PEA15, PPP1CA and TUFT1 were downloaded from http://www.oncolnc.org/ (17.01.2021) and the expression was classified as high (upper median) or low (lower median).
Statistics
Data are represented as mean ± standard deviation (SD) of at least three independent experiments. Statistical significance was determined with GraphPad Prism (GraphPad Software, Version 8) by two-tailed Student's t-tests, ordinary one-way ANOVA or by two-way ANOVA with Dunnett's or Sidaks multiple comparisons test. For experiments analyzing sorafenib-induced apoptosis and cell viability, statistical significance was determined by two-way ANOVA with Tukey's or Dunnett's multiple comparisons test as indicated.
Results
Ago-RIP sequencing identifies new miR-449a-5p target genes
Functional microRNAs are incorporated into the Ago-RISC complex and bind to the mRNAs of their target genes, which are subsequently translationally inhibited or degraded [21]. To identify direct target genes of miR-449a-5p, we transfected HLE cells with miR-449a-5p mimics. After cell lysis (total lysate = input, inputmiR, inputctrl) we immunoprecipitated the Ago complexes from total lysates with Ago antibodies (AgomiR, Agoctrl) and from controls with IgG antibodies (IgGmiR, IgGctrl) to remove unspecific background. The RNA of the input, Ago and IgG fractions of three independent Ago immunoprecipitation experiments were analyzed by high-throughput sequencing (Ago-RIP-Seq). To statistically evaluate the Ago-RIP-Seq results, three comparisons were established to identify direct miR-449a-5p target genes (Fig. 1A). Potential miR-449a target genes were defined by positive log2FC values in comparison 1 and comparison 2 and negative log2FC in comparison 3. Ago-RIP sequencing yielded 182 potential direct miR-449a-5p target genes that fulfilled these conditions in all three replicates (Fig. 1B, Supplementary Table 1). Target gene filtering was conducted to further prioritize the identified miR-449a targets for functional analyses (Fig. 1B). In a first cut-off 65 genes were selected with a Wald-test p-value smaller than 0.05. As an additional selection criterion, the expression of target genes was required to be higher in liver cancer tissue than in the adjacent liver tissue according to TCGA datasets. This prioritization yielded 57 direct miR-449a targets. As a result of manual literature search, 13 out of the 57 genes were identified as potential tumorigenic oncogenes not yet described as potential miR-449a-5p target genes. To validate these findings, we quantified the expression of seven genes (PEA15, PPP1CA, TUFT1, FOSL1, TSPAN14, NDC1 and CFL1) by quantitative Real-Time PCR using input fractions of HLE miR-449a and HLE miR-control cells. 48 h after transfection mRNA expression levels were reduced in all seven cases (Fig. 1C). In the following, we focused on PEA15, PPP1CA and TUFT1 because they showed a strong regulation in qRT-PCR and literature research of these genes indicated promising approaches for the treatment of hepatocellular carcinoma.
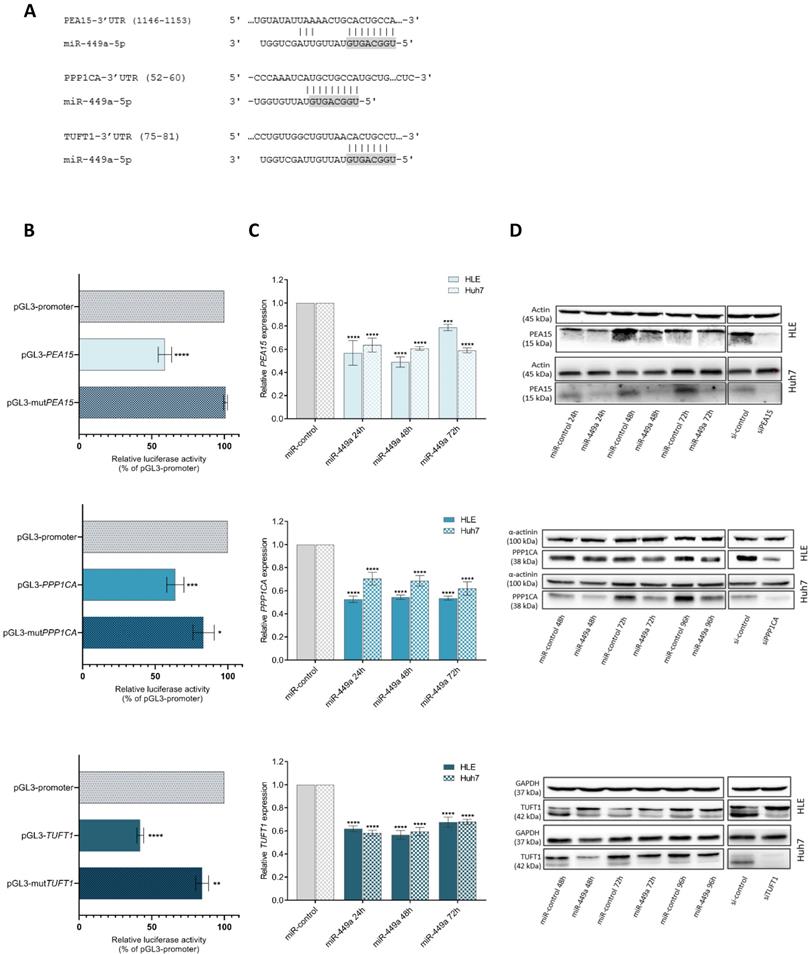
PEA15, PPP1CA and TUFT1 are direct target genes of miR-449a-5p
Next, we aimed to validate the predicted regulation of PEA15, PPP1CA and TUFT1 by miR-449a-5p by performing luciferase reporter assays. For this, we used vectors harbouring the 3'UTRs of PEA15, PPP1CA or TUFT1 including either intact or mutated binding sites for miR-449a-5p that were predicted by TargetScan [22] or IntaRNA [23]. Fig. 2A shows the respective miR-449a-5p binding site with the strongest predicted binding energy for the interaction with PEA15, PPP1CA and TUFT1 3'UTR. Cotransfection of miR-449a with pGL3-wildtype 3'UTR of the candidate genes significantly reduced luciferase activity compared to the empty pGL3 promoter vector while there was no change upon cotransfection with pGL3-mutPEA15 and only a minor reduction upon cotransfection with pGL3-mutPPP1CA and pGL3-mutTUFT1 (Fig. 2B). Furthermore, we analyzed the expression of PEA15, PPP1CA and TUFT1 in HLE and Huh7 cells at different times after miR-449a-5p transfection. The mRNA levels of PEA15, PPP1CA and TUFT1 were reduced by miR-449a-5p at all stages (Fig. 2C). This downregulation was also observed on protein levels (Fig. 2D). For functional analyses, siRNA pools against PEA15, PPP1CA and TUFT1 were established (Suppl. Fig. S1). Western blot analyses after siPool transfection of HLE and Huh7 cells showed a reduced protein expression of PEA15, PPP1CA and TUFT1 (Fig. 2D). Together our results indicate that PEA15, PPP1CA and TUFT1 are direct target genes of miR-449a-5p.
Ago-RIP sequencing identifies new miR-449a-5p target genes. (A) Comparisons calculated to detect potential direct miR-449a-5p target genes and transcriptional changes considering the concept of linear contrasts. (B) Workflow of miR-449a-5p target gene filtering and definition of potential direct miR-449a-5p targets. C = Contrast, FC = fold change. (C) Validation of miR-449a-5p targets in HLE cells by quantitative Real-Time PCR. Relative expression of target RNAs was normalized to miR-control treated HLE cells. ****p<0.0001; ordinary one-way ANOVA with Dunnett's multiple comparisons test.
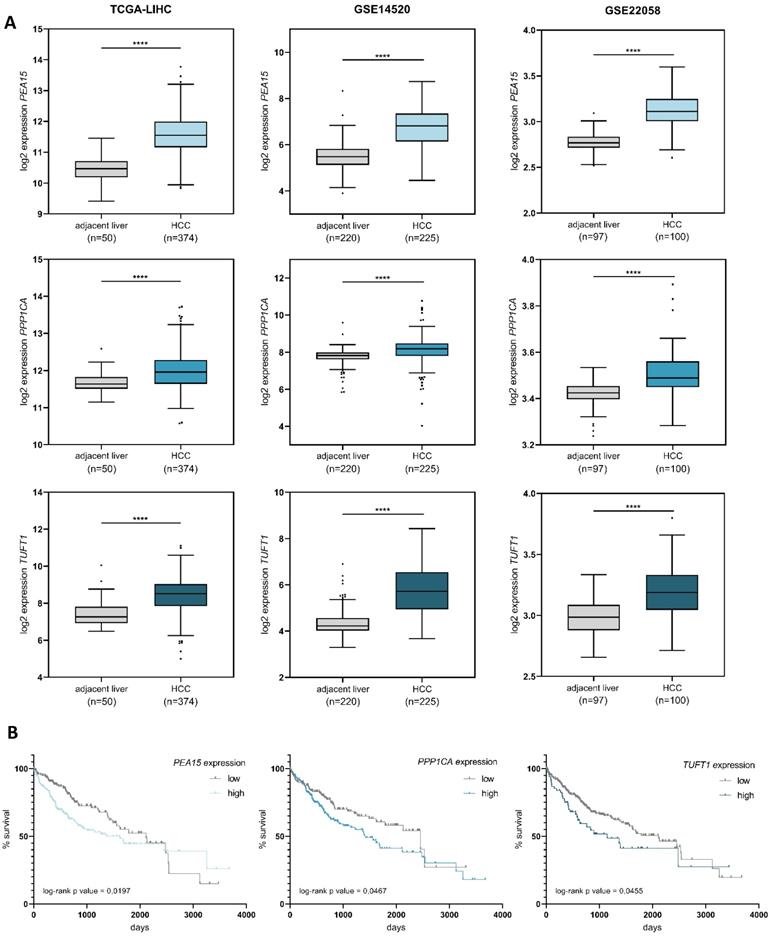
Overexpression of PEA15, PPP1CA and TUFT1 correlates with a poor survival prognosis of HCC patients
To determine the relevance of PEA15, PPP1CA and TUFT1 in hepatocellular carcinoma in vivo, we analyzed three publically available expression datasets of primary HCCs. In all three datasets the expression levels of PEA15, PPP1CA and TUFT1 were significantly increased in liver cancer tissue compared to adjacent non-tumorous liver tissue (Fig. 3A). Furthermore, we observed that patients with high expression of PEA15, PPP1CA or TUFT1 had a trend towards an unfavourable survival prognosis (Fig. 3B). Our findings demonstrate that PEA15, PPP1CA and TUFT1 are frequently overexpressed in HCC and that patients with hepatocellular carcinoma may benefit from the repression of these genes.
PEA15, PPP1CA and TUFT1 are direct target genes of miR-449a-5p. (A) Represented are predicted binding sites in the 3'UTR of PEA15, PPP1CA and TUFT1 with the greatest binding energy for miR-449a-5p. Seed regions of miR-449a are highlighted in gray. (B) Firefly luciferase activity was measured and normalized to renilla luciferase activity. (C, D) RNA (C) and protein (D) expression of PEA15, PPP1CA and TUFT1 was analyzed at three different times after transfection of HLE and Huh7 cells with miR-449a-5p or after transfection with siRNA pools. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; two-way ANOVA with Dunnett's multiple comparisons test.
Overexpression of PEA15, PPP1CA and TUFT1 correlates with poor survival of HCC patients. (A) Expression levels of PEA15, PPP1CA and TUFT1 were analyzed using three public HCC data sets (TCGA-LIHC, GSE14520, GSE22058). PEA15, PPP1CA and TUFT1 expression was significantly higher in HCC tissue than in adjacent non-tumorous liver tissue. Tukey box-and-whisker plot. ****p<0.0001; two-tailed Student's t test (B) Expression values of PEA15, PPP1CA and TUFT1 and survival data of the TCGA-LIHC cohort were retrieved from OncoLnc [51]. Patients were grouped into low or high mRNA expression levels with different percentiles (PEA15 = 50th percentile, PPP1CA = 40th percentile, TUFT1 = 80th percentile). Kaplan-Meier with log-rank test.
Knockdown of PEA15, PPP1CA and TUFT1 exerts distinct tumor suppressive functions
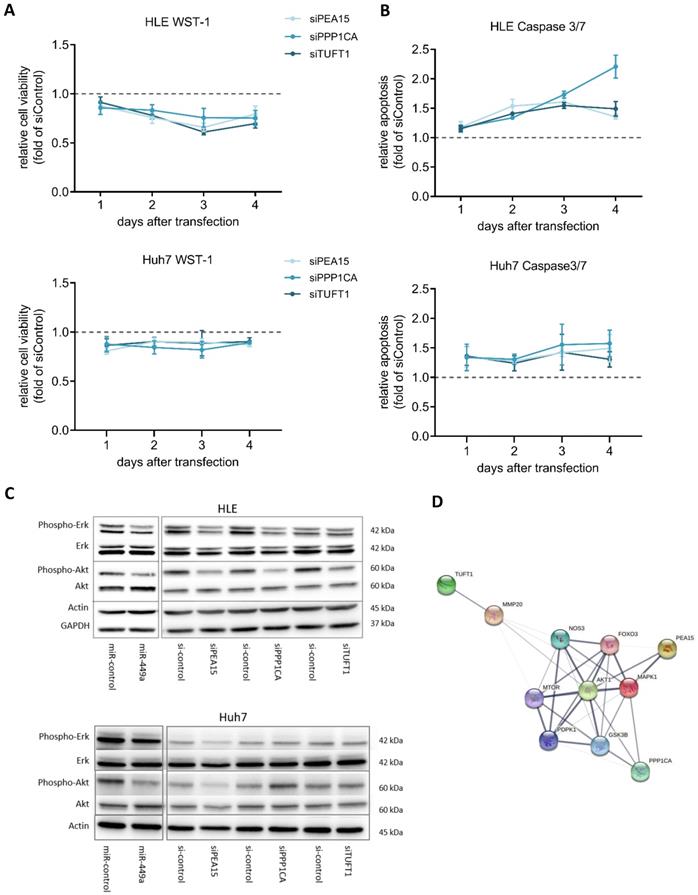
Next, we analyzed the functional effects of PEA15, PPP1CA and TUFT1 by performing cell viability and apoptosis measurements. For this, HCC cell lines HLE and Huh7 were transiently transfected using specific siRNA pools against PEA15, PPP1CA and TUFT1. Knockdown of PEA15, PPP1CA and TUFT1 decreased cell viability (Fig. 4A) and increased apoptosis (Fig. 4B) in each case, whereby knockdown of PPP1CA showed the strongest induction of apoptosis. To further clarify the influence of miR-449a-5p and its target genes PEA15, PPP1CA and TUFT1 on the regulation of HCC related pathways, western blot analyses were performed (Fig. 4C). AKT and ERK 1/2 are active in their phosphorylated state enhancing growth and survival of hepatocellular carcinoma [24]. In this study, transfection of miR-449a-5p led to a reduced phosphorylation of AKT and ERK 1/2 in HLE cells, whereas the total concentration of AKT and ERK 1/2 remained unchanged. In addition, phosphorylation of AKT and ERK 1/2 was also decreased after siRNA induced knockdown of PEA15, PPP1CA and TUFT1 with the exception that downregulation of TUFT1 only reduced AKT phosphorylation (Fig. 4C). In Huh7 cells, miR-449a overexpression decreased AKT phosphorylation and the knockdown of PEA15 led to a reduced AKT and ERK signalling (Fig. 4C). A STRING functional protein association network [25] confirmed the interaction of PEA15, PPP1CA and TUFT1 with AKT and ERK (MAPK1) whereby PEA15 indicated the strongest interaction (Fig. 4D). In summary, knockdown of all three validated miR-449a target genes exerted tumor suppressive functions and has an impact on AKT and ERK signaling, although the extent of the observed effects differed.
miR-449a-5p overexpression and the repression of its target genes PEA15, PPP1CA and TUFT1 prevent tumor angiogenesis
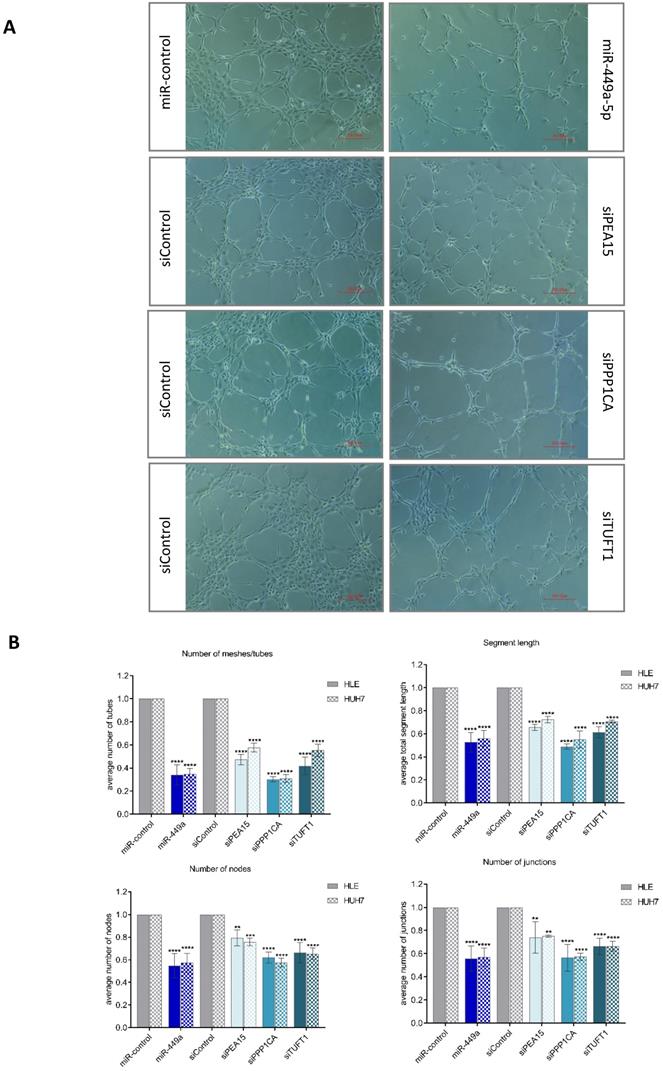
To investigate the impact of miR-449a-5p and its target genes on tumor angiogenesis, we established a co-culture system with endothelial cells (HUVECs) using a 0.4 µm polycarbonate membrane. We used phenotypic changes of HUVECs to evaluate the effects of transiently transfected liver cancer cells on angiogenesis in the co-culture system. HUVECs, co-cultured with HLE or Huh7 cells that were transfected with a negative control, formed a tight cluster of capillary-like tubes and created a mesh-like structure on matrigel. In contrast, after 6 h of co-culture with miR-449a-5p transfected cells, HUVECs elongated, lost contact with each other and the capillary-like tubular structure was less dense, as shown in Fig. 5A. HLE and Huh7 cells transfected with siRNA pools against PEA15, PPP1CA or TUFT1 caused similar morphological changes of HUVECs, whereby transfection of siPPP1CA indicated the strongest anti-angiogenic impact. Quantitative analysis of capillary-like tubular structures confirmed the visual results. Quantification showed that the average number of tubes, junctions and nodes as well as the total segment length were significantly decreased when HUVECs were co-cultured with miR-449a or siRNA transfected HLE cells (Fig. 5B). An indirect co-culture of HUVECs with transfected Huh7 cells showed similar results. As observed in microscopic examination, transfection of miR-449a-5p as well as knockdown of PPP1CA quantitatively caused the most anti-angiogenic effects (Fig. 5B). Altogether, our results provide evidence for the prevention of tumor angiogenesis through the repression of PEA15, PPP1CA and TUFT1 demonstrating the importance of their downregulation by miR-449a-5p.
miR-449a-5p increases sorafenib efficacy of hepatocellular carcinoma cells via downregulation of PEA15, PPP1CA and TUFT1
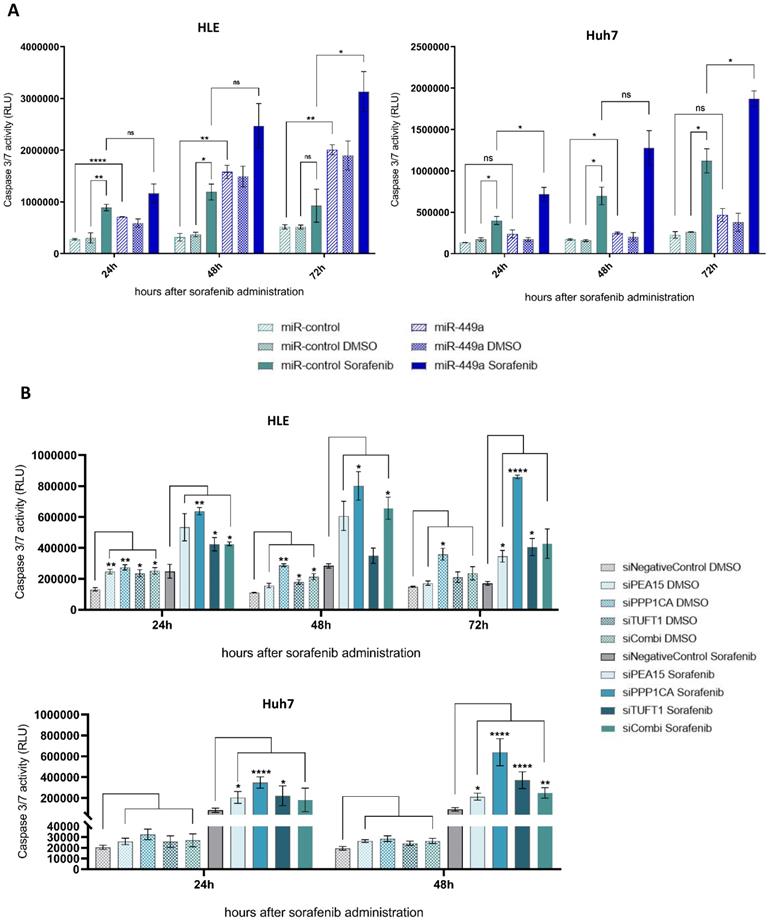
The fact that an increased expression of angiogenesis-related genes as well as an upregulation of MAPK/ERK signaling is associated with sorafenib resistance [26, 27] led us to investigate the impact of miR-449a-5p and its target genes on the effect of sorafenib in hepatocellular carcinoma. First, cell viability and apoptosis of HLE and Huh7 cells were measured at different times after miR-449a-5p overexpression and additional sorafenib treatment. HCC cells transfected with miR-449a-5p and treated with sorafenib exhibited an increased apoptosis at all times compared to cells only treated with sorafenib (Fig. 6A). 72 hours after sorafenib administration, miR-449a-5p significantly tripled sorafenib-induced apoptosis of HLE cells and significantly doubled sorafenib-induced apoptosis of Huh7 cells. Although miR-449a-5p did not significantly enhance the antiproliferative effects of sorafenib, examination of cell viability demonstrated less absorbance in case of HCC cells additionally transfected with miR-449a-5p compared to HCC cells only treated with sorafenib (Suppl. Fig. S2A). Since previous results indicated that knockdown of PEA15, PPP1CA and TUFT1 by miR-449a-5p exerts tumor suppressive functions, we further analyzed the impact of miR-449a-5p target genes on sorafenib efficacy. Therefore, we transiently transfected HLE and Huh7 cells with siRNAs against PEA15, PPP1CA and TUFT1 either alone or in combination (siCombi) and investigated apoptosis and cell viability after sorafenib treatment or DMSO vehicle control administration (Fig. 6B). In total, the apoptosis of HCC cells treated with sorafenib was stronger compared to HCC cells without sorafenib administration at all times. Furthermore, siRNA-mediated knockdown of PEA15, PPP1CA and TUFT1 greatly enhanced the apoptotic effects of sorafenib, whereby siPPP1CA showed the most significant impact (Fig. 6B). Cell viability reflected these results and demonstrated a significant decrease of cell proliferation after additional knockdown of PEA15, PPP1CA and TUFT1 compared to single sorafenib treatment (Suppl. Fig. S2B). Here again, downregulation of PPP1CA caused the most antiproliferative impact. Altogether, the results indicate that miR-449a-5p sensitized hepatocellular carcinoma cells to sorafenib by downregulation of PEA15, PPP1CA and TUFT1, whereby knockdown of PPP1CA had the greatest influence. On top of that, the observed impacts of miR-449a-5p and its target genes on the effects of sorafenib-mediated apoptosis were indicative for the strong tumor suppressive capability of miR-449a and its promising potential in a combination therapy with sorafenib.
miR-449a and knockdown of PEA15, PPP1CA and TUFT1 exert distinct tumor suppressive functions. (A,B) HLE and Huh7 cells were transfected with 3 nM siRNA pools against the expression of either PEA15, PPP1CA or TUFT1. (A) Cell viability was analyzed by WST-1 assay and normalized to si-control (dotted line). (B) Apoptosis was analyzed by caspase 3/7 assay and normalized to cell viability and si-control (dotted line) (C) HLE and Huh7 cells were transfected with miR-449a-5p mimic or siRNA pools against the expression of either PEA15, PPP1CA or TUFT1. 48 h after transfection, protein expression of p-ERK 1/2, ERK 1/2, p-Akt and Akt was analyzed by western blotting with Actin and GAPDH as loading controls. Gels were processed in parallel. (D) Protein association network in STRING illustrating the interaction with MAPK1 (ERK) and AKT1. Line thickness indicates the strength of data support [25].
miR-449a-5p and knockdown of PEA15, PPP1CA and TUFT1 reduce endothelial cell tube formation. (A) Representative images of capillary-like tubular structures of human umbilical vein endothelial cells (HUVEC) on Matrigel when co-cultured with miR-449a-5p/miR-control or siRNA/si-control transfected HLE or Huh7 cells. Angiogenic changes were imaged using an inverted light microscope. Scale bar = 200.00 µm (B) Angiogenic parameters (number of tubes, segment length, number of junctions and number of nodes) were quantified with Angiogenesis Analyzer from ImageJ. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; two-way ANOVA with Sidak's multiple comparisons test.
miR-449a-5p and knockdown of PEA15, PPP1CA and TUFT1 increase sorafenib efficacy of hepatocellular carcinoma cells. (A) HLE and Huh7 cells were transfected with miR-449a-5p mimic/ miR-control and treated with 10 µM sorafenib or DMSO vehicle control. 24 h, 48 h and 72 h after sorafenib treatment tumor cell apoptosis and cell viability were measured. Apoptosis was normalized to cell viability (Suppl. Fig. S2A); two-way ANOVA with Tukey's multiple comparisons (square brackets). (B) HLE and Huh7 cells were transfected with siRNA pools, either alone or in combination (siCombi) and treated with 10 µM sorafenib or DMSO vehicle control. 24 h, 48 h and 72 h after sorafenib treatment tumor cell apoptosis and cell viability were measured. Apoptosis was normalized to cell viability (Suppl. Fig. S2B); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; two-way ANOVA with Dunnett's multiple comparisons test (square brackets). RLU = relative light units.
Discussion
In this study, we comprehensively investigated the impact of miR-449a-5p and its target genes PEA15, PPP1CA and TUFT1 on the effects of sorafenib treatment in hepatocellular carcinoma. Specifically, we examined whether miR-449a-5p is a suitable candidate for a microRNA/ sorafenib combination therapy. Several studies have reported that microRNAs play an emerging role in drug resistance of hepatocellular carcinoma [15, 28, 29] suggesting that a microRNA/ sorafenib combination therapy is an attractive option for HCC therapy. Here, we demonstrated that miR-449a-5p strongly potentiates sorafenib-induced apoptosis of hepatocellular carcinoma cells by downregulating PPP1CA in particular.
As we have previously shown, miR-449a and several other microRNAs are epigenetically deregulated in HCC, so that the role of microRNAs in tumorigenesis has come into focus [10]. In addition, we have demonstrated that miR-449a inhibits proliferation and induces apoptosis in vitro and suppresses tumor growth in vivo in hepatocellular carcinoma [10, 14]. Similar tumor suppressive effects of miR-449a have also been observed in breast [30], lung [31], gastric [32] and prostate cancer [33]. In this study, we confirmed the tumor suppressive potential of miR-449a-5p and demonstrated its anti-angiogenic effect. We further deciphered the large network of genes and signaling pathways that are regulated by miR-449a-5p by Ago-RIP sequencing and, thereby, contributed to the knowledge on its tumor suppressive potential. Seven significantly downregulated mRNAs were detected and showed a strong regulation in qRT-PCR. Literature research led us to focus our study on PEA15, PPP1CA and TUFT1 since they represent promising targets for the treatment of HCC. This is the first study that deciphered the miR-449a-5p targetome via Ago-RIP sequencing and that demonstrated a negative impact of miR-449a-5p on angiogenesis in hepatocellular carcinoma.
Our study provides a link between PEA15, PPP1CA, TUFT1 and miR-449a-5p. Performing luciferase reporter assays and western blottings, we demonstrated that PEA15, PPP1CA and TUFT1 are direct target genes of miR-449a. We revealed overexpression of PEA15, PPP1CA and TUFT1 in three different gene expression datasets of HCC providing evidence for an oncogenic role of these genes in HCC. Other studies have likewise reported an upregulation of PEA15 [26] and TUFT1 [34] in HCC tissues. In addition, we showed that HCC patients with high levels of PEA15, PPP1CA or TUFT1 have a shorter survival than patients with a low expression of these genes. Overexpression of PEA15, PPP1CA and TUFT1 and a related poorer outcome have also been observed in other tumor entities [35-37]. Taken together, PEA15, PPP1CA and TUFT1 appear to act as oncogenes and, therefore, their inhibition or downregulation may improve HCC therapy.
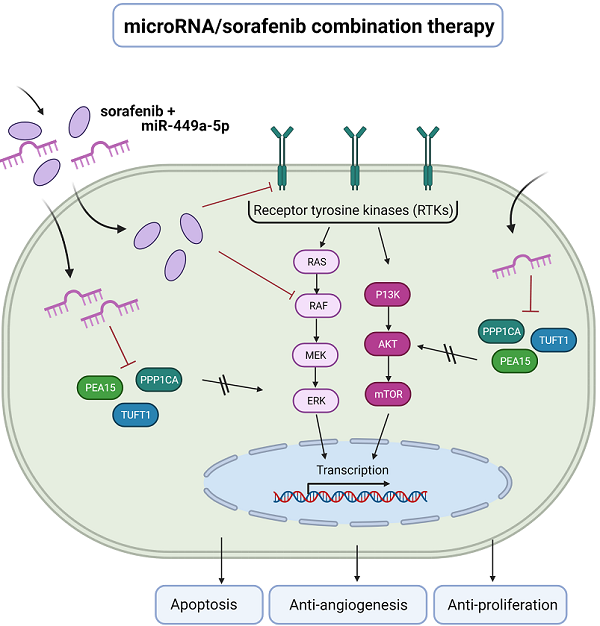
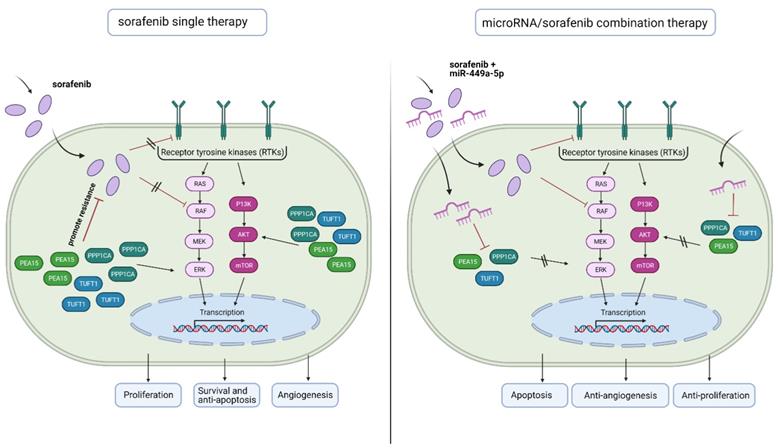
Possible roles in tumorigenesis are already known for each of the three examined genes. PEA15 encodes a death effector domain-containing phosphoprotein that functions as a negative regulator of apoptosis [38] and has been described to be involved in cell proliferation, migration and upregulating the ERK/MAPK signaling pathway [26,39]. However, PEA15 has a dual role in tumor regulation depending on its phosphorylation status and the cellular environment [39, 40]. It has been revealed that phosphorylation of PEA15 has promoted the proliferation and invasion of gastric cancer cells via ERK phosphorylation [35], whereas the unphosphorylated state has inhibited the ERK and EGFR phosphorylation, thus inhibiting proliferation, invasion and metastasis of breast and ovarian cancer [39]. PPP1CA encodes the subunit of serine/threonine specific phosphatase PP1 and is involved in multiple cellular functions including proliferation, invasion, cell survival and differentiation [41, 42]. As a B-Raf activating phosphatase it also plays a role in the upregulation of ERK/MAPK signaling [43, 44]. TUFT1 encodes an acidic protein that is involved in angiogenesis, in the adaptation of hypoxia and promotes tumor growth and metastasis of HCC by activating the PI3K/AKT pathway [34, 45, 46]. Interestingly, many studies have provided evidence that an increased expression of angiogenesis and hypoxia-related genes as well as an upregulation of ERK and AKT signaling is also associated with sorafenib resistance in hepatocellular carcinoma (Fig. 7) [5, 8, 26, 27]. In addition, it is already known that PEA15 confers resistance to sorafenib in HCC [26].
In line with these observations, we showed that knockdown of PEA15, PPP1CA and TUFT1 increases apoptosis, decreases cell viability and significantly reduces angiogenesis in HLE and Huh7 cells. In addition, downregulation of PEA15, PPP1CA and TUFT1 decreased AKT signaling and knockdown of PEA15 and TUFT1 also reduced ERK signaling. These results indicate that miR-449a-5p and its target genes play a role in counteracting sorafenib resistance (Fig. 7). Consistent with our findings, we demonstrated that miR-449a-5p overexpression as well as downregulation of PEA15, PPP1CA and TUFT1 significantly potentiated sorafenib-induced apoptosis, whereby knockdown of PPP1CA had the greatest impact. Beyond this, Wei et al. have observed significantly altered miRNA expression in a variety of drug-resistant HCC cells, compared to those in drug-sensitive cells, suggesting that miRNAs may promote individualized HCC therapy [15]. In this study, miR-449a-5p enhanced the efficacy of sorafenib via silencing PEA15, PPP1CA and TUFT1. These results are in line with Yang et al. who have reported that the miR-449a-5p related miR-34 reduces cell viability, promotes cell apoptosis and enhances sorafenib-induced apoptosis in HCC cell lines [47]. Thus, our data suggest that targeting PEA15, PPP1CA and TUFT1 by miR-449a-5p overexpression may have significant implications in potentiating the effects of sorafenib therapy.
Schematic diagram depicting the benefit of microRNA/ sorafenib combination therapy. (Left) Sorafenib resistance in hepatocellular carcinoma is associated with an increased expression of angiogenesis and hypoxia-related genes (TUFT1) as well as an upregulation of ERK and AKT signaling (PEA15, PPP1CA). (Right) A microRNA-449a/ sorafenib combination therapy increases sorafenib efficacy via downregulation of miR-449a target genes PPP1CA, PEA15 and TUFT1. This downregulation results in an increased apoptosis and a decreased angiogenesis and tumor cell proliferation. Created with BioRender.com.
Various studies have strongly suggested that counterbalancing the expression of microRNAs in drug resistant cells can re-sensitize cancer cells to therapeutic agents [29]. However, there is no clinical trial for microRNA/sorafenib combination therapy yet. For further development of microRNA therapies, an extensive characterization of candidate microRNAs and detailed knowledge of their specific target, regulated pathways and functional effects is essential [14]. Accordingly, we analyzed the miR-449a-5p targetome and its effect on sorafenib efficacy in HCC.
Recently, immunotherapy (atezolizumab plus bevazizumab) has been FDA approved as additional first-line therapy in advanced HCC [4]. Even though immunotherapy is a promising therapy option for advanced HCC, it can unbalance the immune system and result in a wide range of immune-related adverse events [48]. Studies have shown that HCCs belonging to the immune-excluded class, characterized by Wnt/ß-Catenin mutations, are proposed to be primarily resistant to immunotherapy [4, 49]. Furthermore, Pfister et al. recently revealed that immunotherapy did even impair overall survival in patients with non-viral HCC [50]. That is why sorafenib continues to be an important first-line therapy option especially in non-viral related HCC. The combination of sorafenib with miR-449a may extend the overall survival of HCC patients and reactivates sorafenib's attractiveness in advanced hepatocellular carcinoma.
Conclusions
In conclusion, we here present the first evidence that miR-449a-5p increases the efficacy of sorafenib in HCC cells. Furthermore, our data suggest that counterbalancing microRNA expression may have significant implications in potentiating the effect of sorafenib therapy. Although further validation is required before miR-449a-5p may enter the routine clinical scenario. Our study provides evidence that miR-449a-5p is a promising candidate for a microRNA/sorafenib combination therapy. Therefore, miR-449a-5p may be a novel option for the treatment of patients with advanced HCC.
Abbreviations
Ago-RIP: Argonaute-2-RNA-immunoprecipitation; AKT: AKT Serine/Threonine Kinase 1; C1/C2/C3: Contrast 1/2/3; ERK: extracellular-signal regulated kinases; FDA: Food and Drug administration; HCC: Hepatocellular carcinoma; MEK: Mitogen-activated protein kinase kinase; miR-449a: microRNA-449a-5p; miRNA/miR: microRNA; PEA15: Proliferation And Apoptosis Adaptor Protein 15; PPP1CA: Protein Phosphatase 1 Catalytic Subunit Alpha; Raf-1: Raf-1 Serine/Threonine Kinase; RISC: RNA-induced silencing complex; RLU: relative light units; Seq: Sequencing; TUFT1: Tuftelin 1; VEGFR: Vascular Endothelial Growth Factor Receptor; 3'UTR: 3' untranslated region.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank the German Cancer Aid (Deutsche Krebshilfe) for supporting this work and Claudia Davenport for critical reading of the manuscript.
Funding
This work was supported by grants from the German Cancer Aid (70113683).
Availability of data and materials
The public datasets analyzed during the current study are available in the repositories listed below:
• Gene Expression Omnibus
GSE14520 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520
GSE22058 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058
• The Cancer Genome Atlas
TCGA-LIHC https://portal.gdc.cancer.gov/projects/TCGA-LIHC
• OncoLnc
OncoLNC-LIHC http://www.oncolnc.org/
Authors' contributions
BS, TRks, TI and BSch conceived the general experimental scheme. TRks, AS, NH, ME, VS and ODB designed and performed experiments. RIP-Seq experiments were designed and piloted by ODB and TRks. The manuscript was written by TRks and revised by BS. The authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018Nov;68(6):394-424
2. Galle PR, Forner A, Llovet JM, Mazzaferro V, Piscaglia F, Raoul J-L. et al. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. Journal of Hepatology. 2018Jul;69(1):182-236
3. Li D, Sedano S, Allen R, Gong J, Cho M, Sharma S. Current Treatment Landscape for Advanced Hepatocellular Carcinoma: Patient Outcomes and the Impact on Quality of Life. Cancers. 2019Jun18;11(6):841
4. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021Dec;7(1):6
5. Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F. et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Sig Transduct Target Ther. 2020Dec;5(1):87
6. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenet. 2019Dec;11(1):25
7. Longley D, Johnston P. Molecular mechanisms of drug resistance. J Pathol. 2005Jan;205(2):275-92
8. Zhai B. Mechanisms of resistance to sorafenib and the corresponding strategies in hepatocellular carcinoma. WJH. 2013;5(7):345
9. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005Jun;435(7043):834-8
10. Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H. et al. Histone Deacetylases Activate Hepatocyte Growth Factor Signaling by Repressing MicroRNA-449 in Hepatocellular Carcinoma Cells. Gastroenterology. 2012Sep;143(3):811-820.e15
11. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods: SALIMINEJAD et al. J Cell Physiol. 2019May;234(5):5451-65
12. Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009Jan;136(2):215-33
13. Miska EA. How microRNAs control cell division, differentiation and death. Current Opinion in Genetics & Development. 2005Oct;15(5):563-8
14. Sandbothe M, Buurman R, Reich N, Greiwe L, Vajen B, Gürlevik E. et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. Journal of Hepatology. 2017May;66(5):1012-21
15. Wei L, Wang X, Lv L, Liu J, Xing H, Song Y. et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019Dec;18(1):147
16. Llovet JM, Hilgard P, de Oliveira AC, Forner A, Zeuzem S, Galle PR. et al. Sorafenib in Advanced Hepatocellular Carcinoma. n engl j med. 2008 13
17. Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D. et al. Sorafenib Blocks the RAF/MEK/ERK Pathway, Inhibits Tumor Angiogenesis, and Induces Tumor Cell Apoptosis in Hepatocellular Carcinoma Model PLC/PRF/5. Cancer Research. 2006Dec15;66(24):11851-8
18. Pickl JMA, Tichy D, Kuryshev VY, Tolstov Y, Falkenstein M, Schüler J. et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget. 2016Sep13;7(37):59589-603
19. Meier J, Hovestadt V, Zapatka M, Pscherer A, Lichter P, Seiffert M. Genome-wide identification of translationally inhibited and degraded miR-155 targets using RNA-interacting protein-IP. RNA Biology. 2013Jun;10(6):1017-29
20. Ching KA, Wang K, Kan Z, Fernandez J, Zhong W, Kostrowicki J. et al. CELL INDEX DATABASE (CELLX): A WEB TOOL FOR CANCER PRECISION MEDICINE. Pac Symp Biocomput. 2015:10-9
21. Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. RNA. 2019Jan;25(1):1-16
22. Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015Aug12;4:e05005
23. Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Research. 2017Jul3;45(W1):W435-9
24. Huang S-Z, Wei M-N, Huang J-R, Zhang Z-J, Zhang W-J, Jiang Q-W. et al. Targeting TF-AKT/ERK-EGFR Pathway Suppresses the Growth of Hepatocellular Carcinoma. Front Oncol. 2019Mar15;9:150
25. Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J. et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research. 2009Jan1;37(Database):D412-6
26. Quintavalle C, Hindupur SK, Quagliata L, Pallante P, Nigro C, Condorelli G. et al. Phosphoprotein enriched in diabetes (PED/PEA15) promotes migration in hepatocellular carcinoma and confers resistance to sorafenib. Cell Death Dis. 2017Oct;8(10):e3138-e3138
27. Karashima T, Fukuhara H, Tamura K, Ashida S, Kamada M, Inoue K. et al. Expression of angiogenesis-related gene profiles and development of resistance to tyrosine-kinase inhibitor in advanced renal cell carcinoma: Characterization of sorafenib-resistant cells derived from a cutaneous metastasis: Resistance to sorafenib in renal cell carcinoma. Int J Urol. 2013Sep;20(9):923-30
28. Zhang M, Zhang H, Hong H, Zhang Z. MiR-374b re-sensitizes hepatocellular carcinoma cells to sorafenib therapy by antagonizing PKM2-mediated glycolysis pathway. 2019 Apr 1;9(4):765-778.
29. Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019Oct;7(20):577-577
30. Xu B, Zhang X, Wang S, Shi B. MiR-449a suppresses cell migration and invasion by targeting PLAGL2 in breast cancer. Pathology - Research and Practice. 2018May;214(5):790-5
31. Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR-449a Suppresses Tumor Growth, Migration, and Invasion in Non-Small Cell Lung Cancer by Targeting a HMGB1-Mediated NF-kB Signaling Pathway. 2020;9.
32. Li X, Li H, Zhang R, Liu J, Liu J. MicroRNA-449a Inhibits Proliferation and Induces Apoptosis by Directly Repressing E2F3 in Gastric Cancer. 2015;10.
33. Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H. et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009Apr;28(14):1714-24
34. Dou C, Zhou Z, Xu Q, Liu Z, Zeng Y, Wang Y. et al. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca2+/PI3K/AKT pathway. Oncogene. 2019Feb;38(8):1239-55
35. Jiang X, Zhang C, Li W, Jiang D, Wei Z, Lv M. et al. PEA-15 contributes to the clinicopathology and AKT-regulated cisplatin resistance in gastric cancer. ONCOLOGY REPORTS. 2019 11
36. Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T. et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011Sep;105(6):833-41
37. Liu W, Zhang L, Jin Z, Zhao M, Li Z, Chen G. et al. TUFT1 is expressed in breast cancer and involved in cancer cell proliferation and survival. Oncotarget. 2017Aug24;8(43):74962-74974
38. Pastorino S, Renganathan H, Caliva MJ, Filbert EL, Opoku-Ansah J, Sulzmaier FJ. et al. The death effector domain protein PEA-15 negatively regulates T-cell receptor signaling. FASEB j. 2010Aug;24(8):2818-28
39. Tang B, Liang W, Liao Y, Li Z, Wang Y, Yan C. PEA15 promotes liver metastasis of colorectal cancer by upregulating the ERK/MAPK signaling pathway. Oncol Rep. 2019Jan;41(1):43-56
40. Sulzmaier FJ, Opoku-Ansah J, Ramos JW. Phosphorylation is the switch that turns PEA-15 from tumor suppressor to tumor promoter. Small GTPases. 2012Jul;3(3):173-7
41. Wu J-G, Wang J-J, Jiang X, Lan J-P, He X-J, Wang H-J. et al. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015Oct;18(4):729-39
42. Xiao L, Gong L-L, Yuan D, Deng M, Zeng X-M, Chen L-L. et al. Protein phosphatase-1 regulates Akt1 signal transduction pathway to control gene expression, cell survival and differentiation. Cell Death Differ. 2010Sep;17(9):1448-62
43. Chen M, Wan L, Zhang J, Zhang J, Mendez L, Clohessy JG. et al. Deregulated PP1α phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat Commun. 2018Dec;9(1):159
44. Sun H, Ou B, Zhao S, Liu X, Song L, Liu X. et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019Oct;48:236-47
45. Liu H, Zhu J, Mao Z, Zhang G, Hu X, Chen F. Tuft1 promotes thyroid carcinoma cell invasion and proliferation and suppresses apoptosis through the Akt-mTOR/GSK3β signaling pathway. Am J Transl Res. 2018Dec15;10(12):4376-4384
46. Ria R, Simeon V, Melaccio A, Di Meo G, Trino S, Mazzoccoli C. et al. Gene expression profiling of normal thyroid tissue from patients with thyroid carcinoma. Oncotarget. 2016May17;7(20):29677-88
47. Yang F, Li Q, Gong Z, Zhou L, You N, Wang S. et al. MicroRNA-34a Targets Bcl-2 and Sensitizes Human Hepatocellular Carcinoma Cells to Sorafenib Treatment. Technol Cancer Res Treat. 2014Feb;13(1):77-86
48. Cui T, Liu Y, Wang J, Liu L. Adverse Effects of Immune-Checkpoint Inhibitors in Hepatocellular Carcinoma. OTT. 2020 Nov;Volume 13:11725-40
49. Pinyol R, Sia D, Llovet JM. Immune Exclusion-Wnt/CTNNB1 Class Predicts Resistance to Immunotherapies in HCC. Clin Cancer Res. 2019Apr1;25(7):2021-3
50. Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M. et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature. 2021Apr;592(7854):450-456
51. Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016Jun13;2:e67
Author contact
![]() Corresponding author: Dr. Britta Skawran, Department of Human Genetics, Hannover Medical School, Carl-Neuberg-Straße 1, 30625 Hannover, Germany; +49 511 532-4544; skawran.brittade
Corresponding author: Dr. Britta Skawran, Department of Human Genetics, Hannover Medical School, Carl-Neuberg-Straße 1, 30625 Hannover, Germany; +49 511 532-4544; skawran.brittade

 Global reach, higher impact
Global reach, higher impact