3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):174-183. doi:10.7150/jca.65031 This issue Cite
Review
The roles of long non-coding RNAs in lung cancer
1. Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China.
2. Clinical Research Center for Wound Healing in Hunan Province, Changsha 410013, Hunan, China.
* These authors contributed equally.
Received 2021-7-16; Accepted 2021-11-5; Published 2022-1-1
Abstract
Lung cancer is the most common malignancy, being a serious threat of human lives. The incidence and mortality of lung cancer has been increasing rapidly in the past decades. Although the development of new therapeutic modes, such as target therapy, the overall survival rate of lung cancer remains low. It is urgent to advance the understanding of molecular oncology and find novel biomarkers and targets for the early diagnosis, treatment, and prognostic prediction of lung cancer. Long non-coding RNAs (lncRNAs) are non-protein coding RNA transcripts that are more than 200 nucleotides in length. LncRNAs exert diverse biological functions by regulating gene expressions at transcriptional, translational, and post-translational levels. In the past decade, it has been shown that lncRNAs are extensively involved in the pathogenesis of various diseases, including lung cancer. In this review, we highlighted the lncRNAs characterized in lung cancer and discussed their translational potential in lung cancer clinics.
Keywords: LncRNAs, lung cancer, tumor biomarkers, MALAT1, and HOTAIR
1. Introduction
Lung cancer is the most common malignancy with the highest incidence and mortality among human cancers. Pathologically, lung cancer is sorted into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), and the NSCLC accounts for approximately 85% [1]. According to the pathological characteristics, NSCLC is subdivided into three types: lung adenocarcinoma (LAD), large cell carcinoma (LCC) and lung squamous cell carcinoma (LSCC) [2, 3]. In recent years, targeted therapy against lung cancer is widely used in clinics, such as tyrosine kinase inhibitors (TKIs) and epidermal growth factor receptor (EGFR) inhibitors [4, 5], but the five-year survival rate of lung cancer is still low due to the late-stage diagnosis and metastasis, as well as drug resistance [6].
Cancer is a complex disease derived from accumulations of genetic and epigenetic alterations, including gene amplification, mutation and abnormal expression, as well as histone methylation and chromosomal modifications [7-9]. A wealth of compelling evidence has suggested that aberrantly expressed non-long coding RNAs (lncRNAs) may be important molecules in the pathogenesis and progression of cancers, including lung cancer, and be potential biomarkers for diagnosis, treatment and prognosis of cancers, as well as individualized therapies [10, 11].
The involvement of lncRNAs in the development and progression of lung cancer has attracted the eyeballs of scientists in lung cancer research, and many lncRNAs have been identified and extensively investigated in lung cancer thus far [12-14]. Reviews published in lncRNAs in lung cancer include articles focused on single lncRNA in multiple tumors [15-18], as well as briefly on the lncRNA profiles in lung cancer [19, 20]. LncRNAs that may act in signaling transduction, diagnosis, therapeutics and prognosis are also comprehensively discussed in review articles [13, 21, 22]. Table 1 summarizes the lncRNAs that function in etiology, diagnosis, treatment and prognosis of lung cancer. This present article is aimed to review the lncRNAs in lung cancer from a different angle, highlighting important oncogenic and tumor suppressive lncRNAs characterized in lung cancer. The lncRNAs discussed in this article include lncRNAs MALAT1, HOTAIR, H19, ANRIL, AFAP1-AS1, UCA1, MEG3, GAS5, TUG1, etc. (Table 2).
2. LncRNA biology and function
Long non-coding RNAs (lncRNAs) are non-protein coding RNA transcripts that are more than 200 nucleotides in length. Only 1.5% of nucleic acids in a human genome are used for protein encoding; other 98.5% of the genome does not encode proteins [23]. LncRNAs as non-protein-coding RNAs were initially considered to be the by-products of the transcription process [24]. With the in-depth of knowledge of lncRNAs, the mystery is gradually unveiled. LncRNAs are biologically functional molecules; and according to the relative location to protein encoding genes in the genome, lncRNAs are classified as sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intron lncRNAs, intergenic lncRNAs, and enhancer intergenic lncRNAs [25-27].
To date, lncRNAs have been recognized as key regulatory factors in cancer development and progression. They can regulate gene expression at the epigenetic, transcriptional, translational and post-transcriptional levels [27, 28]. The most important feature of lncRNA-mediated regulatory network is that lncRNAs can act as a scaffold through which lncRNAs interact with various signaling molecules and regulatory factors. Depending on the type and number of their bond partners, lncRNAs perform a variety of regulatory functions, including gene expression, histone methylation, genomic imprinting, and chromatin modifications [29, 30]. LncRNAs may also regulate gene expression through the function as a co-factor of transcriptional factor and a regulator of RNA polymerase II activity or the transcription machinery [31, 32]. Via specific complementary interactions with target sequences, lncRNAs can also regulate the post-transcriptional processing and translation of mRNAs, such as capping, splicing, editing, transportation and stability [27, 31]. Therefore, lncRNAs are involved in various physiological and pathological processes of the body, including tumorous and non-neoplastic diseases [26, 27, 33].
LncRNAs involved in etiology, diagnosis, treatment and prognosis of lung cancer.
| LncRNAs | Involved function | References |
|---|---|---|
| MALAT1 | Prognosis | [1] |
| Treatment | [2, 3] | |
| HOTAIR | Etiology | [4] |
| Prognosis | [5, 6] | |
| Treatment | [7, 8] | |
| H19 | Etiology | [9, 10] |
| Prognosis | [11, 12] | |
| Diagnosis | [11] | |
| ANRIL | Prognosis | [13] |
| Treatment | [13] | |
| AFAP1-AS1 | Prognosis | [14, 15] |
| UCA1 | Prognosis | [16] |
| Treatment | [17, 18] | |
| MEG3 | Prognosis | [16, 19] |
| Treatment | [20] | |
| GAS5 | Prognosis | [21] |
| Treatment | [22, 23] | |
| TUG1 | Prognosis | [24] |
| Treatment | [25] |
Oncogenic and tumor suppressive lncRNAs in lung cancer.
| LncRNAs | Chromosome Location | Expression | Key Factors | Function | References |
|---|---|---|---|---|---|
| MALAT1 | 11q13.1 | Up | SR, PC2, hnRNP C | Promote cell proliferation, migration, invasion, cell cycle, and EMT; inhibit DNA damage, apoptosis, autophagy | [43, 45, 46, 50, 51, 55, 56] |
| HOTAIR | 12q13.13 | Up | PRC2, LSD1 | Promote viability, proliferation, cell cycle, migration, invasion, autophagy, and EMT; suppress apoptosis | [62-64, 66, 67, 69, 70] |
| H19 | 11p15.5 | Up | c-Myc, p53, miR-675 | Suppress apoptosis; regulate CSCs characteristics; enhance proliferation, differentiation, migration, invasion, and EMT | [73-77] |
| ANRIL | 9p21.3 | Up | PRC2 | Enhance cell proliferation, migration, invasion; suppress apoptosis | [81, 83, 84] |
| AFAP1-AS | 4p16.1 | Up | / | Correlate with TNM stages and tumor size | [87, 89, 90] |
| UCA1 | 10p13.12 | Up | / | Promotes cell proliferation migration, and EMT | [96, 97] |
| MEG3 | 14q32.2 | Down | P53 | Inhibit cell viability, proliferation, autophagy, and chemoresistance; induce apoptosis | [98-101] |
| GAS5 | 1q25.1 | Down | P53, E2F1, miR-21 | Induce apoptosis, drug resistance | [105-107, 111] |
| TUG1 | 22q12.2 | Down | PRC2 | Suppress cell proliferation, migration; inhibit cell cycle | [112, 116, 119-122] |
Aberrant expression of lncRNAs is associated with various cancers. Through regulation of gene expression and signaling transduction, lncRNAs function as oncogenes or tumor suppressors, extensively involved in the development and progression of cancers. They can affect various aspects of cell activities, such as growth and proliferation, survival, migration and invasion, as well as genomic stability. Thus it is increasingly evidenced that lncRNAs play a key role in tumor growth, lymph node/distant metastasis, and patient survival [22], and the single nucleotide polymorphisms (SNP) of lncRNAs are identified as risk factors and correlated to tumorigenesis and metastasis of cancers [34, 35]. More interestingly, some lncRNAs are found to be increased in the plasma of cancer patients, yielding the possibility as diagnostic markers in blood [36, 37]. Therefore, the identification and characterization of lncRNAs have opened a new window in the management of cancers. They may serve as biomarkers of cancers for the development of novel diagnostic tools and prediction of prognosis and as potential targets for novel strategies of cancer therapy. LncRNAs are also involved in therapeutic resistance (e.g. chemo- or radiological resistance) of cancers and may thus be is potential targets to improve the treatment efficacy of cancers [38, 39]. In summary, lncRNAs are important players in the development and progression of cancers, including lung cancer; the discovery and characterization of novel lncRNAs provide new platforms for the development of novel management strategies of cancers.
3. Oncogenic lncRNAs in lung cancer
3.1. Metastasis-Associated Lung Adenocarcinoma Transcript (MALAT1)
MALAT1 is an 8.7 kb intergenic lncRNA located on chromosome 11q13.1. MALAT1 is expressed in a variety of human tissues and is evolutionarily conserved in mammals [40]. MALAT1 is involved in post-transcriptional regulation of gene expression and mRNA splicing, and is associated with the development of a variety of tumors, including lung cancer, liver cancer, prostate cancer, colon cancer, uterus cancer, ovarian cancer, breast cancer, neuroblastoma, and hematological malignancies [41-45].
MALAT1 is an oncogenic lncRNA promoting cancer cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT) and chemoresistance [43, 45, 46]. In NSCLC tissues, the expression of MALAT1 is higher than that in adjacent normal tissues, and the MALAT1 expression is correlated with the overall survival of NSCLC [47], and in lung cancer patients, MALAT1 may negatively regulate the myeloid-derived suppressor cells (MDSCs) [48]. In cultured NSCLC cells, silencing of MALAT1 inhibits cell proliferation and colony formation [49].
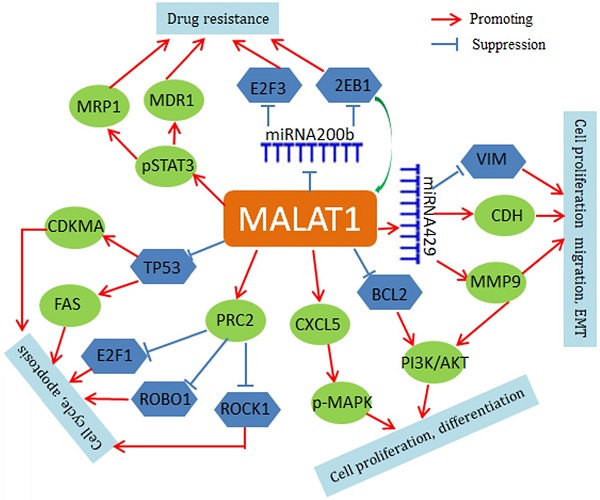
Mechanistically, MALAT1 negatively regulates p53 promoter activity in NSCLC cell lines (A549 and H1299), and MALAT1 depletion leads to up-regulation of both p21 and FAS and cell cycle arrest in G1 [50, 51]. MALAT1 also modulates VIM and CDH1 expression and is involved in the regulation of phosphorylation of AKT1, RPS6KB1, MTOR, CXCL5, MAPK8, MAP2K1/2, and MAPK3/1 [52-54]. In addition, MALAT1 may contribute to chemoresistance by driving expression of MDR1 (ABCB1) and MRP1 (ABCC1) through the phosphorylation activation of STAT3 [55]. MALAT1 may also regulate expression of important genes through a small non-coding RNA-mediated mechanism. For instance, MALAT1 increases zinc finger E-box binding homeobox 1 (ZEB1) expression by sponging miR-200a in A549 cells and promotes the cell proliferation [56]. MALAT1 also inhibits miR-200b function in DTX (docetaxel)-resistant lung adenocarcinoma cells. In cytoplasm, MALAT1 weakens the binding of miR-200b to E2F transcription factor 3(E2F3) and ZEB1 mRNAs, thus leading to increase of E2F3 and ZEB1 protein expression and chemoresistance of lung adenocarcinoma cells [57]. The regulatory roles of MALAT1 are summarized in Figure 1.
In summary, MALAT1 plays an important role in the development and progression of lung cancer through multiple mechanisms and thus may serve as a potential biomarker and target for treatment of lung cancer. Further study in clinical translation of MALAT1 is warranted.
3.2. HOX Transcript Antisense RNA (HOTAIR)
HOTAIR is located on chromosome 12q13.13 in humans and has a length of 2.2 kb containing 6 exons [58]. There are four HOX gene clusters (HOXA, HOXB, HOXC, and HOXD) and 39 HOX gene family members in the genome [59]. The HOTAIR is a transcript of the antisense strand of HOXC, specifically located between HoxC11 and HoxC12, and may regulate gene expression in HOX loci in a cis- or trans-action manner. Abnormal expression of HOTAIR has been reported in a variety of cancerous tissues, such as lung cancer, pancreatic cancer, breast cancer, colorectal cancer, liver cancer and gastric cancer [60, 61].
Regulatory network of MALAT1 in lung cancer. MALAT1 regulates FAS and CDKMA through inhibition of TP53. Through a miRNA429-mediated mechanism, MALAT1 regulates VIM and MMP9. MALAT1 activates PRC2 expression to inhibit E2F1, ROBO1 and ROCK1. MALAT1 also up-regulates phospho-STAT3, ABCB1 and MRP1. In addition, MAIAT1 can acts as a ceRNA for regulation of miRNAs, such as miR-429 and miR-200b.
In lung cancer, HOTAIR expression is significantly higher in tumor tissues than in the adjacent non-tumor tissues, and the HOTAIR expression was correlated with advanced pathological stage, lymph node metastasis, and poor prognosis, being a negative prognostic factor [62, 63]. In vitro, HOTAIR regulates apoptosis and cell cycle and is involved in cisplatin resistance of human lung adenocarcinoma cells [64].
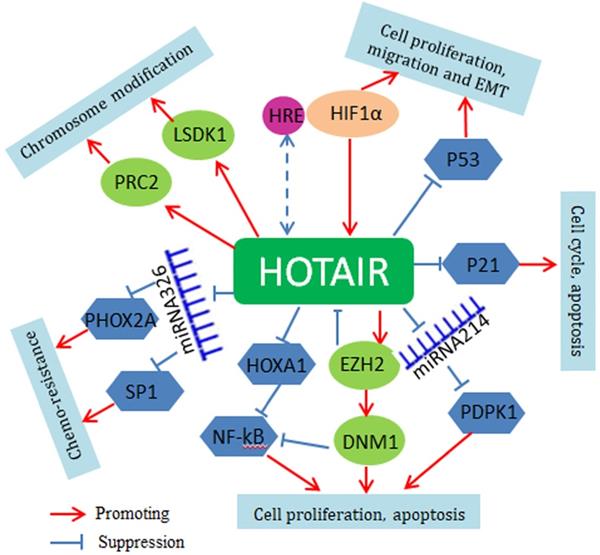
Mechanistically, HOTAIR serves as a bridge scaffold for histone modification complexes mediating histone methylation and chromosomal remodeling. Polycomb repressive complex 2 (PRC2) is a histone methyltransferase implementing epigenetic silencing. The 5′ end of HOTAIR binds to PRC2, regulating its occupancy and histone H3 lysine 27 trimethylation at different genes in the genome. The 3' end of HOTAIR can bind to the LSD1/CoREST/REST complex, regulating lysine 4 demethylation [65]. Through histone H3 lysine 27 trimethylation of p53, HOTAIR suppresses p53 expression and promotes cell proliferation and invasion [66]. HOTAIR can also activate the Wnt/β-catenin signaling pathway to promote tumorigenesis [67]. Very recently, it is found that HOTAIR may function through small non-coding RNAs. For instance, HOTAIR interacts with miR-34a-5p to mediate drug sensitivity in lung cancer cells [68]. HOTAIR also regulates miR-613 and miR-221 and affects the apoptosis, tumorigenesis and metastasis of NSCLC cells [69, 70]. At upstream, HOTAIR is the direct target of HIF-1a and is upregulated under hypoxic conditions; there is a hypoxia-responsive element (HRE) in the HOTAIR promoter region where HIF-1a binds [71]. The regulatory mechanisms of HOTAIR are summarized in Figure 2.
In summary, HOTAIR has become as an important novel master regulator of gene expression and lung cancer development and possesses tremendous potentials in the management of this malignancy. Materialization of HOTAIR's clinical potentials in lung cancer, however, requires further investigations.
3.3. H19
H19 is a 2.3 kb intergenic and maternally-expressed lncRNA. The H19 gene is located on chromosome 11p15.5, which includes five exons and four introns. H19 is the first imprinted genes identified and evolutionarily conserved in mammals. H19 plays an important role in embryonic development and tumorigenesis, and is associated with multiple cancers, such as lung cancer, bladder cancer, ovarian cancer and pancreatic cancer [72].
Regulatory network of HOTAIR in lung cancer. HOTAIR regulates the growth and proliferation of lung cancer cells by inhibition of p53 and p21. HOTAIR-mediated epigenetic gene expression is dependent on its function as a scaffold for PRC2 and LSD1regulating PTEN, HOXD10, β-catenin and Wif1. Through the methylation of HOXA1, HOTAIR activates the NF-kB signaling pathway. HOTAIR also acts as a ceRNA for regulation of miRNAs, such as miR-329 and miR-214.
H19 expression is significantly higher in lung cancer tissues than in adjacent normal tissues, and interestingly, H19 is elevated in the plasma of lung cancer patients [73]. In lung cancer patients, H19 expression is associated with advanced tumor-node-metastasis (TNM) stages, reduced disease-free survival (DFS) and poor prognosis; and in the NSCLC, H19 interacts with microRNA-p21 promoting cancer progression and worse prognosis [73, 74].
In culture cells, H19 promotes cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT), but inhibits apoptosis. H19 can upregulate zinc finger E-box binding Homeobox1 and 2 (ZEB1 and ZEB2) through inhibition of miR-200a function and thus promotes cell proliferation, invasion and EMT [75]. H19 also induces cell proliferation through promoting the expression of proto-oncogene LIN-28 and inhibiting the homologous gene B (LIN-28B) in lung cancer cells A549 and H1299 [76]. In addition, H19 promotes the migration and invasion of NSCLC cells through regulation of cellular signaling pathway proteins, such as metastasis associated in colon cancer1 (MACC1), epidermal growth factor receptor (EGFR), β-catenin, and extracellular-signal-regulated kinase1/2 (ERK1/2) [77].
H19 expression is regulated by c-Myc and p53. The oncogene c-Myc activates the transcription of H19 through binding to its promoter [78, 79]. Tumor suppressor p53 and H19 are mutually counter-regulated, in which p53 represses the H19 expression while the H19 inhibits p53 and p53-dependent gene expression. The p53-H19 interplay appears to play an important role in tumorigenesis and metastasis. In summary, H19 promotes the progression of lung cancer through multiple mechanisms and may also serve as a serological biomarker. Targeting H19 in lung cancer may represent a novel strategy for the diagnosis and management of this malignancy.
3.4. CDKN2B Antisense RNA 1 (CDKN2B-AS1 or ANRIL)
CDKN2B antisense RNA 1 (CDKN2B-AS1), also called ANRIL, is an RNA gene localized on cytogenetic band 9p21.3 with a length of 126.3 kb, consisting of 19 exons [80]. The ANRIL transcript is 3.8 kb. ANRIL is involved in multiple cancers, such as lung cancer, breast cancer, and liver cancer [80-82].
ANRIL expression is increased in NSCLC tumor tissues, and its expression levels are significantly correlated with tumor size, lymph node-metastasis and poor prognosis. In culture cells, siRNA-mediated knockdown of ANRIL expression inhibits the cell proliferation and promote apoptosis [81]. In NSCLC cells, ANRIL promotes proliferation and inhibits apoptosis through suppressing KLF2 and p21 transcription [81]. ANRIL also modulates activity of E2F3 through regulation of miR-449a, leading to cell cycle arrest and senescence of NSCLC cells [83]. Recent studies also showed that ANRIL may drive LAD chemo-resistance through regulating the expression of apoptotic related proteins in LAD cells A549, such as cleaved PARP and Bcl-2 [84]. ANRIL is a downstream effector of c-Myc. The c-Myc directly binds to the E-box in the promoter region of ANRIL and induces ANRIL expression [85]. In brief, ANRIL is involved in the development and progression of lung cancer and further study is warranted.
3.5. Actin Filament Associated Protein 1 Antisense RNA 1 (AFAP1-AS1)
AFAP1-AS1 was initially identified in esophageal cancer. It is a 6.8 kb antisense lncRNA, and its gene is located on chromosome 4p16.1. The exon 2 of AFAP1-AS1 gene overlapped with exons 14, 15 and 16 of AFAP1 gene, and thus regulates the expression of AFAP1. The AFAP1-AS1 is upregulated in many malignant tumors, including lung cancer, HCC, ovarian cancer, gastric cancer, and colorectal cancer [86].
The expression of AFAP1-AS1 is significantly higher in NSCLC tissues than that in adjacent normal tissues, and AFAP1-AS1 expression is positively correlated with tumor pathological grades, TNM stages and distant metastasis of NSCLC, as well as the clinical outcomes of NSCLC patients [87, 88]. AFAP1-AS1 may exert an oncogenic role in the NSCLC cells through epigenetic suppression of p21 expression and serve as a novel prognostic biomarker in human NSCLC [89, 90].
3.6. Urothelial Carcinoma Associated 1 (UCA1)
UCA1 is a 2.3 kb lncRNA located in human chromosome 10p13.12. UCA1 is originally cloned from the human bladder cell lines, and is considered to function as a biomarker for the bladder cancer [91]. Several studies have suggested that UCA1 expression is increased in multiple types of cancers, include lung cancer, breast cancer, gastric cancer, and colorectal cancer [91].
UCA1 transcript is elevated in NSCLC tissues and promotes disease progression, and the UCA1 expression is negatively correlated with the overall survival in NSCLC [92]. Functionally, UCA1 directly regulates miR-193a to increase the expression of HMGB1 that functions as a tumorigenic factor [93, 94]. In addition, UCA1 knockdown upregulates the expression of E-cadherin and decreases the expressions of β-catenin, cyclin D1, and MMP-7, indicating that UCA1 promotes EMT and cell invasion partly through β-catenin [95]. UCA1 may also induce drug resistance. In tamoxifen resistant breast cells, UCA1 is significantly increased, and inhibition of UCA1 improves tamoxifen sensitivity [96, 97].
4. Tumor suppressive lncRNAs in lung cancer
4.1. Maternally Expressed Gene 3 (MEG3)
MEG3 is a RNA transcript with 6.9 kb in length and the gene is located on chromosome 14q32.2. MEG3 is the first lncRNA that has been found to have tumor suppressive function. MEG3 is expressed in many normal tissues, but is downregulated in a variety of human tumor tissues [98]. MEG3 plays a tumor suppressive role by activating the p53; ectopic expression of MEG3 activates p53 and inhibits tumor growth. The regulation of MEG3 on p53 includes two aspects. On the one hand, MEG3 affects the p53 protein expression via acting as a transcriptional synergistic activating factor. On the other hand, MEG3 affects the half-life of the p53 protein, blocking the degradation of p53 mediated by MDM2 [99]. MEG3 also inhibits the expression of apoptosis inhibitory protein B-cell lymphoma-2 (BCL2), but promotes the expression of apoptosis promoting factor BCL2-associated X (Bax), thus inducing cell apoptosis. Therefore, MEG3 demonstrates a suppressor function in multiple types of cancers, including hepatocellular carcinoma, glioma, meningioma, neuroblastoma, bladder cancer and hematological malignancies [98].
In NSCLC tissues, MEG3 is downregulated; and the expression of MEG3 is lower in lung cancer cell lines A549 and HCC823. MEG3 promoter methylation is found in most of NSCLC tumor tissues, which mainly contributes to its downregulation [100]. In NSCLC tissues, MEG3 expression is negatively correlated with advanced pathological stages and tumor size while high expression of MEG3 in NSCLC tissues is associated with better prognosis, serving as a prognostic factor of NSCLC [92]. In NSCLC cells, MEG3 silencing promotes cell proliferation and EMT [99]. In addition, MEG3 interacts with miR-21-5p as a molecular sponge and then regulates the sensitivity of NSCLC cells to cisplatin [101]. In summary, MEG3 plays an important role in the development, progression and drug sensitivity of lung cancer, and maybe a new prognostic marker and potential therapeutic target for this malignancy.
4.2. Growth Arrest Specific 5 (GAS5)
GAS5 gene is located on chromosome 1q25 in humans, and GAS5 lncRNA has a length of 0.65 kb. Evidence to date indicates that GAS5 functions through competitively binding to the glucocorticoid receptor, and thus plays an important role in cell apoptosis [102]. GAS5 is associated with the development of a variety of malignancies, including lung cancer, breast cancer, renal cancer, and prostate cancer [103, 104].
In NSCLC, decreased expression of GAS5 is associated with advanced TNM stages and increased tumor size [105]. In NSCLC cells, increased expression of GAS5 deregulates E2F1 and drives the expression of p21 and p53, thus inhibiting cell proliferation and promoting apoptosis [106]. In addition, GAS5 can deregulate the expression of phospho-EGFR, phospho-MAPK1, phospho-AKT1, and IGF1R; and GAS5 overexpression inversely correlates with the activation of the EGFR pathway [107].
GAS5 can also interact with miRNAs. For instance, miR-21 binds to the putative binding site in GAS5 and regulates its activity; in turn, GAS5 suppresses miR-21 expression as a feedback loop [108]. Additionally, GAS5 can sequester hsa-miR-135b-5p and hsa-miR-23a in NSCLC cells [109, 110]. GAS5 can also regulate radiosensitivity in NSCLC cells through the PTEN signaling pathway [111]. Downregulation of GAS5 promotes resistance to gefitinib in LAD cells [107].
4.3. Taurine Up-regulated gene 1 (TUG1)
TUG1 is a 7.1 kb intergenic lncRNA encoded by the gene on chromosome 22q12.2. TUG1 was first discovered as a crucial player in mouse retinal development [112-114]. To date, TUG1 is found to functions in a variety of tumors, including lung cancer, HCC, breast cancer, ovarian cancer, bladder cancer, gastric cancer and colorectal cancer [115-118].
TUG1 is dysregulated in multiple cancers and affects cell proliferation and survival [119]. TUG1 functions as a tumor suppressor in human glioma through promoting apoptosis and inhibiting cell proliferation [120]. TUG1 also acts as a tumor suppressor in NSCLC. A study analyzed 192 NSCLC and adjacent tissues and found that TUG1 expression was downregulated, and low expression of TUG1 was closely related to high TNM stage, tumor size and poor overall survival rate [112].
TUG1 regulates the expression of growth control genes by binding to polycomb repressive complex 2 (PRC2); the TUG1/PRC2 complex could also bind with hoxb7 promoter and epigenetically activate its expression, thereby activating the Akt and mitogen-activated protein kinase (MAPK) pathways in NSCLC [112, 121]. TUG1 can also regulate the expression of LIMK2b (a splice variant of LIM-kinase 2) through binding to the enhancer of zeste homolog 2 (EZH2), promoting cell growth and chemoresistance of SCLC [122]. TUG1 also competitively binds with transcription factors through the ceRNA function pattern [112-114]. In brief, TUG1 is a novel player in lung cancer and may be a potential marker for the management of this malignant disease.
5. Conclusive Remarks
LncRNAs have become the key regulatory factors in development and progression of lung cancer, functioning as oncogenes (e.g., MALAT1, HOTAIR, H19 and ANRIL) or tumor suppressors (e.g., MEG3, GAS5, and TUG1). These lncRNAs are up or down regulated in lung cancer tissues and significantly correlated with pathological stages, metastasis and patient survival. Targeted expression or silencing of the lncRNAs in lung cancer cells affect the cell proliferation, migration and invasion, and induce EMT changes and drug resistance. Some lncRNAs, such as H19, are detected and elevated in the circulation blood of lung cancer patients. Therefore, lncRNAs hold a great potential as biomarkers and targets in the management of lung cancer. However, most lncRNAs identified thus far lack lung cancer specificity, and are found to be aberrantly expressed in multiple cancers. This intrinsic limitation may restrict their values in clinical practice. In addition, lncRNAs function in a variety of cell activities and are mechanistically very complex in pathogenic process of lung cancer. Vast efforts are warranted to elucidate the function of lncRNAs in lung cancer and bring them into clinical applications. In addition, only a small portion of lncRNAs have been identified and characterized; most lncRNAs encoded by the genome remain unknown. Efforts are continuously warranted to discover novel lncRNAs and their function in various cancers and subtypes. Nevertheless, lncRNAs are critical, functional molecules in the development and progression of lung cancer and other malignancies, and further study is warranted and may bring a novel paradigm in lung cancer management in near future.
Abbreviations
AFAP1-AS1: actin filament associated protein 1 antisense RNA1; ANRIL: CDKN2B antisense RNA 1; Bax: Bcl2-associated X; Bcl2: B-cell lymphoma-2; CELF1: Elav-like family member 1; CTL: cytotoxic T lymphocyte; DFS: disease-free survival; EGFR: epidermal growth factor receptor; EMT: epithelial-mesenchymal transition; ERK1/2: extracellular-signal-regulated kinase 1/2; GAS5: growth arrest specific 5; HCC: hepatocellular carcinoma; HOTAIR: HOX transcript antisense RNA; HREs: hypoxia-responsive elements; LAD: lung adenocarcinoma; LCC: large cell carcinoma; LncRNAs: long non-coding RNAs; LSCC: lung squamous cell carcinoma; MACC1: metastasis associated in colon cancer 1; MALAT1: metastasis-associated lung adenocarcinoma transcript 1; MAPK: mitogen-activated protein kinase; MDSCs: myeloid-derived suppressor cells; MEG3: maternally expressed gene 3; NSCLCs: non-small cell lung cancer; PBMCs: peripheral blood mononuclear cells; OS: overall survival; PRC2: polycomb repressive complex 2; SCLC: small cell lung cancer; SNP: single nucleotide polymorphism; SOX7: SRY-box transcription factor 7; TKIs: tyrosine kinase inhibitors; TNM: tumor-node-metastasis; TUG1: taurine upregulated gene 1; UCA1: urothelial carcinoma associated 1; ZEB1: zinc finger E-box binding Homeobox 1; ZEB2: zinc finger E-box binding Homeobox 2.
Acknowledgements
Funding
This work was supported in part by grants from the following sources: the National Natural Science Foundation of China (81972636, 81872281,81772842), the Natural Science Foundation of Hunan Province (2020JJ5336, 2019JJ40175, 2019JJ40183, 2018JJ1013), the Research Project of Health Commission of Hunan Province (20201020, B20180400, B20180582), China Hunan Provincial Science and Technology Department (2018SK7005), Ascend Foundation of National cancer center (NCC2018b68), and Supported by Hunan Cancer Hospital Climb Plan and By the Fundamental Research Funds for the Central Universities of Central South University (2019zzts832, 2019zzts833).
Author Contributions
Zhe Cao and Linda Oyang contributed to drafting and editing of the manuscript. Qianjin Liao and Deliang Cao designed, revised and finalized the manuscript. Xia Luo and Longzheng Xia participated in the drafting and editing manuscript. Jiaqi Hu and Jinguan Lin participated in the revision and coordination. Shiming Tan and Yanyan Tang contributed to literature search. Yujuan Zhou participated in the conception and coordination. All authors contributed toward data analysis, drafting and revising the paper and agreed to be accountable for all aspects of the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117(2):294-9
2. Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am. 2016;25(3):447-68
3. Zhang M, He J, Li T. et al. Accurate Classification of Non-small Cell Lung Cancer (NSCLC) Pathology and Mapping of EGFR Mutation Spatial Distribution by Ambient Mass Spectrometry Imaging. Front Oncol. 2019;9:804
4. Yang X, Yang K, Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non-small cell lung cancer: a systematic review. Curr Oncol Rep. 2014;16(6):390
5. Gelatti A, Drilon A, Santini FC. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer. 2019;137:113-122
6. Lin HT, Liu FC, Wu CY, Kuo CF, Lan WC, Yu HP. Epidemiology and Survival Outcomes of Lung Cancer: A Population-Based Study. Biomed Res Int. 2019;2019:8148156
7. de Alencar V, Formiga MN, de Lima V. Inherited lung cancer: a review. Ecancermedicalscience. 2020;14:1008
8. El-Telbany A, Ma PC. Cancer genes in lung cancer: racial disparities: are there any. Genes Cancer. 2012;3(7-8):467-80
9. Fernández LC, Torres M, Real FX. Somatic mosaicism: on the road to cancer. Nat Rev Cancer. 2016;16(1):43-55
10. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339-46
11. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699-712
12. Ge X, Li GY, Jiang L. et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene. 2019;38(16):3061-3076
13. Jiang L, Li Z, Wang R. Long non-coding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications (Review). Int J Oncol. 2019;55(3):585-596
14. Wang RQ, Long XR, Ge CL. et al. Identification of a 4-lncRNA signature predicting prognosis of patients with non-small cell lung cancer: a multicenter study in China. J Transl Med. 2020;18(1):320
15. Ghafouri-Fard S, Dashti S, Farsi M, Hussen BM, Taheri M. A review on the role of oncogenic lncRNA OIP5-AS1 in human malignancies. Biomed Pharmacother. 2021;137:111366
16. Ghafouri-Fard S, Shoorei H, Bahroudi Z, Abak A, Taheri M. The role of H19 lncRNA in conferring chemoresistance in cancer cells. Biomed Pharmacother. 2021;138:111447
17. Goyal B, Yadav S, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(2):188502
18. Zheng QX, Wang J, Gu XY. et al. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomed Pharmacother. 2021;135:111169
19. Ghafouri-Fard S, Shoorei H, Branicki W, Taheri M. Non-coding RNA profile in lung cancer. Exp Mol Pathol. 2020;114:104411
20. Ye R, Tang R, Gan S. et al. New insights into long non-coding RNAs in non-small cell lung cancer. Biomed Pharmacother. 2020;131:110775
21. Martínez-Terroba E, Dimitrova N. Long noncoding RNA amplified in lung cancer rewires cancer pathways. J Cell Biol. 2020 219(9)
22. Chen Y, Zitello E, Guo R, Deng Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin Transl Med. 2021;11(4):e367
23. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47-62
24. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629-41
25. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904-14
26. Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol. 2017;1008:47-74
27. Bhatti GK, Khullar N, Sidhu IS. et al. Emerging role of non-coding RNA in health and disease. Metab Brain Dis. 2021;36(6):1119-1134
28. Gao N, Li Y, Li J. et al. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol. 2020;10:598817
29. Fernandes N, Buchan JR. RNAs as Regulators of Cellular Matchmaking. Front Mol Biosci. 2021;8:634146
30. Luo J, Qu L, Gao F, Lin J, Liu J, Lin A. LncRNAs: Architectural Scaffolds or More Potential Roles in Phase Separation. Front Genet. 2021;12:626234
31. Bhat SA, Ahmad SM, Mumtaz PT. et al. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016;1(1):43-50
32. Leisegang MS, Fork C, Josipovic I. et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017;136(1):65-79
33. Hu G, Niu F, Humburg BA. et al. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. 2018;9(26):18648-18663
34. Hashemi M, Moazeni-Roodi A, Sarabandi S, Karami S, Ghavami S. Association between genetic polymorphisms of long noncoding RNA H19 and cancer risk: a meta-analysis. J Genet. 2019 98
35. Mathias C, Garcia LE, Teixeira MD. et al. Polymorphism of lncRNAs in breast cancer: Meta-analysis shows no association with susceptibility. J Gene Med. 2020;22(12):e3271
36. Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: An update and review. World J Gastroenterol. 2015;21(34):9863-86
37. Yang Y, Li Y, Yang H, Guo J, Li N. Circulating MicroRNAs and Long Non-coding RNAs as Potential Diagnostic Biomarkers for Parkinson's Disease. Front Mol Neurosci. 2021;14:631553
38. Izadirad M, Jafari L, James AR, Unfried JP, Wu ZX, Chen ZS. Long noncoding RNAs have pivotal roles in chemoresistance of acute myeloid leukemia. Drug Discov Today. 2021
39. Wang Y, Wang Y, Qin Z. et al. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin Drug Metab Toxicol. 2021;17(3):291-306
40. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705-1714
41. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77(15):3965-3981
42. Huang JL, Liu W, Tian LH. et al. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol Rep. 2017;38(3):1353-1362
43. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757-6768
44. He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6-15
45. Sun Y, Ma L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel). 2019 11(2)
46. Schmidt LH, Spieker T, Koschmieder S. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984-92
47. Chen W, Zhao W, Chen S. et al. Expression and correlation of MALAT1 and SOX9 in non-small cell lung cancer. Clin Respir J. 2018;12(7):2284-2291
48. Zhou Q, Tang X, Tian X. et al. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J Cancer. 2018;9(14):2436-2442
49. Gutschner T, Hämmerle M, Eissmann M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180-9
50. Tano K, Onoguchi-Mizutani R, Yeasmin F. et al. Identification of Minimal p53 Promoter Region Regulated by MALAT1 in Human Lung Adenocarcinoma Cells. Front Genet. 2017;8:208
51. Yang Q, Chen W, Xu Y, Lv X, Zhang M, Jiang H. Polyphyllin I modulates MALAT1/STAT3 signaling to induce apoptosis in gefitinib-resistant non-small cell lung cancer. Toxicol Appl Pharmacol. 2018;356:1-7
52. Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121(1):101-8
53. Tang Y, Xiao G, Chen Y, Deng Y. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29(8):725-735
54. Liu C, Li H, Jia J, Ruan X, Liu Y, Zhang X. High Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Expression Promotes Proliferation, Migration, and Invasion of Non-Small Cell Lung Cancer via ERK/Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway. Med Sci Monit. 2019;25:5143-5149
55. Fang Z, Chen W, Yuan Z, Liu X, Jiang H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed Pharmacother. 2018;101:536-542
56. Feng C, Zhao Y, Li Y, Zhang T, Ma Y, Liu Y. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch Bronconeumol. 2019;55(12):627-633
57. Chen J, Liu X, Xu Y. et al. TFAP2C-Activated MALAT1 Modulates the Chemoresistance of Docetaxel-Resistant Lung Adenocarcinoma Cells. Mol Ther Nucleic Acids. 2019;14:567-582
58. Tang Q, Hann SS. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol Biochem. 2018;47(3):893-913
59. Rinn JL, Kertesz M, Wang JK. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311-23
60. Gupta RA, Shah N, Wang KC. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071-6
61. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90
62. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464
63. Ren MM, Xu S, Wei YB. et al. Roles of HOTAIR in lung cancer susceptibility and prognosis. Mol Genet Genomic Med. 2020;8(7):e1299
64. Liu Z, Sun M, Lu K. et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8(10):e77293
65. Tsai MC, Manor O, Wan Y. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689-93
66. Zhai N, Xia Y, Yin R, Liu J, Gao F. A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. Onco Targets Ther. 2016;9:5713-5720
67. Zhan S, Wang K, Song Y. et al. Long non-coding RNA HOTAIR modulates intervertebral disc degenerative changes via Wnt/β-catenin pathway. Arthritis Res Ther. 2019;21(1):201
68. Zheng F, Li J, Ma C. et al. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J Cell Mol Med. 2020;24(10):5578-5592
69. Jiang C, Yang Y, Yang Y. et al. Long Noncoding RNA (lncRNA) HOTAIR Affects Tumorigenesis and Metastasis of Non-Small Cell Lung Cancer by Upregulating miR-613. Oncol Res. 2018;26(5):725-734
70. Sun YJ, Li J, Chen CH. Effects of miR-221 on the apoptosis of non-small cell lung cancer cells by lncRNA HOTAIR. Eur Rev Med Pharmacol Sci. 2019;23(10):4226-4233
71. Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36(12):9179-88
72. Alipoor B, Parvar SN, Sabati Z, Ghaedi H, Ghasemi H. An updated review of the H19 lncRNA in human cancer: molecular mechanism and diagnostic and therapeutic importance. Mol Biol Rep. 2020;47(8):6357-6374
73. Luo J, Li Q, Pan J, Li L, Fang L, Zhang Y. Expression level of long noncoding RNA H19 in plasma of patients with nonsmall cell lung cancer and its clinical significance. J Cancer Res Ther. 2018;14(4):860-863
74. Zhou Y, Sheng B, Xia Q, Guan X, Zhang Y. Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther. 2017;24(8):317-324
75. Zhao Y, Feng C, Li Y, Ma Y, Cai R. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460(1-2):1-8
76. Ren J, Fu J, Ma T. et al. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle. 2018;17(11):1372-1380
77. Wang L, Sun Y, Yi J. et al. Targeting H19 by lentivirus-mediated RNA interference increases A549 cell migration and invasion. Exp Lung Res. 2016;42(7):346-353
78. Barsyte-Lovejoy D, Lau SK, Boutros PC. et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330-7
79. Cui J, Mo J, Luo M. et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(10):12400-9
80. Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963-9
81. Nie FQ, Sun M, Yang JS. et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268-77
82. Drak Alsibai K, Vacher S, Meseure D. et al. High Positive Correlations between ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF Gene Cluster Overexpression in Multi-Tumor Types Suggest Deregulated Activation of an ANRIL-ARF Bidirectional Promoter. Noncoding RNA. 2019 5(3)
83. Ren XS, Yin MH, Zhang X. et al. Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 2014;344(2):195-203
84. Xu R, Mao Y, Chen K, He W, Shi W, Han Y. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget. 2017;8(24):39177-39184
85. Lu Y, Zhou X, Xu L, Rong C, Shen C, Bian W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther. 2016;9:3077-84
86. Zhang F, Li J, Xiao H, Zou Y, Liu Y, Huang W. AFAP1-AS1: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2018 51(1)
87. Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother. 2015;75:8-11
88. Yu H, Xu Q, Liu F, Ye X, Wang J, Meng X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J Thorac Oncol. 2015;10(4):645-54
89. Leng X, Ding X, Wang S. et al. Long noncoding RNA AFAP1-AS1 is upregulated in NSCLC and associated with lymph node metastasis and poor prognosis. Oncol Lett. 2018;16(1):727-732
90. Yin D, Lu X, Su J. et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92
91. Fan Y, Shen B, Tan M. et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281(7):1750-8
92. Wang M, Ma X, Zhu C. et al. The prognostic value of long non coding RNAs in non small cell lung cancer: A meta-analysis. Oncotarget. 2016;7(49):81292-81304
93. Nie W, Ge HJ, Yang XQ. et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99-106
94. Wu H, Zhou C. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. 2018;496(2):738-745
95. Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20(13):2819-24
96. Liu H, Wang G, Yang L, Qu J, Yang Z, Zhou X. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/β-Catenin Pathway. PLoS One. 2016;11(12):e0168406
97. Li Z, Yu D, Li H, Lv Y, Li S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54(3):1033-1042
98. Ghafouri-Fard S, Taheri M. Maternally expressed gene 3 (MEG3): A tumor suppressor long non coding RNA. Biomed Pharmacother. 2019;118:109129
99. Lu KH, Li W, Liu XH. et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461
100. Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J Biol Chem. 2017;292(1):82-99
101. Wang P, Chen D, Ma H, Li Y. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Onco Targets Ther. 2017;10:5137-5149
102. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832(10):1613-23
103. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195-208
104. Guo C, Song WQ, Sun P, Jin L, Dai HY. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci. 2015;22:100
105. Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093
106. Yu Y, Hann SS. Novel Tumor Suppressor lncRNA Growth Arrest-Specific 5 (GAS5) In Human Cancer. Onco Targets Ther. 2019;12:8421-8436
107. Dong S, Qu X, Li W. et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8:43
108. Cao L, Chen J, Ou B, Liu C, Zou Y, Chen Q. GAS5 knockdown reduces the chemo-sensitivity of non-small cell lung cancer (NSCLC) cell to cisplatin (DDP) through regulating miR-21/PTEN axis. Biomed Pharmacother. 2017;93:570-579
109. Mei Y, Si J, Wang Y. et al. Long Noncoding RNA GAS5 Suppresses Tumorigenesis by Inhibiting miR-23a Expression in Non-Small Cell Lung Cancer. Oncol Res. 2017;25(6):1027-1037
110. Xue Y, Ni T, Jiang Y, Li Y. Long Noncoding RNA GAS5 Inhibits Tumorigenesis and Enhances Radiosensitivity by Suppressing miR-135b Expression in Non-Small Cell Lung Cancer. Oncol Res. 2017;25(8):1305-1316
111. Chen L, Ren P, Zhang Y, Gong B, Yu D, Sun X. Long non-coding RNA GAS5 increases the radiosensitivity of A549 cells through interaction with the miR-21/PTEN/Akt axis. Oncol Rep. 2020;43(3):897-907
112. Zhang EB, Yin DD, Sun M. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5(5):e1243
113. Ma PJ, Guan QK, Meng L, Qin N, Zhao J, Jin BZ. Long non-coding RNA TUG1 as a potential prognostic biomarker in human cancers: a meta-analysis. Oncotarget. 2017;8(37):62454-62462
114. Ghaforui-Fard S, Vafaee R, Taheri M. Taurine-upregulated gene 1: A functional long noncoding RNA in tumorigenesis. J Cell Physiol. 2019;234(10):17100-17112
115. Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49(4):471-5
116. Liu H, Zhou G, Fu X. et al. Long noncoding RNA TUG1 is a diagnostic factor in lung adenocarcinoma and suppresses apoptosis via epigenetic silencing of BAX. Oncotarget. 2017;8(60):101899-101910
117. Zhang E, He X, Yin D. et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7(2):e2109
118. Guo C, Qi Y, Qu J, Gai L, Shi Y, Yuan C. Pathophysiological Functions of the lncRNA TUG1. Curr Pharm Des. 2020;26(6):688-700
119. Zhou H, Sun L, Wan F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol Lett. 2019;18(5):4393-4402
120. Li J, Zhang M, An G, Ma Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp Biol Med (Maywood). 2016;241(6):644-9
121. Khalil AM, Guttman M, Huarte M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667-72
122. Niu Y, Ma F, Huang W. et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):5
Author contact
![]() Corresponding authors: Qianjin Liao or Deliang Cao, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel:86-731-88651681; Fax:86-731-88651999; Email:march-oncom. Or 1814589590com
Corresponding authors: Qianjin Liao or Deliang Cao, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel:86-731-88651681; Fax:86-731-88651999; Email:march-oncom. Or 1814589590com

 Global reach, higher impact
Global reach, higher impact