3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):124-133. doi:10.7150/jca.63578 This issue Cite
Research Paper
Akkermansia muciniphila administration exacerbated the development of colitis-associated colorectal cancer in mice
1. Gannan Medical University, Ganzhou, Jiangxi, 341000, China.
2. The Fifth People's Hospital of Jinan, Jinan, Shandong, 250000, China.
3. School of Basic Medicine, Gannan Medical University, Ganzhou, Jiangxi 341000, China.
4. Center for Immunology, Key Laboratory of Prevention and Treatment of Cardiovascular and Cerebrovascular Diseases, Ministry of Education, Gannan Medical University, Ganzhou, Jiangxi 341000, China.
Received 2021-6-6; Accepted 2021-11-12; Published 2022-1-1
Abstract
Colorectal cancer (CRC) is one of the most common digestive tract malignancies and inflammation and gut microbiota are well-known key factors to influence CRC development. Akkermansia mucinipila is an important gram-negative anaerobic bacterium that can degrade mucin in gut. Previous studies suggested that A. muciniphila may affect inflammation and cell proliferation, but the relationship between A. muciniphila and CRC is not clarified. Here C57BL/6 mice were administrated with A. muciniphila or PBS and then treated with azoxymethane (AOM)/dextran sodium sulphate (DSS) to induce CRC. The mice receiving A. muciniphila administration had more serious weight loss, shorter colon length and more intestinal tumors than those receiving PBS administration after AOM/DSS treatment. More colon damage and less goblet cells were also observed in A. muciniphila treated mice. Furthermore, A. muciniphila administration induced more Ki67+ proliferating cells, higher PCNA expression and elevated gene expression of proliferation-associated molecules including Snrpd1, Dbf4 or S100A9. At early stage of CRC development, in comparison with controls, the mice receiving A. muciniphila administration also had more body weight loss and shorter colon length, as well as higher gene expression of inflammatory cytokines. Furthermore, the in vitro experimental results showed that the co-culture of colon epithelial cells with A. muciniphila enhanced the cell proliferation and gene expression of proliferation-associated molecules. Therefore, A. mucinipila may promote the formation of CRC in mice through increasing the early level of inflammation and the proliferation of intestinal epithelial cells.
Keywords: Akkermansia mucinipila, Colorectal cancer, Cell proliferation, Inflammatory cytokines
Introduction
Colorectal cancer (CRC) is one of the most common digestive tract malignancies and a major cause of cancer-related morbidity and mortality, which has done great harm to people's health around the world [1]. Despite its severity, the pathogenesis of CRC is still unclear, and the current treatment has not been quite satisfying. Therefore, the studies on the molecular mechanism of CRC development that can provide new ideas and therapeutic targets for its diagnosis and treatment are in urgent need. The cause of CRC is complicated and involves genetic factors, diet, and inflammation. Among them, inflammation is a well-known risk factor. Clinically, patients with inflammatory bowel diseases (IBD) had an increased risk of developing CRC, which was also supported by the studies on CRC pathogenesis utilizing IBD mouse model [2, 3].
Absent in melanoma 2 (AIM-2) is a pattern recognition receptor that can recognize dsDNA. In our previous study and other studies, it was found that AIM2 could suppress colitis-associated colorectal cancer (CAC) induced by azoxymethane (AOM) and dextran sodium sulfate (DSS) treatment. The underlying mechanism can be divided into two parts: on the one hand, AIM2 suppressed overt proliferation by controlling the proliferation of intestinal epithelial cells as well as the expansion of intestinal stem cells [4-6]. On the other hand, AIM2 suppressed colorectal tumorigenesis by modulating the gut microbiota [6]. The genomic sequencing of mouse microbiota showed that Aim2-/- mice harbored increased levels of Akkermansia muciniphila, Anaeroplasma, and decreased levels of Anaerostipes, Bifidobacterium, Flexispira, Prevotella and Paraprevotella species relative to WT mice. More importantly, the susceptibility to CRC could be reduced in Aim2-/- mice by co-housing them with WT mice, indicating that WT mouse might harbor some protective microbiota in their intestine, and there's potential gut microbiota alteration in Aim2-/- mice which led to hyper-susceptibility to CRC [6]. However, it's still unclear how the alteration of gut microbiota affected the proliferation of intestinal stem cells as well as the progression of CRC.
A. muciniphila, a gram-negative anaerobic bacterium from phylum Verrucomicrobia, can be isolated from the human intestine and other animals. It was previously shown that A. muciniphila can produce acetate and propionate during its growth on mucin [7, 8]. A. muciniphila engraftment in the mucosal layer of intestinal mucosa triggers the host's metabolic and immune responses. The number of A. muciniphila is often considered as an indicator in monitoring host's metabolic status due to the bacterium's ability to increase the thickness of intestinal mucosa and to enhance the intestinal barrier function. Other than that, A. muciniphila can produce short-chain fatty acids (SCFAs) that are in favor of the host and other gut microbiota, thus improving intestinal homeostasis [9].
The relationship between A. muciniphila and CRC remains controversial. A study showed that the abundance of A. muciniphila in CRC patients was 4 times higher than that of healthy subjects [10]. In addition, A. muciniphila was also found to be significantly increased in mucosal biopsy samples of CRC patients [11]. Another study showed that the number difference of A. muciniphila between CRC patients and non-CRC patients was indistinctive [12]. In the meantime, it was showed decreased number of A. muciniphila in CRC patients with severe symptoms, indicating that A. muciniphila did not contribute to CRC development [12, 13]. As for animal studies, it was found that enrichment of A. muciniphila in mice developed CAC [14]. Interestingly, a study found that mice with intestinal epithelial cells (IEC)-specific deficiency in Pten, a potent tumor suppressor, did not spontaneously develop CRC, which was correlated with diminished A. muciniphila, indicating the role of A. muciniphila in promoting CRC [15]. Recently, a study showed that the treatment of vitamin D can alleviate CRC development and gut microbiota sequencing results showed increased number of A. muciniphila in mice with less symptoms of CRC [16]. In conclusion, it is still baffling how the number of A. muciniphila affect CRC development.
Some other studies used oral administration of A. muciniphila methods to determine the role of A. muciniphila more directly in intestinal inflammation and tumorigenesis. During Salmonella typhimurium infection, A. muciniphila were shown to exacerbate S. typhimurium-induced intestinal inflammation with reduced goblet cell numbers [17]. After introducing A. muciniphila to intestine specific conditional Apc mutant mice that spontaneously develop tumors in the colon, significantly increased tumor burden was observed in those mice, suggesting that A. muciniphila might promote tumorigenesis in the colon [18]. These results suggest that A. muciniphila may promote inflammation and tumorigenesis in the intestine. However, it is still unclear whether or how A. muciniphila influences CRC development.
In our study, mice were orally gavaged by A. muciniphila before AOM/DSS treatment to develop CAC, then we evaluated the inflammation status and neoplastic development of those mice. In vitro study, A. muciniphila and human CRC cell line were co-cultured, then we monitored the influence of bacterium on cell proliferation. This study will contribute to our understanding on host interaction with specific intestinal bacteria and provide new ideas and theoretical basis for the prevention and treatment of CRC.
Materials & Methods
Mice
C57BL/6 mice were from Nanjing Animal Model Center (Nanjing, Jiangsu, China). They were maintained in a specific pathogen-free facility in the Experimental Animal Center of Gannan Medical University (Ganzhou, Jiangxi, China). This study was approved by the Ethics Committee of Gannan Medical University.
Bacterial infection
A. muciniphila (ATCC#BAA-835) was grown anaerobically in brain heart infusion broth overnight and then sub-cultured. The bacterial solution was collected, and then diluted to a concentration of 1×108 cells of A. muciniphila per 100 µl medium. The mice were orally administrated with PBS or A. muciniphila at the 0, 3, 5, and 7 days after experiments were initiated, and the feces were collected on day 0 and day 14 to validate the colonization of A. muciniphila.
CAC animal model
The CAC animal model was established as previously described [6]. Mice were intraperitoneally injected with AOM (10 mg/kg) and then treated with 3% DSS in drinking water for 5 days at 5 days after AOM injection. The mice were treated with DSS for another two rounds and sacrificed on the 80th day after AOM injection to observe the tumor burden.
RT-qPCR
Total RNA was extracted using Trizol (Invitrogen) following manufacture's instruction, and RNA concentration was measured using Nanodrop. mRNA was reversely transcribed into cDNA. qPCR was performed on an QuantStudio 7 Flex real-time PCR instrument with SYBR Green kit (ThermoFisher, Fremont, CA, USA) using corresponding primers (the sequences of the target gene were shown in Supplementary Table 1). The gene expression was quantitated by PCR with gene-specific primers. The relative gene expression was expressed as 2-ΔΔCT.
Western-blot
Proteins were extracted from colon tissues using RIPA lysis buffer with protease inhibitors. The samples were resolved in SDS-PAGE and transferred onto NC membranes, Membranes were blocked in 5% milk for 1 hour and then incubated with anti- PCNA (CST, #13110, 1:1000 dilution) and anti-β-actin (Proteintech, #66009-1-Ig, 1:1000 dilution) antibodies overnight at 4 °C. After wash, membranes were incubated with Anti-rabbit (BOSTER, #BA1054) or anti-mouse (BOSTER, #BA1050) secondary antibody at room temperature for 1h. Finally, after adding ECL (Thermo, #A38555) substrates, the protein bands were analyzed by Chemdoc (Bio-rad).
Hematoxylin & Eosin (H&E) staining and Immunohistochemistry
Colon tissues were embedded in paraffin, sectioned (4 μm), and stained using regular H&E methods. The inflammatory cells per crypt in slides were counted under the light microscopes. Histological lesion were scored by a histologist blindly based on the extent and severity of inflammation, ulceration, and hyperplasia of the mucosa as previously described [19]. Specifically, scores for inflammation were as follows: 0 = normal (within normal limits); 1 = mild (small, focal, or widely separated, limited to lamina propria); 2 =moderate (multifocal or locally extensive, extending to submucosa); 3 = severe (transmural inflammation with ulcers covering >20 crypts). Scores for ulceration were as follows: 0 = normal (no ulcers); 1 = mild (1-2 ulcers involving up to a total of 20 crypts); 2 = moderate (1-4 ulcers involving a total of 20-40 crypts); 3 = severe (>4 ulcers or over 40 crypts). Scores for Mucosal hyperplasia were as follows: 0 = normal (within normal limits); 1 = mild (crypts 2-3 times normal thickness, normal epithelium); 2 = moderate (crypts 2-3 times normal thickness, hyperchromatic epithelium, reduced goblet cells, scattered arborization); 3 = severe (crypts >4 times normal thickness, marked hyperchromasia, few to no goblet cells, high mitotic index, frequent arborization). Scoring for extent of lesions: 0 = normal (0% involvement); 1 = mild (up to 30% involvement); 2 = moderate (30%-70% involvement); 3 = severe (over 70% involvement).
Paraffin-embedded tissue sections were re-hydrated, and their antigen retrieval was performed using citric acid sodium buffer. Sections were then incubated overnight at 4 °C with a primary antibody targeting the Ki67 (Sigma, #SAB4501880, 1:800 dilution). Sections were then washed and subsequently incubated with goat anti-rabbit IgG-peroxidase (Sigma, #A9169, 1:1000 dilution) for 30 min at room temperature. After wash, the sections were stained with DAB. Finally, the sections were dehydrated, made transparent in xylene, and mounted with neutral gum. The Ki67+ cells per crypt in slides were counted under the light microscopes.
AB-PAS staining
This was performed using AB-PAS staining solution kit (Servicebio, Wuhan, China). Specifically, tissues sections were rehydrated, then stained with PAS dye solution C and B for 15 min, respectively. Then sections were then rinsed with running and distilled water. After that, PAS dye solution A was used to stain the sections. Finally, the sections were dehydrated, made transparent in xylene, and mounted with neutral gum. Then the stained goblet cells per crypt were counted under the light microscopes.
MTT assay
MTT kit (Solarbio, #M1020) were utilized. Human colon epithelial cell line SW620 (ATCC, # CCL-227) cells were collected and plated in 96-well plates. At day 1, 2, and 3, cells were incubated with 106 A. muciniphila for 4 hours, then treated with gentamicin (Solarbio, Beijing) to kill extracellular bacteria, and further washed. After that, cells were then incubated with MTT solution for 4h, washed, and incubated with Formazan solution. Finally, OD490 values were measured.
Statistical analysis
Data were analyzed using Graphpad Prism 7.0 software. Data were expressed as Mean ± SEM. The comparison among multiple groups and between two groups were performed by analysis of variance (ANOVA) and unpaired t-test, respectively. P<0.05 was considered statistically different.
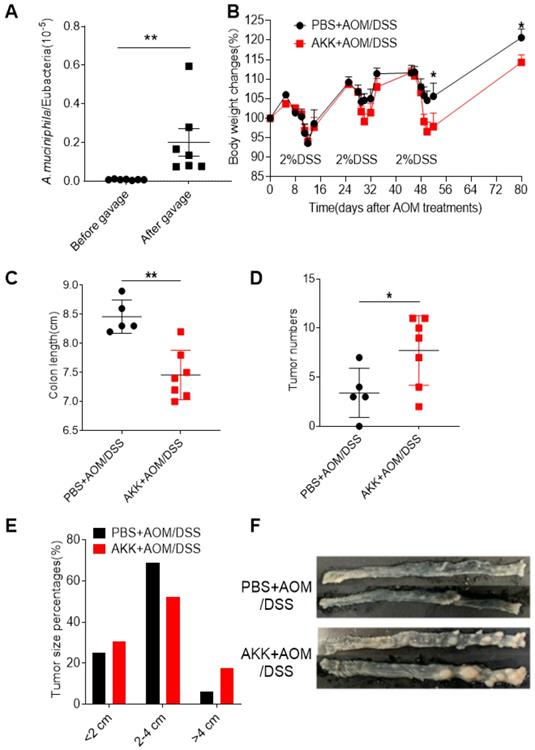
Akkermansia muciniphila administration enhanced the susceptibility of mice to colitis-related colorectal cancer (CAC). C57BL/6 mice were orally administrated with A. muciniphila or PBS and treated with AOM/DSS to induce CRC. Bacterial colonization was validated in the feces collected before and after gavage by quantitative PCR. Mouse body weights were monitored during the AOM/DSS treatment. After 80 days, the mice were sacrificed, and the colons were collected to measure the tumor burden. A. The number of A. muciniphila in the feces of mice before and after gavage; B. body weight change; C. Colon length; D. The number of colon tumors; E. Tumor diameter; F. Representative photos of colon tumors. The data of A-D were represented by the Mean ± SEM, PBS+AOM/DSS, n=5; AKK+AOM/DSS, n=7. *P<0.05, **P<0.01, A, C, D. Unpaired student T test, B.Two-way ANOVA analysis. The experiments were repeated twice independently.
Results
A. muciniphila administration increased host susceptibility to AOM/DSS induced CRC
Gut microbiota plays a key factor in CRC development, but the role of A. muciniphila remains elusive. To directly assess the role of A. muciniphila in CRC, we orally inoculated mice with A. muciniphila or PBS before induction of CRC. Firstly, the feces were collected before and after the A. muciniphila inoculation, and DNA were extracted. The numbers of A. muciniphila were determined by qPCR using bacterium specific primers to validate the bacterial colonization. The results showed that the number of A. muciniphila in the feces significantly increased after A. muciniphila inoculation (Figure 1A), showing that A. muciniphila had been successfully colonized in the experimental mice. After that, AOM/DSS treatment were used to induce CRC and the body weight change was monitored. The AKK+AOM/DSS group mice that received A. muciniphila inoculation and AOM/DSS treatment lost more body weights on second and third rounds of DSS treatment than PBS+AOM/DSS group mice that received PBS inoculation and AOM/DSS treatment (Figure 1B). At 80 days post-AOM treatment, it was found that the colons of the mice in AKK+AOM/DSS group were significantly shorter than those in the PBS+AOM/DSS group (Figure 1C), and the mice in AKK+AOM/DSS group developed significantly increased numbers and size of tumors compared to mice in PBS+AOM/DSS group (Figure 1D-F). These results suggesting that A. muciniphila administration increased host susceptibility to AOM/DSS induced CRC.
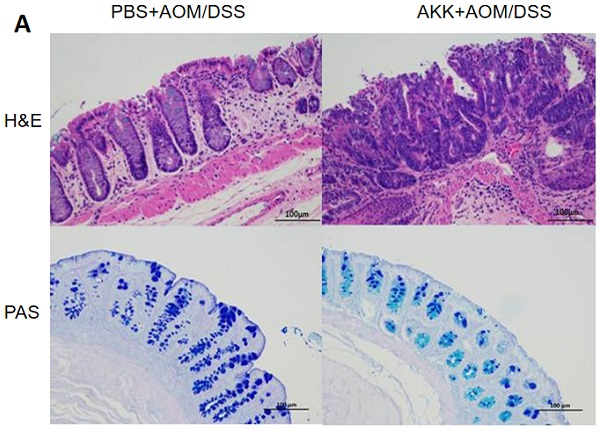
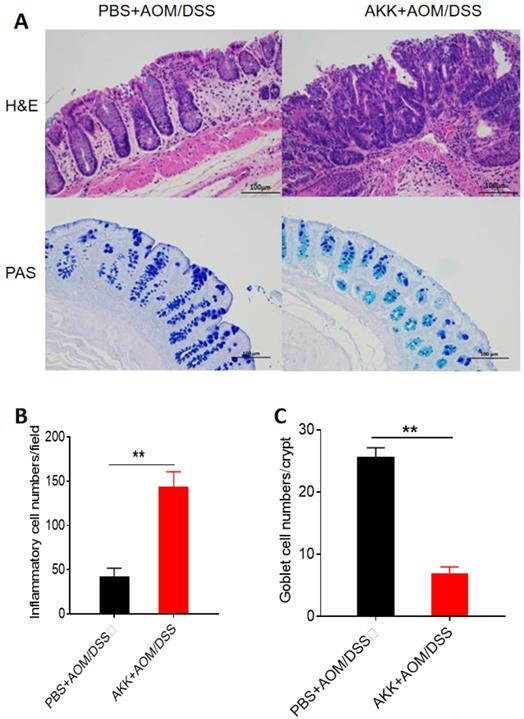
Mouse colon tissue damage and goblet cell staining after A. muciniphila administration. C57BL/6 mice were orally administrated with A. muciniphila or PBS and treated with AOM/DSS to induce CRC. The mice were sacrificed on the 80th day after AOM treatment, and the colons were collected for histochemical staining. A. H&E staining and PAS staining, scale bar: 100µm; B. The average numbers of inflammatory cells in each crypt. C. The average number of goblet cells in each crypt. **P<0.01, B, C. Unpaired student T test. The experiments were repeated twice independently.
A. muciniphila promoted inflammatory cell infiltration, reduced the number of goblet cells, and promoted the proliferation of intestinal epithelial cells
To further evaluate the tissue damage, we performed H&E staining in the colons at 80 days post AOM treatment. The AKK+AOM/DSS group mice presented more tissue damage and infiltration of inflammatory cells in the colons than PBS+AOM/DSS group mice (Figure 2A, B). A. muciniphila can degrade mucin mainly secreted by goblet cells in the intestine, and we performed staining analysis of goblet cells on colon tissues. The results showed that the goblet cells number in the AKK+AOM/DSS group was significantly reduced in comparison with the controls (Figure 2A, C).
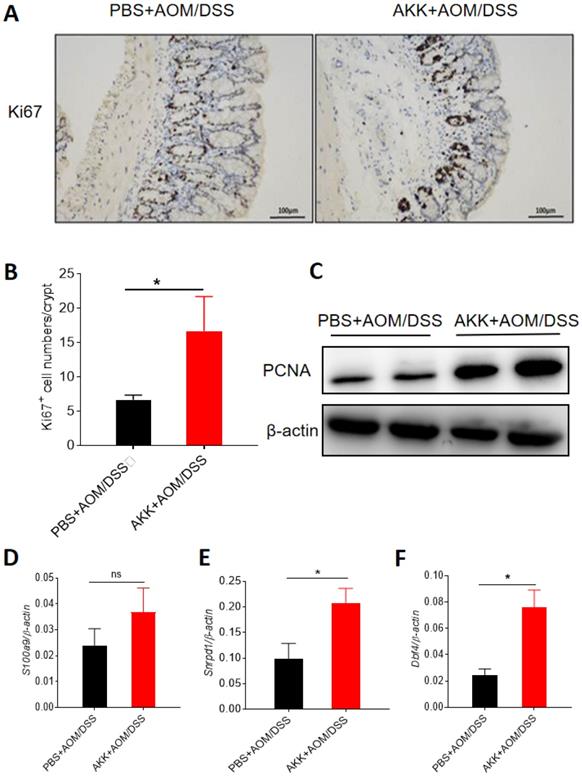
To further study the role of A. muciniphila colonization in the pathogenesis of CRC, we investigated the proliferation of the intestinal epithelial cells through immunohistochemical staining for Ki67. The numbers of Ki67-positive cells in the colons of AKK+AOM/DSS group were significantly higher than those in PBS+AOM/DSS group, indicating that A. muciniphila can promote the proliferation of intestinal epithelial cells (Figure 3A-B). Consistently, we measured the expression of PCNA, a marker for cell proliferation, in colon samples by Western-blot, and the results showed that AKK+AOM/DSS group had a higher level of PCNA protein expression than controls (Figure 3C). Calprotectin (S100 calcium binding protein A9, S100a9), small nuclear ribonucleoprotein D1 polypeptide (Snrpd1) and Dbf4 zinc finger protein (DBF4 zinc finger, Dbf4) are molecules associated with the proliferation of intestinal epithelial cells[6]. We used RT-qPCR to analyze the expression levels of these genes, and the results showed that the colonic expression level of Snrpd1 and Dbf4 were significantly elevated in AKK+AOM/DSS group relative to PBS+AOM/DSS group. The gene expression of S100a9 tended to increase in AKK+AOM/DSS group as well (Figure 3D-F). These results suggest that A. muciniphila can promote the proliferation of intestinal epithelial cells.
Proliferation of mouse colonic cells was increased after A. muciniphila administration. C57BL/6 mice were orally administrated with A. muciniphila or PBS and treated with AOM/DSS to induce CRC. The mice were sacrificed at day 80, and colon tissues were collected. Ki67 staining was applied using IHC method, and RT-qPCR and Western-blot were used to measure the expression of proliferation-related molecules. A. Ki67 staining results, scale bar: 100 um; B. The average numbers of Ki67+ cells in each crypt. C. Immunoblot analysis of PCNA in colon tissues; D. Colonic S100a9 gene expression; E, Snrpd1 gene expression. F, Dbf4 gene expression. (C-E) Data were represented by Mean ± SEM. PBS+AOM/DSS, n=5; AKK+AOM/DSS, n=7. *P<0.05, ns: no significantly different, B, D, E-F. Upaired student T test. The experiments were repeated twice independently.
A. muciniphila administration exacerbated the inflammation during CRC development. C57BL/6 mice were orally administrated with A. muciniphila or PBS and treated with AOM/DSS. After the first round of DSS treatment (day 14 after AOM injection), the mice were sacrificed to collect the colon tissues. The gene expression of pro-inflammatory cytokines were analyzed by RT-qPCR. A. Mouse body weight changes; B. Representative colon pictures; C. Colon length; D. Representative colon H&E staining pictures; E. Histological lesion scores; F. Colonic IL-6 gene expression; G. KC gene expression; H. TNF-α gene expression; I. MCP-1 gene expression. The data were represented by the Mean ± SEM. PBS+AOM/DSS, n=6; AKK+AOM/DSS, n=4. *P<0.05, **P<0.01, ***P<0.001.ns: no significantly different, A.Two-way ANOVA analysis, C-I. Unpaired student T test. The experiments were repeated twice independently.
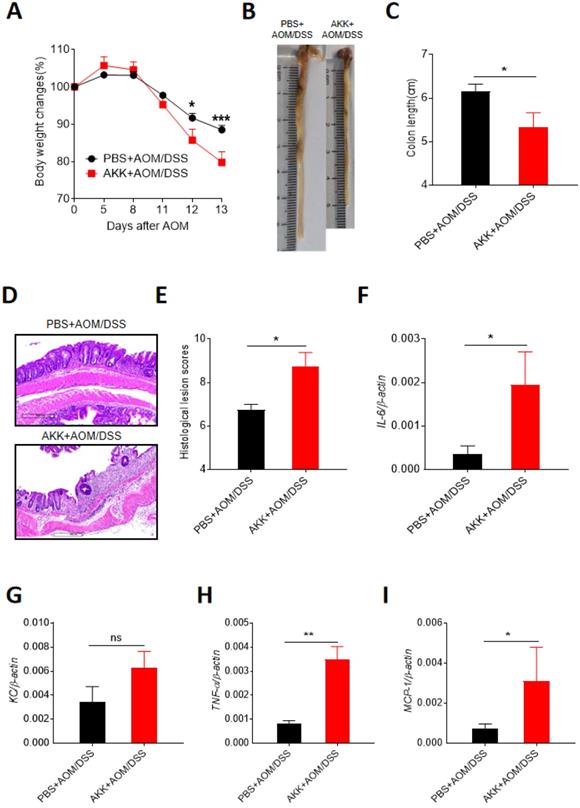
A. muciniphila administration aggravated inflammation in mice early stage after AOM/DSS treatment
To further study the possible role of A. muciniphila administration in inducing CRC development. We investigated the colonic changes in 14 days post-AOM injection which was right after AOM treatment and the first round of DSS treatment. Compared to PBS+AOM/DSS group, the mice in AKK+AOM/DSS group lost more body weights (Figure 4A), had significantly shorter colon lengths (Figure 4B-C), and higher level of histological lesion (Figure 4D-E), further suggesting that A. muciniphila could increase the host susceptibility to CRC development. Inflammation is an important risk factor in the formation of CRC. We measured the expression levels of multiple classical inflammatory cytokines by RT-qPCR, including Interleukin-6 (IL-6), Tumor necrosis factor alpha (TNF-α), Monocyte chemotactic protein 1 (MCP-1), and Keratinocyte chemoattractant (KC). The results showed that the expression levels of all the inflammatory cytokines except KC were elevated in AKK+AOM/DSS group relative to controls (Figure 4F-I). These results indicated that A. muciniphila could promote the proliferation of intestinal epithelial cells, possibly by aggravating colonic inflammation.
A. muciniphila administration promoted the proliferation of intestinal epithelial cell in vitro
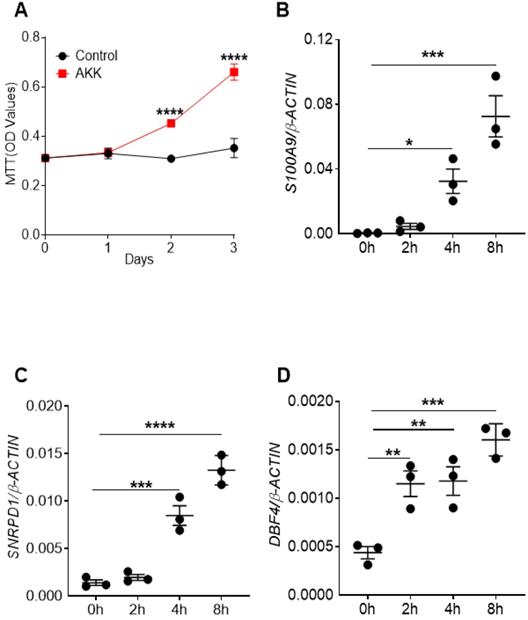
To directly study the possible mechanism of A. muciniphila in promoting the proliferation of intestinal epithelial cells, human colon epithelial cell line SW620 cells were co-cultured with or without A. muciniphila for 4 hours following gentamycin treatment to kill extracellular bacteria for 3 days. Cell proliferation levels were measured by MTT assay on the 0, 1, 2, and 3 days after co-culture. The results showed the cell proliferation rates of the co-culture group was higher than that of the control group at 2 and 3 days after co-culture (Figure 5A). In addition, in this co-culture experiment, cells were collected after gentamicin treatment at 0, 2, 4, 6, and 8 hours respectively. Then RT-qPCR was used to analyze the expression of genes that were associated with cell proliferation. Consistent with the results of MTT assay, the gene expression levels of S100A9, SNRPD1 and DBF4 were elevated in co-culture group than controls (Figure 5B-D). These results suggest that A. muciniphila could promote the proliferation of intestinal epithelial cells in vitro.
Cell proliferation rates and gene expression levels of proliferation-related factors after co-culture of A. muciniphila and colon epithelial cells. The colon epithelial cell line SW620 were co-cultured with A. muciniphila or PBS for 4 hours and later gentamycine was used to kill extracellular bacteria. The cell proliferation was measured by MTT assay at 0, 1, 2, and 3 days after co-culture. A. OD value after co-culture of A. muciniphila and colon epithelial cellsThe colon epithelial cell line SW620 was incubated with A. muciniphila or PBS. At 0, 2, 4, 6, and 8 hours after the co-culture of colon epithelial cell line SW620 and A. muciniphila, cells were washed and and the gene expression of proliferation-related molecules were quantitated by RT-qPCR analysis. B. S100A9 gene expression. C. SNRPD1 gene expression. D. DBF4 gene expression. Data were represented by Mean ± SEM. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, A. Two-way ANOVA analysis, B-D. One-way ANOVA analysis followed by Dunnett's post hoc test. The experiments were repeated twice independently.
Discussion
The dysregulation of gut microbiota is considered as a risk factor for CRC. The substantial evidences have proved that alteration of gut microbiota is related to the imbalance in immune response, which is essential for tumor development and progression [20-24]. Gut microbiota such as Escherichia coli, Fusobacterium and A. muciniphila play critical roles in the development of CRC through regulating host immune response [25, 26].
A. muciniphila is a commensal bacterium mainly colonized in the mucus layer of human intestine, which is associated with intestinal barrier function due to its ability to degrade mucin. Previous studies have shown that the number of A. muciniphila in the gut was inversely correlated with diabetes, obesity, and other diseases [27-29]. Yet other studies suggest that A. muciniphila might promote the development of CRC. However, the live A. muciniphila's effect on CRC development has not been directly investigated.
In our study, mice were orally administered with live A. muciniphila or PBS and treated with AOM/DSS to induce CRC. The role of A. muciniphila in CRC development was determined by monitoring tumor burden, inflammation status, and proliferation of intestinal epithelial cells. More body weight loss, shorter colon length and greater tumor burden in mice administrated with A. muciniphila than controls were observed, indicating their hyper-susceptibility to CRC. Furthermore, more severe tissue damage and higher gene expression of inflammatory cytokines in mice receiving A. muciniphila administration were found.
Since A. muciniphila can degrade mucins that play a critical role in maintaining mucosal integrity, we speculate that A. muciniphila may damage the mucosal barrier, thus leading to invasion of gut microbes and secretion of pro-inflammatory cytokines, ultimately overactive immune response. Excessive pro-inflammatory cytokines combined with the carcinogen AOM accelerated the transformation of normal tissues to dysplastic tissue, inducing more tumor burden in mice administrated with A. muciniphila. In addition, A. muciniphila promoted the gene expression levels of molecules that modulate proliferation in intestinal epithelial cells. Furthermore, the in vitro experiments also showed that the expression of proliferation-related genes in intestinal epithelial cells were up-regulated after cells were co-cultured with A. muciniphila. In conclusion, our in vivo and in vitro results suggest that A. muciniphila may promote the CRC development by exacerbating the inflammation and promoting the proliferation of intestinal epithelial cells at the early stage of CRC.
The roles of A. muciniphila in CRC were controversial. There were multiple possible reasons under this discrepancy. First, different bacterial administration procedure such as viability, bacterial numbers, frequency, could all affect the colonization effect of A. muciniphila administration and the development of CRC. Unlike a recent study using dead A. muciniphila [30], we used live A. muciniphila for our administration. Live bacteria could produce various virulence factors, possiblely leading to over-activation of the immune response. Secondly, the gut microbiota, a diverse and dynamic ecological community, can be affected by many factors, such as mouse facilities. The composition of mouse gut microbiota before A. muciniphila gavage could significantly affect CRC development. Finally, the role of A. muciniphila in different CRC models could be different. In our study, we chose the classical AOM/DSS-induced CRC model, which could be different from spontaneous mouse CRC models such as APCmin/+ mice. In summary, the influence of A. muciniphila's on CRC development could be different due to different bacterial administration procedure, gut-microbiota composition, and different CRC models.
In conclusion, our study suggests that A. muciniphila may promote the development of colitis-associated CRC by aggravating inflammation at the early stage of CRC and enhancing intestinal epithelial cell proliferation. However, further studies were required to dissect the specific mechanism of A. muciniphila affecting CRC progression.
Supplementary Material
Supplementary table.
Acknowledgements
This work was supported by funds from National Natural Science Foundation of China (31960163), Jinggang Scholar Program of Jiangxi; Natural Science Foundation of Jiangxi Province (20171ACB20024), The Scientific and Technological Innovation Team Project and Key Project of Cardiocerebrovascular Disease of Gannan Medical University (TD201703; XN201906) (All to Zhiping Liu).
Author Contributions
FW performed experiments, data analysis and interpretation, drafted the manuscript; LH revised the manuscript. KC, QX, LX performed part of experiments; ZL designed the study and revised the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Chen GY, Núñez G. Inflammasomes in intestinal inflammation and cancer. Gastroenterology. 2011;141:1986-99
3. Singh R, Mishra MK, Aggarwal H. Inflammation, Immunity, and Cancer. Mediators of inflammation. 2017;2017:6027305
4. Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y. et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nature medicine. 2015;21:906-13
5. Chen J, Wang Z, Yu S. AIM2 regulates viability and apoptosis in human colorectal cancer cells via the PI3K/Akt pathway. OncoTargets and therapy. 2017;10:811-7
6. Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A. et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45-58
7. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov, sp. nov, a human intestinal mucin-degrading bacterium. International journal of systematic and evolutionary microbiology. 2004;54:1469-76
8. Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP. et al. Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle. Applied and environmental microbiology. 2017;83(18):e01014-17
9. Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best practice & research Clinical gastroenterology. 2017;31:637-42
10. Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS one. 2013;8:e70803
11. Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F. et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. The ISME journal. 2012;6:1858-68
12. Lopez-Siles M, Enrich-Capó N, Aldeguer X, Sabat-Mir M, Duncan SH, Garcia-Gil LJ. et al. Alterations in the Abundance and Co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects. Frontiers in cellular and infection microbiology. 2018;8:281
13. Farhana L, Antaki F, Murshed F, Mahmud H, Judd SL, Nangia-Makker P. et al. Gut microbiome profiling and colorectal cancer in African Americans and Caucasian Americans. World journal of gastrointestinal pathophysiology. 2018;9:47-58
14. Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY. et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692-13
15. Howe C, Kim SJ, Mitchell J, Im E, Kim YS, Kim YS. et al. Differential expression of tumor-associated genes and altered gut microbiome with decreased Akkermansia muciniphila confer a tumor-preventive microenvironment in intestinal epithelial Pten-deficient mice. Biochimica et biophysica acta Molecular basis of disease. 2018;1864:3746-58
16. Zhou X, Chen C, Zhong YN, Zhao F, Hao Z, Xu Y. et al. Effect and mechanism of vitamin D on the development of colorectal cancer based on intestinal flora disorder. Journal of gastroenterology and hepatology. 2020;35:1023-31
17. Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PloS one. 2013;8:e74963
18. Dingemanse C, Belzer C, van Hijum SA, Günthel M, Salvatori D, den Dunnen JT. et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis. 2015;36:1388-96
19. Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J. et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649-60
20. Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y. et al. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS microbiology ecology. 2004;50:25-36
21. Samanta AK, Torok VA, Percy NJ, Abimosleh SM, Howarth GS. Microbial fingerprinting detects unique bacterial communities in the faecal microbiota of rats with experimentally-induced colitis. Journal of microbiology (Seoul, Korea). 2012;50:218-25
22. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844-54.e1
23. Garagnani P, Pirazzini C, Franceschi C. Colorectal cancer microenvironment: among nutrition, gut microbiota, inflammation and epigenetics. Current pharmaceutical design. 2013;19:765-78
24. Kundu JK, Surh YJ. Inflammation: gearing the journey to cancer. Mutation research. 2008;659:15-30
25. Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PloS one. 2013;8:e56964
26. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207-15
27. Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM. et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Scientific reports. 2015;5:16643
28. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60
29. Belzer C, de Vos WM. Microbes inside-from diversity to function: the case of Akkermansia. The ISME journal. 2012;6:1449-58
30. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nature medicine. 2017;23:107-13
Author contact
![]() Corresponding author: Zhiping Liu, School of Basic Medicine, Gannan Medical University, Ganzhou, Jiangxi, 341000, China. E-mail: Zhiping.Liuedu.cn.
Corresponding author: Zhiping Liu, School of Basic Medicine, Gannan Medical University, Ganzhou, Jiangxi, 341000, China. E-mail: Zhiping.Liuedu.cn.

 Global reach, higher impact
Global reach, higher impact