Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):225-242. doi:10.7150/jca.63608 This issue Cite
Research Paper
Epigenetic modifications inhibit the expression of MARVELD1 and in turn tumorigenesis by regulating the Wnt/β-catenin pathway in pan-cancer
1. Department of Gastrointestinal Medical Oncology, Harbin Medical University Cancer Hospital, Harbin, Heilongjiang Province, China.
2. Translational Medicine Research and Cooperation Center of Northern China, Heilongjiang Academy of Medical Sciences, Harbin, Heilongjiang Province, China.
3. Department of Ultrasound, Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, China.
Received 2021-6-7; Accepted 2021-11-21; Published 2022-1-1
Abstract
MARVEL domain-containing 1 (MARVELD1) is one of the MARVEL domain-containing proteins. Expression of MARVELD1 in tumor and non-tumor tissues, the relationship between its expression and cancer prognosis, and upstream regulation of MARVELD1 were examined using pan-cancer data from The Cancer Genome Atlas. MARVELD1 expression was significantly downregulated in tissues used for pan-cancer analysis compared to that in normal tissues. Low expression of MARVELD1 was associated with poor disease outcomes in pan-cancer. Colon cancer patients with low expression of MARVELD1 had worse progression free survival and overall survival than those with high expression levels in our cohort. Hypermethylation and histone modification in the MARVELD1 promoter locus synergistically affected its expression in pan-cancer. The function of MARVELD1 in colon cancer remains to be studied. Gene Ontology enrichment analysis revealed that MARVELD1 may modulate processes associated with inhibition of tumorigenesis in colon cancer. Both upstream transcription factors and downstream functional enrichment of MARVELD1 were related to the Wnt/β-catenin signaling pathway. Overexpression of MARVELD1 inhibited the expression of β-catenin and its entry into the nucleus. MARVELD1 also inhibited the proliferation, migration, and invasion of colon cancer cells. With Wnt/β-catenin activator LiCl treatment, rescue experiments demonstrated that the role of MARVELD1 in colon cancer progression was dependent on the Wnt/β-catenin pathway. These results indicate that MARVELD1 acts as a tumor suppressor and inhibits tumorigenesis via the Wnt/β-catenin pathway.
Keywords: MARVELD1, prognosis, epigenetic modification, pan-cancer, Wnt pathway
Introduction
Epigenetics, a branch of genetics, is defined as the study of heritable alterations in gene expression that do not cause permanent changes in the DNA sequence, including DNA methylation, histone modification, chromosome remodeling, and non-coding RNA regulation [1]. Genetic and epigenetic aberrations, leading to the activation of oncogenes and inactivation of tumor suppressor genes, have been considered to play major roles in the pathogenesis of various cancers [2, 3].
MARVEL domain-containing 1 (MARVELD1) is a member of the MARVEL (myelin and lymphocyte protein (MAL) and related proteins for vesicle trafficking and membrane link) domain-containing protein family. MARVELD1 is located on human chromosome 10q24 (a locus associated with multiple cancers), and it is downregulated in multiple primary tumors derived from the ovary, vulva, uterus, cervix, breast, testis, kidney, bladder, and liver [4, 5]. MARVEL domain-containing proteins have attracted increasing interest because they exhibit tumor suppressor activities and are commonly decreased via promoter methylation in breast, cervical, prostate, hepatocellular, esophageal, and gastric carcinomas or cell lines [6-9]. MARVELD1 inhibits cell proliferation, tumor growth, and chemosensitivity of HCC cells by increasing p53 and p16 both in vitro and in vivo [10]. In lung cancers, the expression of MARVELD1 significantly correlated with diagnostic histopathology and degree of malignancy as it was inactivated via DNA hypermethylation and histone deacetylation synergistically but suppressed the epithelial-mesenchymal transition [10-11]. However, the function of MARVELD1 in colon cancer is unknown. Further studies are required to elucidate the upstream regulation and biological functions of MARVELD1 in tumor progression.
This study was conducted to evaluate the 1) expression of MARVELD1 in human cancer tissues and adjacent normal tissues, 2) relationship between its expression and disease prognosis, and 3) upstream regulation of MARVELD1 using pan-cancer data from The Cancer Genome Atlas (TCGA). In our cohort of patients with colon cancer, low expression of MARVELD1 indicated poor prognosis. Therefore, we aimed to elucidate the downstream regulatory functions of MARVELD1 and the correlation among MARVELD1 and key genes of important pathways in colon cancer. Collectively, our findings indicate that DNA methylation and histone modification synergistically affect the expression of MARVELD1, which inhibited tumor occurrence and progression via Wnt/β-catenin signaling.
Materials and Methods
Acquisition of Public Date
Genome-wide expression profiles of MARVELD1 as well as other genes and the clinicopathological progression were downloaded from TCGA (https://tcga-data.nci.nih.gov/). MARVELD1 expression levels were dichotomized using the median expression as the cutoff to define “high value” at or above the median versus “low value” below the median. The log-rank test was used to examine the survival difference between different patient groups.
The DNA methylation profile was measured experimentally using the Illumina Infinium HumanMethylation450 (450K) platform. It was downloaded from TCGA database. Beta values were obtained from Johns Hopkins University and University of Southern California TCGA genome characterization center. DNA methylation values, described as beta values, were recorded for each array probe in each sample via BeadStudio software (Illumina). DNA-methylation beta values were continuous variables between 0 and 1, representing the ratio of the intensity of the methylated bead type to the combined locus intensity. Thus, higher beta values represent higher levels of DNA methylation and lower beta values represent lower levels of DNA methylation.
Transcription Factor Prediction
Transcriptional factors were predicted using JASPAR software (jaspar.binf.ku.dk). Geneontology term enrichment (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were analysed for these TFs using DAVID Functional Annotation Bioinformatics Microarray Analysis (https://david.ncifcrf.gov/) (12, 13). ChIP-seq data were downloaded from the ENCODE database (https://www.encodeproject.org/).
Functional Enrichment Analysis
Pairwise Pearson correlation analysis was performed to determine the expression of MARVELD1 and all other genes. Only positively correlated genes with an R ≥ 0.6 and a significant correlation (P < 0.05) were retained. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using DAVID Functional Annotation Bioinformatics Microarray Analysis (https://david.ncifcrf.gov/).
Ethics approval, Patient consent, and Tissue samples
Colon cancer tissues and the corresponding adjacent normal colon tissues, for qRT-PCR, were collected from patients at the Third Affiliated Hospital of Harbin Medical University. The specimens were frozen in liquid nitrogen and stored at -80 °C until use.
Tissue samples from advanced or metastatic colon cancer patients, diagnosed between January 2008 and December 2012, and from pancreatic cancer patients, diagnosed between January 2010 and December 2015, were obtained for immunohistochemistry at the Third Affiliated Hospital of Harbin Medical University. These samples met the following requirements: (1) the paraffin-embedded pathological tissue specimens were well preserved, and (2) the basic information and clinicopathological data were complete. (3) All patients have complete follow-up records. (4) All patients received chemotherapy after surgery. The chemotherapy regimen for pancreatic cancer is based on gemcitabine. The chemotherapy regimen for colon cancer is fluorouracil and oxaliplatin (patients with stage I colon cancer do not need chemotherapy after surgery and were not included in this study).
The studies involving human participants were reviewed and approved the Research Ethics Committee of the Third Affiliated Hospital of Harbin Medical University. All patients provided written informed consent, and the specimens were handled in accordance with accepted ethical standards.
Immunohistochemistry
The samples were cut into sections (5 μm), and the sections were de-paraffinized in xylene, rehydrated in ethanol, and washed in phosphate-buffered saline (PBS). The sections were incubated with anti‐MARVELD1 antibody (TA351385, OriGene) overnight at 4 °C followed by incubation with a secondary antibody (Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) for 30 min at room temperature. The samples were observed and photographed using a light microscope (Olympus). Three pathology professionals scored the expression of MARVELD1 in the tissues was scored by immunoreactive score (IRS), and 8 points were selected as the cut-off value, and they were divided into high expression group and low expression group. According to the patient's follow-up information, the correlation between MARVELD expression and the patient's prognosis was analyzed. All patients provided written informed consent, agreed to use their organization for this study, and the specimens were handled in accordance with accepted ethical standards.
Cell Culture
All cell lines were obtained from the Chinese Type Culture Collection, Chinese Academy of Sciences. The cells were cultured in RPMI-1640 (Gibco, Rockville) or DMEM (Gibco, Rockville) medium, that were supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of penicillin/streptomycin at 37 °C in an atmosphere containing 5% CO2. Cells in the logarithmic growth phase were used. The Wnt/β-catenin activator LiCl was obtained from Sigma-Aldrich (Munich, Germany). HCT116 and HT29 cells with MARVELD1 overexpression or control transfection were cultured in medium with or without 20 mmol/L LiCl.
Plasmid Transfection and Lentivirus Infection
MARVELD1 ORF region was cloned into the vector pcDNA3.1/flag (Invitrogen, Carlsbad). The cells were transfected with the MARVELD1 construct using Lipofectamine 2000 (11668-019, Invitrogen) according to the manufacturer's protocol. The negative control and MARVELD1 over-expressing lentiviruses/flags were purchased from Invitrogen. Next, lentiviruses infected 5 × 105 cells in a 6-well plate with 4-6 µg/mL polybrene (107689, Sigma). After 24 h, the infected cells were subjected to selection with 4 µg/ml puromycin (540411, Calbiochem) and cultured for 3 days. Stable overexpression cell lines were identified using qRT-PCR and western blot assays.
Transfection of Small Interfering RNAs
A scrambled negative control small interfering RNA (siRNA) and MARVELD1 siRNAs were purchased from Invitrogen. The cells were transfected with the siRNAs using Lipofectamine 2000 according to the manufacturer's protocol. The sequences were as follows: siMARVELD1#1: 5′-AUUGGAACCAGGCUUCUGGTT-3′; siMARVELD1#2: 5′-CCAGAAGCCUGGUUCCAAUTT-3′.
Cell Viability Assays
The viability of treated cells was measured using a Cell Counting Kit-8 (CCK-8; CK04; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's instructions. The cells (1 × 103) were cultured at 37 °C in a 96-well plate. Cell viability was assessed at 24, 48, and 72 h. After 10 μL CCK-8 solution was added to each well, the plates were incubated at 37 °C for 1 h. A microplate reader was used to measure the absorbance of each cell suspension at 450 nm. RPMI-1640 medium containing 10% FBS and 10% CCK-8 was used as the control.
Colony Formation Assays
Cells were plated in a 6-well plate with 1,000 cells/well and cultured in RPMI-1640 medium containing 10% FBS for 2 weeks. Cell colonies were washed with PBS, fixed with 4% paraformaldehyde, stained with 0.5% crystal violet (332488, Sigma-Aldrich) for 30 min. The cells counted under a microscope for quantitative analysis.
Invasion Assays
The transwell chambers were first coated with Matrigel solution. HCT116 and HT29 cells lines in serum-free RPMI-1640 medium were placed in the BRAND® insert with Matrigel (BR782806, Sigma-Aldrich). RPMI-1640 medium containing 20% FBS was added to the lower chamber. After incubating at 37 °C for 24 h, the invading cells that had penetrated through the lower surface of the membrane were fixed with 4% paraformaldehyde for 30 min. The cells were stained with 0.5% crystal violet for 30 min and were counted in six randomly chosen fields under a light microscope.
Wound Healing Assay
The cells were plated in 6-well culture plates and cultivated to achieve over 90% confluence in RPMI-1640 medium containing 10% FBS. A vertical wound per well was created using a 10 µL pipette tip. After 2 washes with PBS to eliminate cell debris, the cells were re-incubated in RPMI-1640 containing 5% FBS. Images were recorded at the indicated time to measure the size of the remaining wound.
RNA Preparation and qRT-PCR
Total RNA was extracted using TRIzol Reagent (15596018, Invitrogen) according to the manufacturer's protocol. The Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) was used to reverse transcribe RNA into complementary DNA. mRNA expression was quantified via real-time PCR using FastStart Universal SYBR Green Master Mix (04913850001, Roche) with gene-specific primers, with GAPDH as the internal control. The results were determined using the comparative CT (2-ΔΔCT) method. The forward (F) and reverse (R) primer sequences were as follows: MARVELD1-F: 5′ -ACTGAGAAGTCCCGCTGTTACAG-3′; MARVELD1-R: 5′-GGGATGCTGGGAATCTTAAGG-3′; GAPDH-F: 5′-CATGTTCGTCATGGGTGTGAA-3′; GAPDH-R: 5′-GGCATGGACTGTGGTCATGAG-3′.
Western Blot Analysis
Total cell lysates were harvested from cultured cells using ice-cold lysis buffer. Nuclear and Cytoplasmic Protein Extraction Kit (WLA020a, Wanleibio) was used to extract the nuclear and cytoplasmic proteins. Protein concentrations were evaluated using a protein assay kit (5000001, Bio-Rad), and equal amounts of protein were separated using 10% SDS-PAGE, followed by electroblotting onto PVDF membranes, which were blocked with 5% non-fat milk in PBS with 0.1% Tween-20 (Tween20-PBS) for 2 h at room temperature. The membranes were immunoblotted with primary antibodies against MARVELD1 (TA351385, origin), β-catenin (Proteintech), Lamin B1 (13435S; Cell Signaling Technology), flag tag (66008-3-lg, Proteintech), and GAPDH (TA-08, ZSGB-BIO) overnight at 4 °C. After washing with Tween20-PBS, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Proteintech) for 1 h. Immunoreactive proteins were detected using a chemiluminescence solution (Thermo Fisher Scientific).
Animal Experiments
The animal study was reviewed and approved by the Medical Experimental Animal Care Commission of Harbin Medical University. Written informed consent was obtained from the owners for the participation of their animals in this study. Four-week-old Balb/c mice were obtained from Hunan Slac Jingda Laboratory Animal Co, Ltd. Approximately 2 × 106 stably overexpressing cells, i.e., HCT116-controls or HCT116-flagMARVELD1, in 200 μL of serum-free RPMI-1640 medium were injected directly into the right back of mice using an 1-mL syringes and 20-gauge needles. The tumor volume was calculated using the following formula: (length × width2)/2. The mice were euthanized, and the tumor weight was examined.
Statistical Analysis
The expression and epigenetic modifications of MARVELD1 in cancer tissues compared to those in adjacent non-cancerous tissues were examined by a t-test. The Kaplan-Meier method and a log-rank test were used to evaluate the difference in survival between patients with high and low MARVELD1 expression. The differences among groups in the results of in vitro and in vivo experiments were also analyzed using Student's t-test. All statistical tests were two-tailed, and P < 0.05 indicated statistical significance. Statistical analyses were performed using R.3.5.3 software.
Results
MARVELD1 expression was downregulated and positively associated with prognosis of patients in pan-cancer database
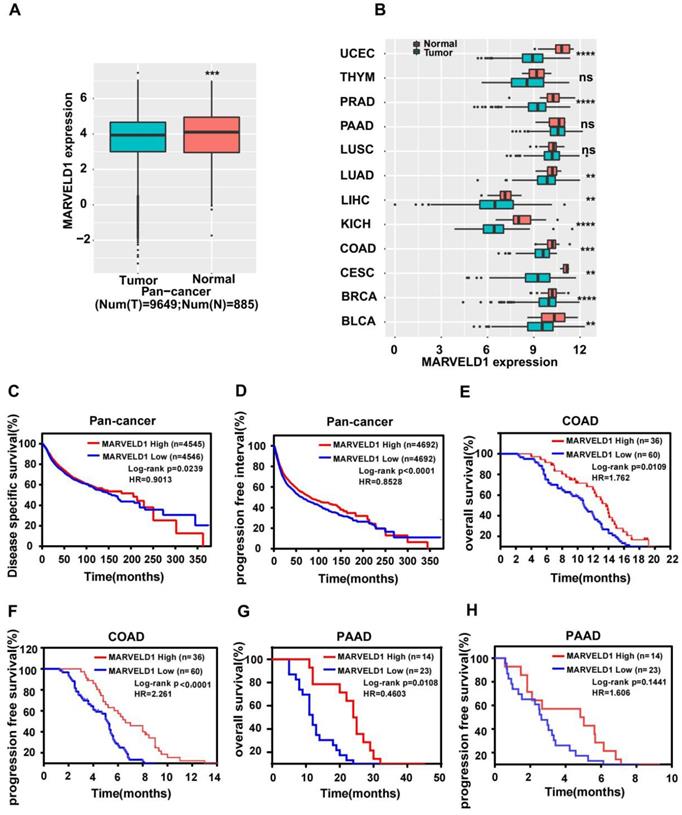
TCGA data were downloaded and analyzed to assess the expression of MARVELD1 in human cancers. Data from 10,534 patients representing 33 cancer types were obtained from TCGA pan-cancer database and were used to validate the expression of MARVELD1. The expression of MARVELD1 was significantly downregulated in cancer tissues compared to that in normal tissues in pan-cancer (Figure 1A; Supplementary Table 1). MARVELD1 expression was significantly lower in most types of cancers, including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), endocervical adenocarcinoma (CESC), colon adenocarcinoma (COAD), kidney Chromophobe (KICH), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD), and uterine corpus endometrial carcinoma (UCEC), than in their respective non-cancer tissues (Figure 1B, Supplementary Table 1). This result was consistent with previous reports. Moreover, in our cohort, the expression of MARVELD1 was lower in colon cancer samples than in their corresponding non-cancer counterparts (Supplementary Figure 1A,B). The results indicate that MARVELD1 was mainly located in the nucleus in colon cancer, which was consistent with previous reports [5]. Kaplan-Meier analysis and the log-rank test were used to explore the relationship between MARVELD1 expression and survival status in patients using pan-cancer database samples. The results revealed that patients with low MARVELD1 expression had poor disease-specific survival (DSS) and progression-free interval (PFI) than those with high expression levels (Figure 1C,D). Colon cancer and pancreatic cancer were analyzed in our cohort. In the colon cancer group, there were 60 and 36 patients with low and high expression of MARVELD1, respectively (Supplementary Figure 1C). Downregulation of MARVELD1 in colon cancer was associated with both poor overall survival (OS) and progression free survival (PFS) unlike in cancers with high expression of MARVELD1 (Figure 1E,F, Supplementary Table 2). Low expression of MARVELD1 in pancreatic cancer was associated with poor OS unlike in cancers with high expression levels of MARVELD1 in our cohort (Figure 1G,H, Supplementary Table 3).
Expression of MARVEL domain-containing 1 (MARVELD1) in tumor and non-tumor tissues, the relationship between its expression and cancer prognosis in human cancers. (A) MARVELD1 expression is lower in the cancer cohort than in the corresponding normal tissues as observed in the data from The Cancer Genome Atlas (TCGA). (B) MARVELD1 expression is lower in UCEC, PRAD, LUAD, LIHC, KICH, COAD, CESC, BRCA, and BLCA than in the corresponding normal tissues. (C,D) Low expression of MARVELD1 correlates with poor disease specific survival (DSS) and progression free interval (PFI) according to TCGA pan-cancer database. (E,F) Low expression of MARVELD1 in COAD correlates with poor overall survival (OS) and progression free survival (PFS) in our cohort. (G,H) Low expression of MARVELD1 in PAAD correlates with poor OS in our cohort. *p< 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, no significance.
Epigenetic modification of MARVELD1
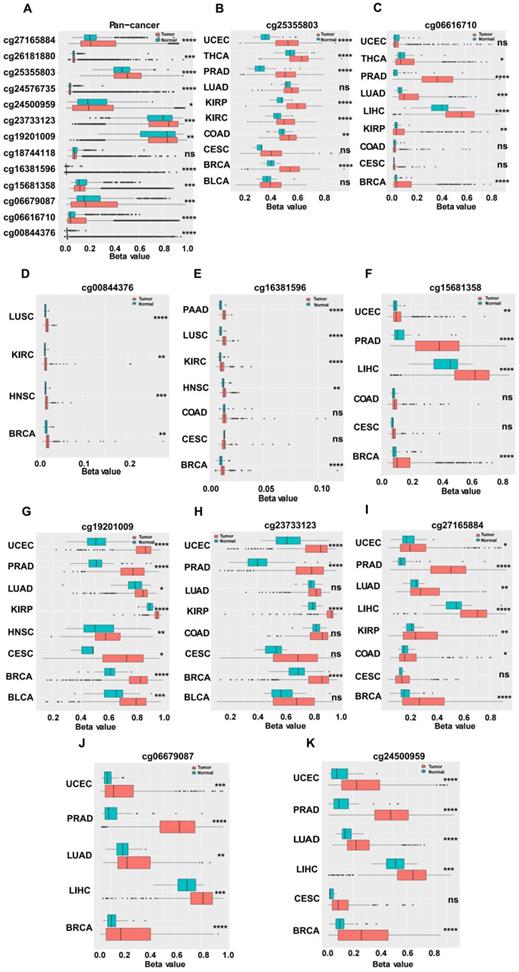
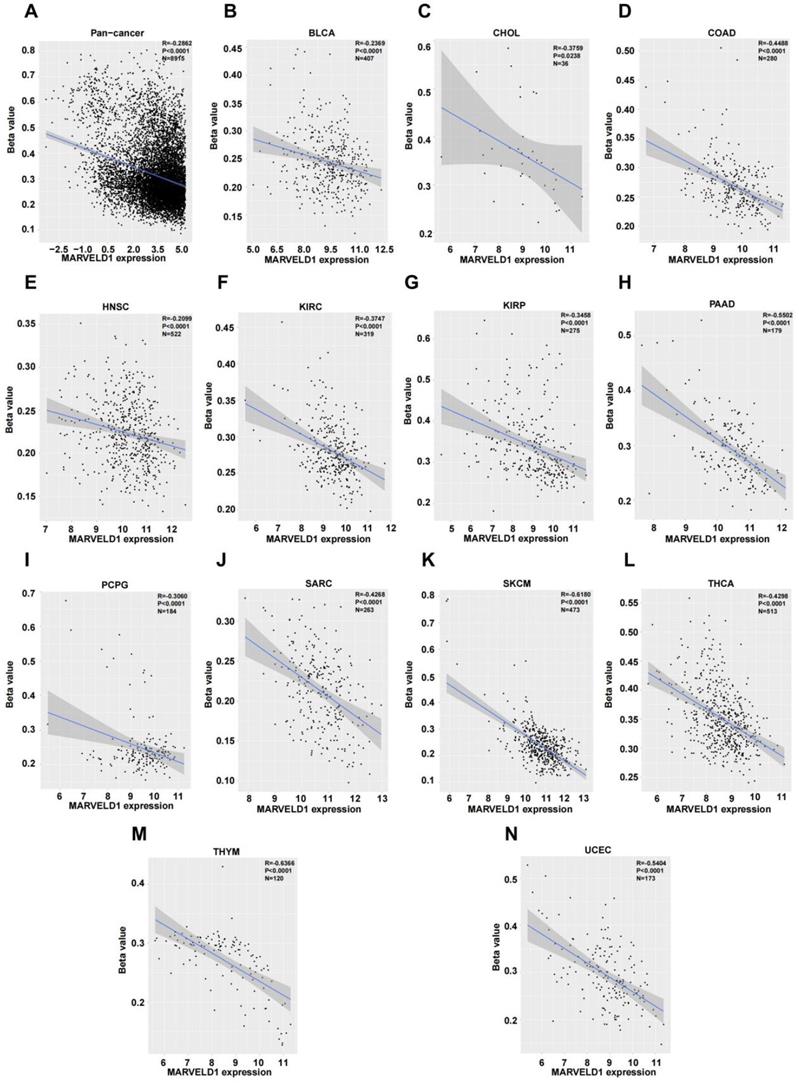
We investigated the upstream regulation of MARVELD1. As epigenetic regulation had been identified as a common type of control in gene expression [14]. In this study, we used TCGA pan-cancer database to further analyze the correlation between the expression of MARVELD1 and epigenetic modifications. Data from the 450K platform and ENCODE were downloaded from TCGA portal to predict 13 methylation sites in the promoter locus of MARVELD1. These sites included cg27165884, cg26181880, cg25355803, cg24576735, cg24500959, cg23733123, cg19201009, cg18744118, cg16381596, cg15681358, cg06679087, cg06616710, and cg00844376. As expected, significant hypermethylation of the 13 MARVELD1 methylation sites was observed in the tissues from the pan-cancer data compared to the non-cancer tissues from TCGA (Figure 2A). Every methylation site in the MARVELD1 promoter locus was significantly hypermethylated than in its non-cancer counterpart in most types of cancers (Figure 2B-K). Data from 19,639 patients representing 33 cancer types obtained from TCGA were also examined to analyze the relationship between the methylation status of MARVELD1 promoter locus and its corresponding expression. The results indicated that MARVELD1 expression was inversely proportional to the DNA methylation level found in the pan-cancer data (Figure 3A). Moreover, MARVELD1 promoter locus hypermethylation inversely and significantly affected the expression of MARVELD1 in BLCA, cholangio carcinoma (CHOL), COAD, head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), sarcoma (SARC), skin cutaneous melanoma (SKCM), thyroid carcinoma (THCA), thymoma (THYM), and UCEC (Figure 3B-N). These specific data, including the number of patients and the P values, are shown in Tables 1 and 2.
In addition to DNA methylation, other epigenetic modifications, including H3K27ac and H3K4me3, were also studied to examine their roles in regulating MARVELD1 expression. The results indicated that H3K27ac and H3K4me3 were enriched in the MARVELD1 promoter locus in ten cancer cell lines representing 10 cancer types, namely U87, MCF-7, LoVo, LNCaP, IMR90, HCT116, HCC1954, H128, Caco-2, and A549 (Figure 4A,B). These results indicated that the low expression of MARVELD1 could be attributed to a biphasic regulation of DNA methylation and histone modification. The lower levels of MARVELD1 expression were related to the comprehensive regulation of epigenetic factors, such as DNA methylation and histone acetylation.
Methylation of MARVELD1 locus between cancer and normal tissues in Pan-cancer types
| Cancer type | cg site | Number of tumor and normal tissues | P value |
|---|---|---|---|
| Pan-cancer | cg18744118 | Num(T)=8916;Num(N)=723 | ns |
| cg23733123 | Num(T)=8916;Num(N)=723 | *** | |
| cg25355803 | Num(T)=8916;Num(N)=723 | **** | |
| cg27165884 | Num(T)=8916;Num(N)=723 | **** | |
| cg06679087 | Num(T)=8916;Num(N)=723 | *** | |
| cg06616710 | Num(T)=8916;Num(N)=723 | **** | |
| cg00844376 | Num(T)=8915;Num(N)=723 | **** | |
| cg24576735 | Num(T)=8915;Num(N)=723 | **** | |
| cg26181880 | Num(T)=8916;Num(N)=723 | *** | |
| cg16381596 | Num(T)=8916;Num(N)=722 | **** | |
| cg15681358 | Num(T)=8916;Num(N)=723 | *** | |
| cg24500959 | Num(T)=8916;Num(N)=723 | * | |
| cg19201009 | Num(T)=8914;Num(N)=723 | ** | |
| cg22756156 | Num(T)=8916;Num(N)=723 | **** |
Notes: * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns: no significance.
Regulation of upstream transcription factors of the MARVELD1 promoter locus
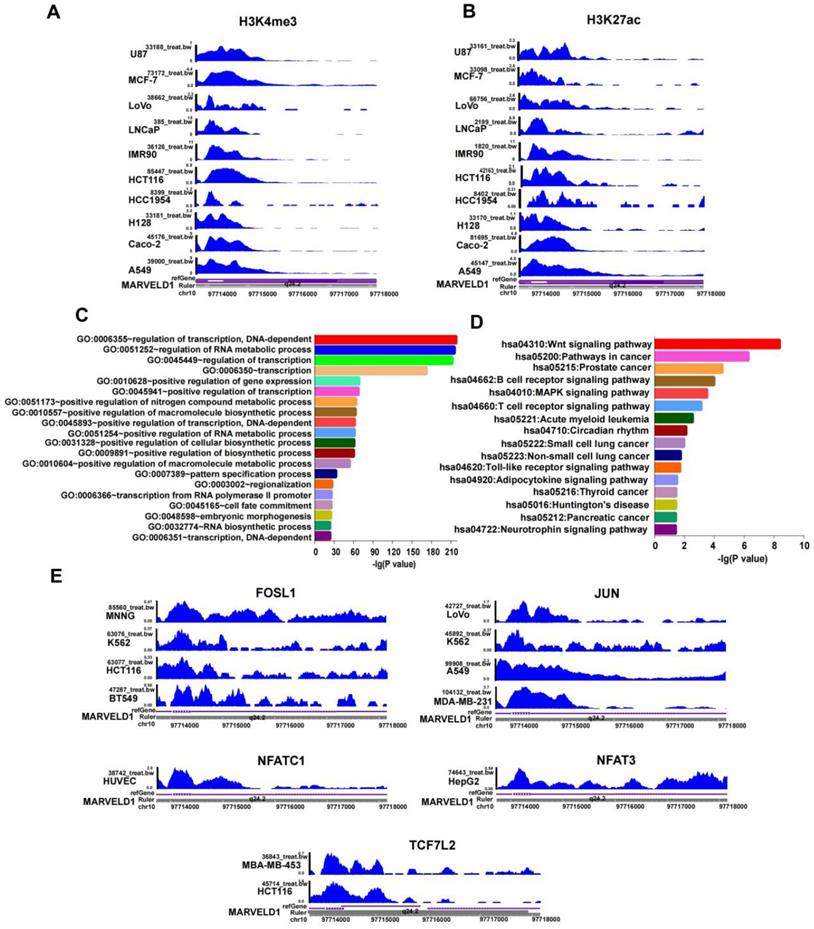
JASPAR and TRANSFAC software were used to predict transcription factor (TF) binding to the MARVELD1 promoter locus. DAVID Functional Annotation Bioinformatics Microarray Analysis analyzed GO and KEGG pathways. The results indicated that transcription factors bound to the promoter region of MARVELD1 were related to the Wnt pathway. We then analyzed the crucial genes of the Wnt pathway according to KEGG, and screened five transcription factors, including FOSL1, JUN, NFATC1, NFAT3, and TCF7L2 (Figure 4C,D). To verify this result, we downloaded ChIP-seq data for FOSL1, JUN, NFATC1, NFAT3, and TCF7L2 from the ENCODE database. The results indicated that FOSL1 binds to the promoter region of MARVELD1 in BT549, HCT116, K562, and MNNG cell lines; JUN in MDA-MB-231, A549, K562, and LoVo cell lines; NFATC1 in HUVEC cell lines; NFAT3 in HepG2 cell lines; and TCF7L2 in HCT116 and MBA-MB-453 cell lines (Figure 4E).
Pivotal biological functions of MARVELD1
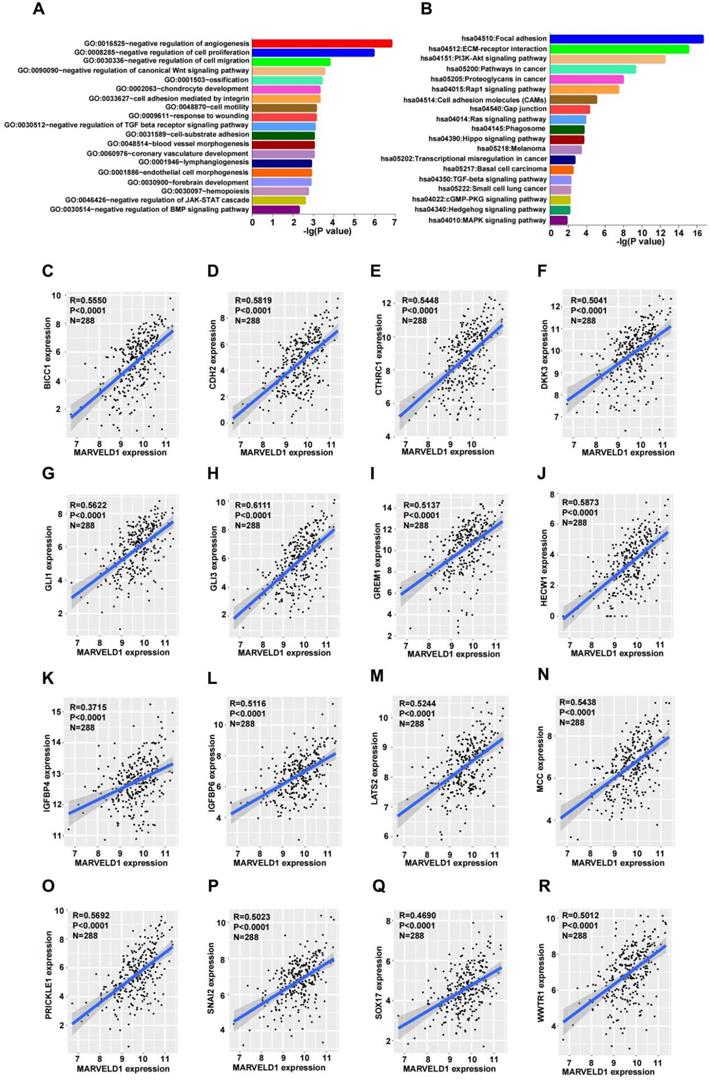
To explore the possible functions of MARVELD1, guilt-by-association analyses were performed using the data from TCGA. Patients with low expression of MARVELD1 had poor prognosis than those with high expression levels in our colon cancer cohort. Therefore, we analyzed the data from TCGA for COAD for MARVELD1 related genes through Pairwise Pearson correlation. GO and KEGG pathways were analyzed for the potential functions and pathways of MARVELD1 and correspond to enriched genes in COAD. Based on this analysis, the main functions of MARVELD1 were to negatively regulate angiogenesis, cell proliferation, cell migration, cell adhesion, and cell motility. Further, the major pathways regulated via MARVELD1 included the canonical Wnt, TGF-beta receptor, JAK-STAT cascade, BMP, Rap1, Ras, Hippo, MAPK, and PI3K-Akt signaling pathways (Figure 5A,B). We analyzed the correlation of MARVELD1 with key genes of essential pathways in COAD. The expression of MARVELD1 was correlated with genes related to the canonical Wnt signaling pathway (BICC1, CDH2, CTHRC1, DKK3, GLI1, GLI3, GREM1, HECW1, IGFBP4, IGFBP6, LATS2, MCC, PRICKLE1, SNAI2, SOX17, and WWTR1) (Figure 5C-R); angiogenesis (COL4A2, DCN, ECSCR, GPR4, and NPR1) (Supplementary Figure 2A-E); BMP signaling pathway (CHRD, FSTL3, FZD1, HTRA3, and SPG20) (Supplementary Figure 2F-J); cell migration (CLIC4, DLC1, DPYSL3, and IGFBP5) (Supplementary Figure 3A-D); cell proliferation (ADAMTS1, ADARB1, BCL6, CD33, CDH13, and COL18A1) (Supplementary Figure 3E-J); JAK-STAT pathway (BGN, FLRT2, LRRC15, PODN, and SOCS5) (Supplementary Figure 4A-E); and TGF-beta receptor pathway (ASPN, CHST11, ENG, and HTRA1) (Supplementary Figure 4F-I).
Methylation of MARVELD1 locus between cancer and normal tissues in different cancer types
| cg site | Cancer type | Number of tumor and normal tissues | P value |
|---|---|---|---|
| cg23733123 | BLCA | Num(T)=413;Num(N)=21 | ns |
| BRCA | Num(T)=790;Num(N)=98 | **** | |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | ns | |
| LUAD | Num(T)=460;Num(N)=32 | ns | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | **** | |
| KIRP | Num(T)=276;Num(N)=45 | **** | |
| cg25355803 | BLCA | Num(T)=413;Num(N)=21 | ns |
| BRCA | Num(T)=790;Num(N)=98 | **** | |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | ** | |
| LUAD | Num(T)=460;Num(N)=32 | ns | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | **** | |
| KIRP | Num(T)=276;Num(N)=45 | **** | |
| THCA | Num(T)=515;Num(N)=56 | **** | |
| KIRC | Num(T)=320;Num(N)=160 | **** | |
| cg27165884 | BRCA | Num(T)=790;Num(N)=98 | **** |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | * | |
| LUAD | Num(T)=460;Num(N)=32 | ** | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | * | |
| KIRP | Num(T)=276;Num(N)=45 | ** | |
| LIHC | Num(T)=379;Num(N)=50 | **** | |
| cg06679087 | BRCA | Num(T)=790;Num(N)=98 | **** |
| LUAD | Num(T)=460;Num(N)=32 | ** | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | *** | |
| LIHC | Num(T)=379;Num(N)=50 | *** | |
| cg06616710 | BRCA | Num(T)=790;Num(N)=98 | **** |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | ns | |
| LUAD | Num(T)=460;Num(N)=32 | *** | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | ns | |
| KIRP | Num(T)=276;Num(N)=45 | ** | |
| LIHC | Num(T)=379;Num(N)=50 | **** | |
| THCA | Num(T)=515;Num(N)=56 | * | |
| cg00844376 | BRCA | Num(T)=790;Num(N)=98 | ** |
| LUSC | Num(T)=372;Num(N)=43 | **** | |
| HNSC | Num(T)=530;Num(N)=50 | *** | |
| KIRC | Num(T)=320;Num(N)=160 | ** | |
| cg16381596 | BRCA | Num(T)=790;Num(N)=98 | **** |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | ns | |
| LUSC | Num(T)=372;Num(N)=43 | **** | |
| PAAD | Num(T)=185;Num(N)=10 | **** | |
| HNSC | Num(T)=530;Num(N)=50 | ** | |
| KIRC | Num(T)=320;Num(N)=160 | **** | |
| cg15681358 | BRCA | Num(T)=790;Num(N)=98 | **** |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| COAD | Num(T)=299;Num(N)=38 | ns | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | ** | |
| LIHC | Num(T)=379;Num(N)=50 | **** | |
| cg24500959 | BRCA | Num(T)=790;Num(N)=98 | **** |
| CESC | Num(T)=309;Num(N)=3 | ns | |
| LUAD | Num(T)=460;Num(N)=32 | **** | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | **** | |
| LIHC | Num(T)=379;Num(N)=50 | *** | |
| cg19201009 | BLCA | Num(T)=413;Num(N)=21 | *** |
| BRCA | Num(T)=790;Num(N)=98 | **** | |
| CESC | Num(T)=309;Num(N)=3 | * | |
| LUAD | Num(T)=460;Num(N)=32 | * | |
| PRAD | Num(T)=499;Num(N)=50 | **** | |
| UCEC | Num(T)=432;Num(N)=46 | **** | |
| KIRP | Num(T)=276;Num(N)=45 | **** | |
| HNSC | Num(T)=530;Num(N)=50 | ** |
Notes: * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns: no significance.
MARVELD1 inhibits tumor growth in vivo and in vitro in colon cancer
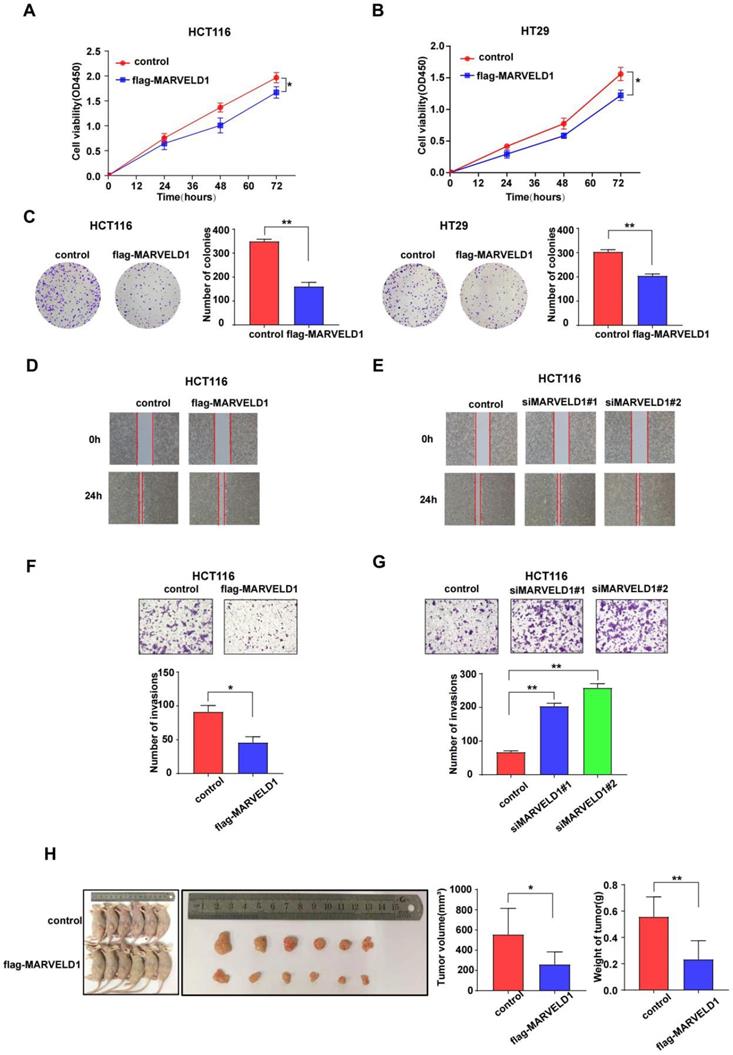
As the main functions and important pathways modulated via MARVELD1 were found in colon cancer when analyzed through GO and KEGG, colon cancer was selected for functional studies to elucidate the potential role of MARVELD1 in inhibiting tumorigenesis. MARVELD1 was stably overexpressed using purified flag-tagged lentivirus. Transient knockdown of MARVELD1 was achieved using two different siRNAs. In the HCT116 and HT29 cell lines, overexpression of MARVELD1 inhibited proliferation and clone formation as observed using the CCK-8 assay and clonogenic assay (all P < 0.05) (Figure 6A-C). In HCT116 and HT29 cell lines, overexpression of MARVELD1 decreased cell migration and invasion (all P < 0.05) (Figure 6D-G, Supplementary Figure 5). HCT116 cells, transfected with either control or overexpression MARVELD1 lentivirus, were examined to understand the role of MARVELD1 in vitro. Therefore, when BALB/c mice were injected with HCT116-controls or HCT116-flag-MARVELD1, the overexpression of MARVELD1 inhibited tumor growth compared with the controls. Moreover, the median tumor weight in the MARVELD1 overexpression group was lower than that in the control group (all P < 0.05) (Figure 6H). Therefore, MARVELD1 acts as a tumor suppressor and inhibits tumor growth in colon cancer.
Hypermethylation in promoter locus of MARVEL domain-containing 1 (MARVELD1) in human cancers compared to their non-cancer counterparts from The Cancer Genome Atlas (TCGA). (A) Methylation is higher in the MARVELD1 promoter locus at 13 methylation sites in human cancers, including cg27165884, cg26181880, cg25355803, cg24576735, cg24500959, cg23733123, cg19201009, cg18744118, cg16381596, cg15681358, cg06679087, cg06616710, and cg00844376. (B-K) Every methylation site in the MARVELD1 promoter locus was significantly hypermethylated than in its non-cancer counterpart in most types of cancers. All statistical tests were two-tailed. *p< 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, no significance.
MARVEL domain-containing 1 (MARVELD1) promoter locus hypermethylation is inversely correlated with MARVELD1 expression in human cancer. (A) MARVELD1 promoter locus hypermethylation is inversely correlated with MARVELD1 expression in a pan‐cancer cohort. (B-N) MARVELD1 promoter locus hypermethylation is inversely and significantly correlated with MARVELD1 expression in BLCA, CHOL, COAD, HNSC, KIRC, KIRP, PAAD, PCPG, SARC, SKCM, THCA, THYM, and UCEC from The Cancer Genome Atlas (TCGA) database. All statistical tests were two-tailed, and P < 0.05 indicated statistical significance.
Histone modification and transcription factor (TFs) bind to the MARVEL domain-containing 1 (MARVELD1) promoter locus are observed in the MARVELD1 promoter locus. (A-B) H3K27ac and H3K4me3 enrichment in the MARVELD1 promoter locus are observed in U87, MCF-7, LoVo, LNCaP, IMR90, HCT116, HCC1954, H128, Caco-2, and A549 cell lines. (C-D) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) are used in function and pathway enrichment analysis for upstream TF binding to the MARVELD1 promoter locus. (E) FOSL1, JUN, NFAT3, NFATC1, TCF7L2 bind to the promoter region of MARVELD1 in most types of cancers cell lines.
MARVEL domain-containing 1 (MARVELD1) is associated with biological processes in colon cancer. (A-B) Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of MARVELD1 in colon cancer. (C-R) MARVELD1 expression correlates with BICC1, CDH2, CTHRC1, DKK3, GLI1, GLI3, GREM1, HECW1, IGFBP4, IGFBP6, LATS2, MCC, PRICKLE1, SNAI2, SOX17, WWTR1 from the Wnt signaling pathway. All statistical tests were two-tailed, and P < 0.05 indicated statistical significance.
MARVEL domain-containing 1 (MARVELD1) inhibites tumor cell proliferation, migration, and invasion, both in vivo and in vitro. (A-C) The CCK-8 and the clonogenic assay indicate that overexpressing MARVELD1 inhibited the proliferation of HCT116 and HT29 cancer cells, respectively. (D-G) MARVELD1 inhibites migration and invasion in HCT116 cancer cell by the wound healing and the transwell assay. (H) The tumorigenesis and tumor tissue of each group of Balb/c mice (n = 6 per group) are shown. Tumor growth and tumor weight were measured for tumors in Balb/c mice induced by HCT116 cells transfected with an overexpression MARVELD1 lentivirus or control. Overexpression of MARVELD1 inhibited tumor growth compared with the controls. The median tumor weight in the MARVELD1 overexpression group was lower than that in the control group. *p< 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, no significance.
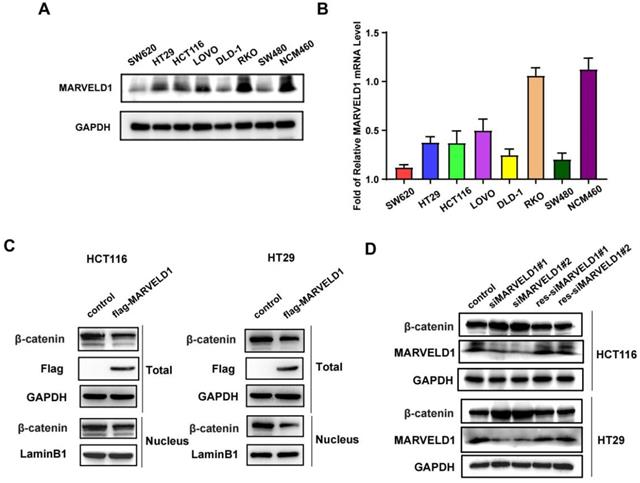
The expression of MARVEL domain-containing 1 (MARVELD1) is downregulated in colon cancer cell lines in comparison to that in normal colon cell and regulates the Wnt/β-catenin pathway in HCT116 and HT29 cells. (A-B) The expression of MARVELD1 is downregulated in SW620, HT29, HCT116, LoVo, DLD-1, RKO, and SW480 colon cancer cell lines in comparison to that in normal colon cells, i.e., NCM460, as observed by western blotting and qRT-PCR. (C) Overexpression of MARVELD1 inhibited β-catenin expression in HCT116 and HT29 cancer cells via western blot in the nucleus and in the total protein. (D) Knockdown of MARVELD1 increased β-catenin expression and then overexpressed, causing the expression of β-catenin to decrease in HCT116 and HT29 cancer cells via western blot.
MARVELD1 suppressed activity of the Wnt/β-catenin pathway in colon cancer cells
The expression of MARVELD1 was downregulated in seven colon cancer cell lines (SW620, HT29, HCT116, LoVo, DLD-1, RKO, and SW480) compared with normal colon cells, NCM460, at both mRNA and protein levels (Figure 7A-B). The activation of Wnt signaling caused the accumulation of β-catenin in the cytoplasm that was eventually translocated to the nucleus, which promotes the occurrence and progression of tumors (15, 16). We isolated the cytoplasm and nucleus from colon cancer cells and found that overexpression of MARVELD1 inhibited the expression of β-catenin in the nucleus and in the total protein (Figure 7C). The expression of MARVELD1 was knockdowned and then rescued, causing the expression of β-catenin to decrease at the protein levels (all P < 0.05) (Figure 7D).
Function of MARVELD1 in colon cancer cells was mediated by the Wnt/β-catenin pathway
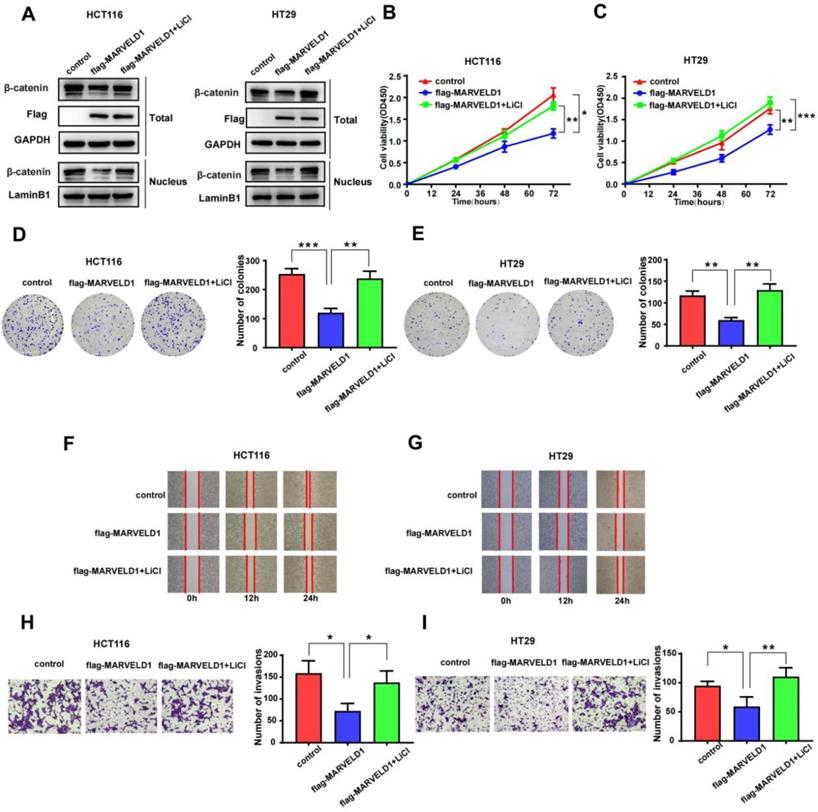
To examine whether the function of MARVELD1 in colon cancer was mediated by the Wnt/β-catenin pathway, we used LiCl to activate the Wnt/β-catenin pathway in a rescue experiment [17]. The role of MARVELD1 overexpression on the inhibition of β-catenin expression was partly reversed by LiCl in the total protein and the nucleus (Figure 8A). CCK-8 assay and clone formation showed that the inhibition of proliferation by MARVELD1 overexpression was partly reversed by LiCl in the HCT116 and HT29 cell lines (Figure 8B-E). The wound healing and the transwell assays showed that the inhibition of migration and invasion by MARVELD1 overexpression were partly reversed by LiCl (Figure 8F-I). All these results suggested that the role of MARVELD1 in colon cancer progression is mediated by the Wnt/β-catenin pathway.
Discussion
The evolutionarily conserved MARVEL domain-containing proteins are downregulated in various types of cancer, and they exert tumor-suppressive functions [6-9]. These proteins also play a role in several biological processes, including cell cycle progression, chemotactic activity, and clathrin-mediated endocytosis [18-20]. MARVELD1, a novel member of this family, is widely expressed in normal human tissues but downregulated through promoter methylation in many primary tumors, including breast, cervical, prostate, hepatocellular, esophageal, gastric, and lung carcinomas or cancer cell lines [5, 21]. In this study, first, we clarified the relationship between MARVELD1 expression and prognosis in pan-cancer from TCGA. Second, we analyzed the regulation upstream through epigenetic modification, and the transcription factors of MARVELD1 from pan-cancer data obtained from TCGA. Third, we elucidated the downstream regulatory functions of MARVELD1, the correlation between MARVELD1 and key genes of important pathways through GO and KEGG analysis in colon cancer. Fourth, we revealed that MARVELD1 regulated the occurrence and progression of tumors by inhibiting WNT/β-catenin signaling in colon cancer.
Rescue experiments of MARVELD1. (A) HCT116 and HT29 cells with MARVELD1 overexpression or control transfection were first incubated with or without 20 mmol/L LiCl and then the inhibition of β-catenin expression was partly reversed by western blotting in the nucleus and in the total protein. (B-E) HCT116 and HT29 cells with MARVELD1 overexpression or control transfection were first incubated with or without 20 mmol/L LiCl and then the inhibition of proliferation was partly reversed by the CCK-8 and clonogenic assays. (F-I) HCT116 and HT29 cells with MARVELD1 overexpression or control transfection were first incubated with or without 20 mmol/L LiCl and then the inhibition of migration and invasion were partly reversed by the wound healing and transwell assays. *p< 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, no significance.
In this study, we showed that the expression of MARVELD1 was downregulated in pan-cancer, which is consistent with previous reports. The expression of MARVELD1 was lower in colon cancer tissues than in their non-cancer tissue counterparts. MARVELD1 was mainly located in the nucleus in colon cancer, which was consistent with previous reports [5]. TCGA data was used to validate the association of low expression of MARVELD1 with poor DSS and PFI in pan-cancer. Different types of cancers were also analyzed in our cohort. The expression of MARVELD1 was negatively correlated with both OS and PFS in COAD, and negatively correlated with OS in PRAD. Patients with non-small cell lung cancer (NSCLC) and liver cancer who had a poor prognosis exhibited low expression of MARVELD1 compared with those patients with high expression levels [11, 22]. These results suggest that MARVELD1 has tumor suppressor activity.
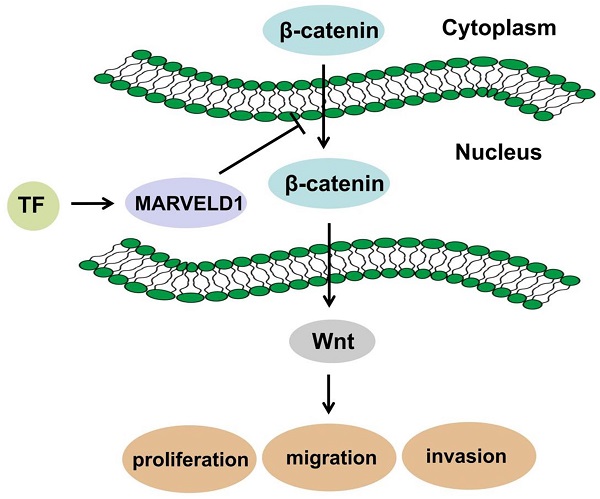
The hypermethylation and demethylation of promoters of cancer-related genes are usually related to cell proliferation. Therefore, hypermethylation of tumor suppressor genes and demethylation of oncogenes contribute toward tumorigenesis [2, 3, 23]. DNA methylation is reduced in the MARVELD1 promoter locus in multiple tumors and cell lines [4]. MARVELD1 is synergistically inactivated through both DNA hypermethylation and histone deacetylation in lung cancer cells [18]. Therefore, it is pertinent to study the epigenetic modifications that regulate MARVELD1 expression using pan-cancer data. Analyzing TCGA and 450K array databases, we predicted 13 methylation sites in the MARVELD1 promoter locus, which were hypermethylated in tissues from the pan-cancer data compared with those in non-cancer tissues. In most types of cancer, every methylation site in the MARVELD1 promoter locus was significantly hypermethylated than in their non-cancer counterparts. Moreover, our predicted methylation sites in pan-cancer were partially consistent with the previously verified methylation sites in lung cancer [21]. We will verify 13 methylation sites of the MARVELD1 promoter locus in future colon cancer studies. In addition to DNA methylation, we also found that H3K27ac and H3K4me3 were enriched in the MARVELD1 promoter locus in ten cancer cell lines, including U87, MCF-7, LoVo, LNCaP, IMR90, HCT116, HCC1954, H128, Caco-2, and A549. Both H3K27Ac and H3K4me3 are generally correlated with gene activation. However, previous research reported that BAMBI, a tumor suppressor, was modified by H3K27ac [24]. ZEB1 has tumor suppressor activity and is down-regulated in tumors, while H3K4me3 is also enriched in the promoter region of ZEB1 [25, 26]. The mechanism of gene transcription and expression is complex and affected by many factors, including DNA methylation and histone modification. Therefore, we hypothesized that MARVELD1, a tumor suppressor, is biphasically regulated via DNA methylation and histone modification, resulting in its reduced expression in cancer tissues. The lower levels of MARVELD1 expression were related to the comprehensive regulation of epigenetic factors, such as DNA methylation and histone acetylation. Transcription factors promote or inhibit gene expression by interacting with cis-acting factors. Transcription factors mainly interact with the transcription regulatory region, including the transcription activation and transcription repression domains. The transcription repression domain inhibits transcription in tumors. Therefore, in addition to studying methylation and histone modification of the MARVELD1 promoter region, we used the JASPAR software to predict upstream transcription factors. We screened five transcription factors, including FOSL1, JUN, NFATC1, NFAT3, and TCF7L2, which also related to the Wnt signaling pathway as shown by GO and KEGG analyses. Visualizing the ChIP‐seq data validated these five transcription factors in different cell lines using the ENCODE database. In the HCT116 cell lines, we predicted two transcription factors, FOSL1 and TCF7L2, bound to the MARVELD1 promoter locus. Previous studies reported that FOSL1 was downregulated in cervical carcinoma, and it inhibited the tumorigenicity of cervical carcinoma [27, 28]. Moreover, loss of the nuclear Wnt pathway effector TCF7L2 promoted migration and invasion of human colorectal cancer cells [29]. Previous reports indicated that both FOSL1 and TCF7L2 had tumor suppressor activity. In colon cancer, transcription factors regulate MARVELD1 expression, and MARVELD1 in turn decreases the cellular level of β-catenin. It means that MARVELD1 helps to construct a negative feedback loop for the Wnt/β-catenin pathway.
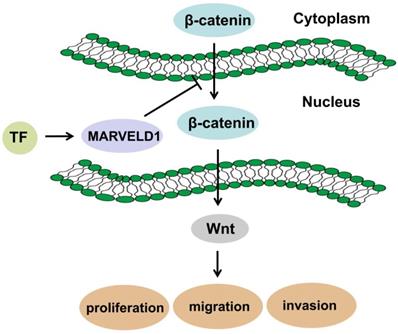
MARVELD1 is a microtubule-associated protein that can significantly inhibit cell proliferation, promote G1 phase cell-arrest, and reduce cell migration in mice [30]. Moreover, MARVELD1 plays a role in the pre-mRNA processing of integrin β1, thereby regulating cell adhesion and cell movement, a process paralleled to nonsense-mediated mRNA decay (NMD) and associated with the potential NMD factor Importin b1 [31-33]. Reducing MARVELD1 levels in lung cancer tissues also reduces the efficiency of NMD by diminishing the link between the UPF1/SMG1 components of the NMD complex and the mRNA containing a premature stop codon (PTC-mRNA) [18, 34]. These reports suggest that the function of MARVELD1 could be related to the NMD regulatory signaling pathway. Low expression of MARVELD1 was significantly associated with poor prognosis of colon cancer. Therefore, we analyzed the potential biological functions and pathways of MARVELD1 in COAD through the GO and KEGG pathway analyses. The results indicated that MARVELD1 also influenced angiogenesis, cell-substrate adhesion, blood vessel morphogenesis, lymphangiogenesis, and endothelial cell morphogenesis. Furthermore, MARVELD1 negatively regulates many tumor-related signaling pathways in COAD, including canonical Wnt, TGF-beta receptor, JAK-STAT cascade, BMP, Rap1, Ras, Hippo, MAPK, and PI3K-Akt signaling pathways. In this study, MARVELD1 inhibited cell proliferation, migration, and invasion in vitro and inhibited proliferation in vivo in colon cancer cells. These results indicate that MARVELD1 is a tumor suppressor in colon cancer. As the upstream transcriptional regulation and downstream functional enrichment of MARVELD1 indicated the Wnt signaling pathway, we analyzed this potential relationship through molecular biology experiments. Studies have shown that the Wnt pathway in cells is mainly composed of the following proteins: β-catenin, adenoma-tous poly posis coli protein (APC), glycogen synthase kinase-3β (GSK-3β) and Axin or conductin. β-catenin is a positive regulator of Wnt pathway, while APC protein, GSK-3β and Axin are negative regulators [35, 36]. The activation of this pathway enables β-catenin to accumulate in the cytoplasm and eventually translocate to the nucleus, where it serves as a co-activator of the TCF/LEF family of transcription factors, which promotes tumor formation and progression [13, 14]. Therefore, we tested whether MARVELD1 inhibited β-catenin translocation to the nucleus. We observed that MARVELD1 negatively regulated β-catenin expression at the protein level. In addition, we observed that overexpression of MARVELD1 downregulated β-catenin expression in the nucleus and in the total protein. To examine whether the function of MARVELD1 in colon cancer was mediated by the Wnt/β-catenin pathway, we used LiCl to activate the Wnt/β-catenin pathway in a rescue experiment [17]. The role of MARVELD1 overexpression on the inhibition of β-catenin expression was partly reversed by LiCl in the total protein and the nucleus in the HCT116 and HT29 cells. LiCl significantly enhanced proliferation, migration, and invasion of colon cancer cells overexpressing MARVELD1. The knockout of MARVELD1 upregulates p-ERK1/2 and cyclinD1 and downregulates p16 and p53 [10]. CyclinD1 is a classic target gene downstream of the Wnt/β-catenin signaling pathway [37-39]. Therefore, we infer that the knockout of MARVELD1 might increase CyclinD1 levels through the Wnt/β-catenin signaling pathway. High MARVELD1 expression reduces integrin b1 levels and inhibits cell migration and EMT in NSCLC cells [11]. The Wnt signaling pathway can induce EMT conversion by inhibiting the degradation of β-catenin [40-42]. In colon cancer, transcription factors regulate MARVELD1 expression, and MARVELD1 subsequently decreases the cellular level of β-catenin. This shows that MARVELD1 helps to construct a negative feedback loop involving the Wnt/β-catenin pathway (Figure 9). Therefore, we conclude that MARVELD1 regulates the occurrence and progression of tumors through the Wnt/β-catenin signaling pathway.
MARVELD1 is also associated with sensitivity to chemotherapy. The overexpression of MARVELD1 enhanced the chemotherapeutic sensitivity of liver cancer cells to epirubicin and 10-hydroxycamptothecin [10]. Low expression of MARVELD1 was also related to ovarian cancer platinum- and taxane-based chemotherapy resistance [43]. In addition, MARVELD1 interacts with catalase (CAT) to regulate the proliferation of epithelial tumor cells of the reproductive system and the sensitivity to chemotherapy drugs by reducing excessive ROS [44].
Schematic diagram of mechanism in which MARVELD1 inhibited Wnt/β-catenin pathway.
Conclusion
Overall, MARVELD1—a tumor suppressor that is epigenetically modified—inhibits the occurrence and progression of tumors through the Wnt/β-catenin signaling pathway.
Abbreviations
DSS: Disease Specific Survival; PFI: Progression free interval; PFS: Progression Free Survival; OS: Overall survival; ACC: Adrenocortical carcinoma; BLCA: Bladder Urothelial Carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangio carcinoma; COAD: Colon adenocarcinoma; DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and Neck squamous cell carcinoma; KICH: Kidney Chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute Myeloid Leukemia; LGG: Brain Lower Grade Glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and Paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin Cutaneous Melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular Germ Cell Tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine Corpus Endometrial Carcinoma; UCS: Uterine Carcinosarcoma; UVM: Uveal Melanoma.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Author Contributions
JCZ and QWL proposed the hypothesis and designed the experiments. JCZ and QWL performed the data analysis and wrote the manuscript. QWL carried out the tissue, immunohistochemical staining, and the data analysis. JCZ performed cell proliferation, migration, invasion assays, and Western blot analysis. QLS performed animal models and RT-PCR. YC and CJL participated in the design of the study. YQZ and YFY conceived the study, participated in its design and helped edit the manuscript.
Funding
This study was supported by funding from The National Natural Science Foundation of China (Grant Nos. 81872427 and 81703000), Applied technology research and development program of Heilongjiang Province (No. GA19C002), National key R&D program of China (No. 2018YFC1313300), China postdoctoral science foundation grant (No. 2018M631956), Haiyan Research Fund (No. JJZD2020-03).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mazzio EA, Soliman KFA. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7:119-30
2. Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181-187
3. Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano Antonio. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. J Cell Physiol. 2007;211(2):287-95
4. Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:599-601
5. Wang S, Li Y, Han F, Hu J, Yue L, Yu Y. et al. Identification and characterization of MARVELD1, a novel nuclear protein that is down-regulated in multiple cancers and silenced by DNA methylation. Cancer Lett. 2009;282:77-86
6. Hatta M, Nagai H, Okino K, Onda M, Yoneyama K, Ohta Y. et al. Down-regulation of members of glycolipid-enriched membrane raft gene family, MAL and BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004;30:53-8
7. Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang Y. et al. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res. 2007;13:5756-62
8. Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H. et al. CMTM3, located at the critical tumor suppressor locus 16q22. 1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69:5194-201
9. Mimori K, Shiraishi T, Mashino K, Sonoda H, Yamashita K, Yoshinaga K. et al. MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene. 2003;22:3463-71
10. Yu Y, Zhang Y, Hu J, Zhang H, Wang S, Han F. et al. MARVELD1 inhibited cell proliferation and enhance chemosensitivity via increasing expression of p53 and p16 in hepatocellular carcinoma. Cancer Sci. 2012;103:716-22
11. Yao Y, Shi M, Liu S, Li Y, Guo K, Ci Y. et al. MARVELD1 modulates cell surface morphology and suppresses epithelial-mesenchymal transition in non-small cell lung cancer. Mol Carcinog. 2016;55:1714-1727
12. Fornes O, Castro-Mondragon J, Khan A, Lee R, Zhang X, Richmond P. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:87-92
13. Liu Y, Chen X, Cheng R, Yang F. Yu M, Wang C, et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer. 2018 17(1),11
14. Jones PA, Ohtani H, Chakravarthy A, De Carvalho D. Epigenetic therapy in immune-oncology.Nat Rev Cancer. 2019;19:151-161
15. Hinze L, Labrosse R, Degar J, Han T, Schatoff EM, Schreek S. et al. Exploiting the Therapeutic Interaction of Wnt Pathway Activation and Asparaginase for Colorectal Cancer Therapy. Cancer Discov. 2020;10:1690-1705
16. Yang S, Liu Y, Li MY, Ng Calvin S H, Yang S, Wang S. et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124
17. Wang W, Liu H, Wang S, Hao X, Li L. A diterpenoid derivative 15-oxospiramilactone inhibits Wnt/β-catenin signaling and colon cancer cell tumorigenesis. Cell Res. 2011;21:730-40
18. Han W, Lou Y, Tang J, Zhang Y, Chen Y, Li Y. et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J. 2001;357:127-35
19. Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S. Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell. 2008;19:4687-93
20. Martín-Belmonte F, Martínez-Menárguez JA, Aranda JF, Ballesta J, de Marco MC, Alonso MA. MAL regulates clathrin-mediated endocytosis at the apical surface of Madin-Darby canine kidney cells. J Cell Biol. 2003;163:155-64
21. Shi M, Wang S, Yao Y, Li Y, Zhang H, Han F. et al. Biological and clinical significance of epigenetic silencing of MARVELD1 gene in lung cancer. Sci Rep. 2014;4:7545
22. Ma W, Shen H, Li Q, Song H, Guo Y, Li F. et al. MARVELD1 attenuates arsenic trioxide-induced apoptosis in liver cancer cells by inhibiting reactive oxygen species production. Annals of Translational Medicine. 2019;7:200-200
23. Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469-76
24. Li J, Wang W, Chen S, Cai J, Ban Y, Peng Q. et al. FOXA1 reprograms the TGF-β-stimulated transcriptional program from a metastasis promoter to a tumor suppressor in nasopharyngeal carcinoma. Cancer Lett. 2019;442:1-14
25. Lindner P, Paul S, Eckstein M, Hampel C, Muenzner J, Erlenbach-Wuensch K. et al. EMT transcription factor ZEB1 alters the epigenetic landscape of colorectal cancer cells. Cell Death Dis. 2020;11:147
26. Almotiri A, Alzahrani H, Menendez-Gonzalez J, Abdelfattah A, Alotaibi B, Saleh L. et al. Zeb1 modulates hematopoietic stem cell fates required for suppressing acute myeloid leukemia. J Clin Invest. 2021;131(1):undefined
27. Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian Q. et al. Fra-1 is downregulated in cervical cancer tissues and promotes cervical cancer cell apoptosis by p53 signaling pathway in vitro. Int J Oncol. 2015;46:1677-84
28. Zhang M, Liang L, He J, He Z, Yue C, Jin X. et al. Fra-1 Inhibits Cell Growth and the Warburg Effect in Cervical Cancer Cells via STAT1 Regulation of the p53 Signaling Pathway. Front Cell Dev Biol. 2020;8:579629
29. Wenzel J, Rose K, Haghighi E, Lamprecht C, Rauen G, Freihen V. et al. Loss of the nuclear Wnt pathway effector TCF7L2 promotes migration and invasion of human colorectal cancer cells. Oncogene. 2020;39:3893-3909
30. Zeng F, Tian Y, Shi S, Wu Q, Liu S, Zheng H. et al. Identification of mouse MARVELD1 as a microtubule associated protein that inhibits cell cycle progression and migration. Mol Cells. 2011;31:267-274
31. Wang S, Hu J, Yao Y, Shi M, Yue L, Han F. et al. MARVELD1 regulates integrin β1-mediated cell adhesion and actin organization via inhibiting its pre-mRNA processing. Int J Biochem Cell Biol. 2013;45:2679-2687
32. Dias SMG, Wilson KF, Rojas KS, Ambrosio ALB, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nature Structural Molecular Biology. 2009;16:930-937
33. Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23:2537-50
34. Hu J, Li Y, Li P. MARVELD1 Inhibits Nonsense-Mediated RNA Decay by Repressing Serine Phosphorylation of UPF1. PLoS One. 2013;8:e68291
35. Pennisi E. How a growth control path takes a wrong turn to cancer. Science. 1998;281(5382):1438-9
36. Peifer M. Beta-catenin as oncogene: the smoking gun. Science. 1997;275(5307):1752-3
37. Borrell-Pagès M, Romero JC, Juan-Babot O, Badimon L. Wnt pathway activation, cell migration, and lipid uptake is regulated by low-density lipoprotein receptor-related protein 5 in human macrophages. Eur Heart J. 2011;32:2841-50
38. Davies CC, Chakraborty A, Cipriani F, Haigh K, Haigh JJ, Behrens A. Identification of a co-activator that links growth factor signalling to c-Jun/AP-1 activation. Nat Cell Biol. 2010;12:963-72
39. Kim B, Mao J, Taketo MM, Shivdasani RA. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 2007;133:529-38
40. Su J, Su B, Xia H, Liu F, Zhao XH, Li J. et al. RORα Suppresses Epithelial-to-Mesenchymal Transition and Invasion in Human Gastric Cancer Cells via the Wnt/β-Catenin Pathway. Front Oncol. 2019;9:1344
41. Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17:126
42. Yang S, Liu Y, Li MY, Ng CSH, Yang S-L, Wang S. et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124
43. János TF, Ágnes Ő, Pete I, Gyula RN, Balázs G. Predictive biomarkers of platinum and taxane resistance using the transcriptomic data of 1816 ovarian cancer patients. Gynecologic Oncology. 2020;156:654-661
44. Zhang C, Han F, Shi M, Sun H, Li Y, Ci Y. et al. MARVELD1 interacting with catalase regulates reactive oxygen species metabolism and mediates the sensitivity to chemotherapeutic drugs in epithelial tumors of the reproductive system. Mol Carcinog. 2019;58:1410-1426
Author contact
![]() Corresponding authors: Yanqiao Zhang, E-mail: yanqiaozhangcom; Yuanfei Yao, E-mail: yaoyuan.feicom.
Corresponding authors: Yanqiao Zhang, E-mail: yanqiaozhangcom; Yuanfei Yao, E-mail: yaoyuan.feicom.

 Global reach, higher impact
Global reach, higher impact