Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):268-277. doi:10.7150/jca.58982 This issue Cite
Review
The functional role of Pescadillo ribosomal biogenesis factor 1 in cancer
1. Department of Gastrointestinal Surgery, The Fourth Affiliated Hospital of China Medical University, Shenyang 110032, China.
2. Department of Gastrointestinal Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo 113-0033, Japan.
3. Cancer Center, The Fourth Affiliated Hospital of China Medical University, Shenyang 110032, China.
Received 2021-2-3; Accepted 2021-7-20; Published 2022-1-1
Abstract
Tumors are neogrowths formed by the growth of normal cells or tissues through complex mechanisms under the influence of many factors. The occurrence and development of tumors are affected by many factors. Pescadillo ribosomal biogenesis factor 1 (PES1) has been identified as a cancer-related gene. The study of these genes may open up new avenues for early diagnosis, treatment and prognosis of tumors. As a nucleolar protein and part of the Pes1/Bop1/WDR12 (PeBoW) complex, PES1 is involved in ribosome biogenesis and DNA replication. Many studies have shown that high expression of PES1 is often closely related to the occurrence, proliferation, invasion, metastasis, prognosis and sensitivity to chemotherapeutics of various human malignant tumors through a series of molecular mechanisms and signaling pathways. The molecules that regulate the expression of PES1 include microRNA (miRNA), circular RNA (circRNA), c-Jun, bromodomain-containing protein 4 (BRD4) and nucleolar phosphoprotein B23. However, the detailed pathogenic mechanisms of PES1 overexpression in human malignancies remains unclear. This article summarizes the role of PES1 in the carcinogenesis, prognosis and treatment of multiple tumors, and introduces the molecular mechanisms and signal transduction pathways related to PES1.
Keywords: PES1, cancer, signaling pathway, treatment, proliferation
Introduction
Tumors are neogrowths formed by the growth of normal cells or tissues through complex mechanisms under the influence of many factors. The global incidence and mortality of cancer are increasing rapidly, and cancer may become the main cause of death in every country/region in the 21st century [1]. After tumor cells proliferate in large quantities, the malignant cells migrate from the original site to other sites through lymph, blood, implantation metastasis or direct metastasis, and then continue to grow in the new site and form the same type of tumor, thereby causing metastasis and invasion [2]. However, the proliferation, metastasis and invasion of tumors are affected by many factors, which provide novel possibilities for tumor treatment.
PES1, also known as Pescadillo or NOP7, is a protein coding gene originally identified in the embryos of zebrafish which is related to DNA replication and ribosome biogenesis [3, 4]. PES1 has been found to play an indispensable role in cell proliferation and may be involved in the process of oncogenic transformation and tumor progression [5]. The PES1 gene encodes a nucleolar protein [5, 6] of 582 amino acids [3] whose localization is regulated by nucleolar phosphoprotein B23 [7]. PES1 is an important part of telomerase assembly in which PES1, telomerase reverse transcriptase (TERT) and telomerase RNA (TR) form a complex. PES1 can directly interact with TERT to control telomerase activity, maintain telomere length and regulate cell senescence [8]. PES1, an essential 60S-preribosomal assembly factor, is also critical for ribosome biogenesis, nucleolar assembly and mammalian cell proliferation [9]. The interaction of Pes1, block of proliferation 1 (Bop1) and WD repeat protein 12 (WDR12) controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit [10]. PES1 mRNA is widely expressed during the first three days of zebrafish embryogenesis, which indicates that it is closely related to the zebrafish embryogenic process, and is also involved in Xenopus pronephros development [3, 11].
A large number of studies have shown that PES1 is related to tumor cell proliferation, invasion and metastasis of multiple types of cancer, including prostate cancer [12, 13], liver cancer [14-18], pancreatic cancer [19], papillary thyroid cancer (PTC) [20], breast cancer [21-24], ovarian cancer [25], gastric cancer (GC) [26], neuroblastoma [27], colon cancer [28], and others. There are also some studies showing that PES1 is related to the prognosis of some cancers [14, 28].
PES1 is involved in a variety of pathophysiology. In addition to sugar metabolism and the cell replication cycle [29], highly expressed PES1 promotes tumor occurrence and progression by promoting tumor cell proliferation, invasion and migration. PES1 exerts its effects through several signaling pathways, including those of phosphatidylinositol 3'-kinase (PI3K)/AKT/glycogen synthase kinase 3β (GSK3β)/cyclin D1, β-Catenin/transcription factor (TCF), c-Jun NH2 terminal kinase (JNK), c-Myc, hypoxia inducible factor (HIF)-1α and vascular endothelial growth factor (VEGF). The purpose of this review is to summarize for the first time the different roles of PES1 in various types or subtypes of tumors and to describe the potential molecular mechanisms and their clinical significance (Tables 1 and 2).
The Structural Characteristics of PES1
The PES1 gene is located at chromosome 22q12.2 and contains 19 exons in humans. The sequence and function of PES1 are well conserved among different species [3]; human PES1 has 40% sequence homology with the Pescadillo homolog of yeast, and 79% sequence homology with the zebrafish Pescadillo homolog [5]. The gene encodes a nucleolar protein which contains 588 amino acids in humans, 582 amino acids in zebrafish and 605 amino acids in yeast. The PES1 protein contains the BRCT domain [30], which can interact with the C-terminus of breast cancer associated gene 1 (BRCA1). The BRCT domain of the PES1 protein is essential for rRNA processing and nucleolar localization [31]. BRCT domains have been found in several proteins related to DNA repair, cell cycle control and recombination [32-34]. There are acidic amino acid clusters of glutamic acid and aspartic acid at the C-terminus of PES1 protein. PES1 protein also has many presumptive tyrosine/serine/threonine phosphorylation sites. Moreover, the PES1 protein contains a conserved site for SUMOylation [5, 21]. PES1, BOP1 and WDR12 form the PEOW complex, which promotes cell proliferation by promoting maturation of 5.8S and 28S ribosomal RNAs and 60S ribosomal subunit maturation. In addition, PES1, TERT and TR form a complex. PES1 can directly interact with TERT to regulate telomerase activity, maintain telomere length and control cellular senescence. PES1 promotes telomerase assembly by inducing a direct interaction between TERT and TR [8].
PES1 regulates a variety of signaling pathways to promote tumor occurrence and development
| Cancer | Related pathways | Role | Function | Refs |
|---|---|---|---|---|
| Breast cancer | - | Oncogene | Proliferation, prognosis | [22, 23] |
| Prostate cancer | - | Oncogene | Proliferation | [12] |
| Hepatocellular carcinoma (HCC) | β-Catenin /TCF; PI3K/AKT/GSK3β/cyclin D1 | Oncogene | Proliferation, tumorigenesis, glycolysis | [16-18] |
| Pancreatic cancer | c-Myc | Oncogene | Proliferation, resistance to chemotherapy drugs | [19] |
| Papillary thyroid cancer (PTC) | - | Oncogene | Proliferation, invasion, migration | [20] |
| Gastric cancer (GC) | HIF-1α | Oncogene | Proliferation | [26] |
| Neuroblastoma | - | Oncogene | Proliferation, prognosis | [27] |
| Ovarian cancer | HIF-1α | Oncogene | Proliferation | [25] |
| Colorectal cancer | JNK | Oncogene | Proliferation | [28] |
The minus sign indicates the related signaling pathway has not been discovered.
Clinical significance of PES1 in various cancers
| Cancer | Clinical significance | Refs |
|---|---|---|
| Breast cancer | Sensitivity to tamoxifen | [23] |
| Prostate cancer | - | - |
| Hepatocellular carcinoma (HCC) | Histologic grade, overall survival, relapse-free survival, tumor size, T classification | [14, 16] |
| Pancreatic cancer | Tumor size, sensitivity to BET inhibitors | [19] |
| Papillary thyroid cancer (PTC) | TNM stage, tumor size, extrathyroidal extension, lymph node metastasis | [20] |
| Gastric cancer (GC) | Tumor size | [26] |
| Neuroblastoma | International Neuroblastoma Staging System (INSS) stage, overall survival, relapse-free survival | [27] |
| Ovarian cancer | - | - |
| Colorectal cancer | Tumor size, sensitivity to anticancer drugs | [28, 59] |
The role of PES1 in different tumors
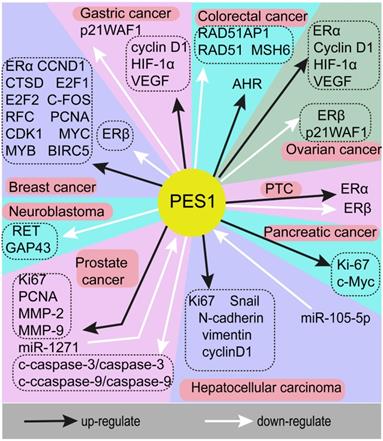
A series of studies has indicated that PES1 is highly expressed in a variety of tumors and plays a crucial role in promoting cancer. This means that PES1 may be a potential key biomarker of adverse consequences and a potential molecular target in cancer treatment. However, the molecular mechanisms underlying the role of PES1 in different tumors are complex. Here, we will describe the tumors related to PES1 and the molecular mechanisms of PES1 in tumors as a starting point from which to explore potential therapeutic targets for patients with malignant tumors. The genes associated with PES1 in different cancers are summarized in Fig. 1.
The genes associated with PES1 in different cancers. Black arrows indicate upregulation, white arrows indicate downregulation. As an oncogene, PES1 exerts a cancer-promoting effect through a variety of molecular mechanisms in the occurrence and development of various cancers.
Breast cancer
PES1 is highly expressed in breast cancer cells and promotes breast tumor growth by regulating the ratio between estrogen receptor (ER)α and ERβ [23, 24]. PES1 was reported to increase the transcription of luciferase reporter gene constructs containing an ER response element (ERE) in response to the ERα-specific agonist propylpyrazoltriol, but in response to the ERβ-specific agonist diarylpropionitrile, ERE reporter gene transcription was reduced. Immunohistochemical (IHC) staining results in 116 breast cancer tissues and 92 adjacent normal breast tissues showed that the expression of PES1 protein was positively correlated with the protein level of ERα and negatively correlated with the protein level of ERβ (P <0.0001). Highly expressed PES1, which was recruited to the promoter regions of ERα and ERβ, increased the homodimerization of ERα while reducing the homodimerization of ERβ and the heterodimerization of ERα-ERβ. PES1 also increased the promoter occupancy of ERα and reduced the promoter occupancy of ERβ. Thus, PES1 differentially regulates the transcriptional activity of ERα and ERβ [35].
Interestingly, after PES1 knockdown, the level of ERα and ERβ protein expression changed, but the level of ERα and ERβ mRNA did not change significantly, which indicated that PES1 regulated the expression of ERα and ERβ at the post-transcriptional level [23]. By regulating the AF2 domains of ERα and ERβ, the 221-322 amino acid region of PES1 upregulated the protein level of ERα, and the 311-358 amino acid region of PES1 downregulated the expression of ERβ protein [23]. After PES1 knockdown, the ubiquitination of endogenous ERα was promoted, while the ubiquitination of endogenous ERβ was inhibited. Therefore, PES1 regulates the stability of ERα and ERβ proteins through the E3 ubiquitin ligase CHIP. However, the expression of some other ER co-factors, including glutamate receptor interacting protein 1 and steroid receptor coactivator 1, regulates ERα and ERβ by simultaneously increasing the transcriptional activity of ERα and ERβ. The other two subtypes of ERβ, ERβ2 and ERβ5 [36, 37], were also downregulated by PES1. MTT analysis and colony formation analysis showed that PES1 knockdown inhibited the proliferation of breast cancer cells, and anchor-independent growth ability test results showed that PES1 knockdown also inhibited the tumorigenicity of breast cancer cells [24].
Moreover, cDNA microarray analysis and RT-PCR results showed that PES1 regulates the expression of four estradiol (E2)-regulated genes, cyclin D1 (CCND1), cathepsin D (CTSD), E2F transcription factor 1 (E2F1) and c-Fos [23]. After PES1 knockdown, the expression of many estrogen-responsive genes related to DNA replication and regulation of the cell cycle decreased, including proliferating cell nuclear antigen (PCNA), minichromosome maintenance genes, replication factor C, cyclin-dependent kinase (CDK) 1, E2F1, E2F2, MYC, MYB, CCND1 and surviving [38, 39]. In contrast, the CDK inhibitor p27Kip1 was significantly upregulated (P<0.05). Similar to ERα and ERβ, PES1 was also recruited to the promoter regions of CTSD, CCND1, E2F1 and CCNE2. Interestingly, PES1 was recruited into the distal enhancers of ERα and ERβ, but not into the distal enhancers of CTSD and CCND1 [40, 41]. The results of electrophoretic mobility shift assays indicated that PES1 interacted with the promoter of the human heme oxygenase-1 [42], which suggests that PES1 may regulate the transcription of genes that respond to estrogen through EREs. Furthermore, among breast cancer patients treated with tamoxifen, those with high PES1 expression tended to have a better prognosis [23].
Prostate cancer
Through the analysis of microarray data in an online database, PES1 was identified as a differentially expressed gene which was upregulated in prostate cancer [12]. PES1-silenced prostate cancer cells showed reduced EdU incorporation rates, increased apoptosis rates and reduced cell migration and invasion capabilities (all p <0.05). Through western blot analysis, it was found that PES1 silencing resulted in the downregulation of Ki67, PCNA, matrix metalloproteinase (MMP)-2 and MMP-9 expression, while the expression of c-caspase-3/caspase-3 and c-caspase-9/caspase-9 was upregulated (all p <0.05). Ki67 and PCNA are related to cell proliferation, while MMP-2 and MMP-9 are involved in migration and invasion [43, 44], and the caspase factors are related to apoptosis. In addition, PES1 silencing resulted in a decrease in PES1 mRNA expression in mice, as well as a decrease in tumor size (p <0.05). The tumor-promoting effect of PES1 in prostate cancer can be inhibited by miR-1271, because miR-1271 targeted PES1 and downregulated its expression [12].
Hepatocellular carcinoma (HCC)
PES1 is an HCC-related protein that is differentially expressed between HCC tissues and adjacent normal tissues. Through western blotting and RT-PCR analyses, Wang et al. found that PES1 was upregulated in HCC tissues and cells and enhanced the proliferation and tumorigenesis of HCC [16, 17]. Observed in the tumor cells of 134 samples, the expression of PES1 in 111 samples was at a high level (111/134, 82.8%), and the overall survival of patients with low PES1 expression was significantly longer than that of patients with high PES1 expression (p<0.05) [16]. Moreover, the expression of PES1 was related to vital state (p<0.001), sex (p<0.05), race (p<0.05) and the largest tumor size (p<0.05) of patients with liver cancer [14, 16]. People with high PES1 expression in liver cancer have a high risk of death. Therefore, PES1 may be a potential biological indicator for judging the prognosis of HCC. PES1 was shown to have moderate value for the diagnosis of liver cancer (AUC=0.724) [14].
PES1 promoted the proliferation, migration, invasion and tumorigenesis of liver cancer cells [14, 16-18]. After knocking down PES1, the expression of Ki67 protein decreased, the number of cells was significantly reduced and the growth of liver cancer cells in mice was decreased [17]. PES1 modulated the proliferation and tumorigenesis of HCC by regulating the PI3K/AKT/GSK3β/cyclin D1 signaling pathway [16]. GLUT1, ENO1 and PKM2 promote glycolysis [45-47], while FBP1 and PCK1 inhibit glycolysis [48, 49]. After knocking down PES1, Fan et al. found that the mRNA levels of GLUT1, ENO1 and PKM2 were downregulated, while the mRNA levels of FBP1 and PCK1were upregulated, and glucose uptake and lactate excretion of liver cancer cells were reduced, which indicated that glycolysis in HCC was reduced [17]. This mechanism may affect the “Warburg effect” of HCC [50, 51]. Moreover, PES1 was upregulated by BRD4 and acts as a key mediator in the anti-tumor process of bromodomain and extra-terminal motif (BET) protein inhibitors in liver cancer [17, 52]. Furthermore, Wei et al. found that CD44 regulates PES1 by regulating the expression of miR-105-5p, thereby promoting the proliferation of liver cancer cells [15]. Wu et al. reported that the protein levels of N-cadherin, snail, vimentin and cyclin D1 were down-regulated after PES1 knockdown [18]. Cyclin D1 is a downstream gene of the β-Catenin/TCF complex, which indicates that PES1 can activate the β- catenin/TCF signaling pathway.
Pancreatic cancer
Through the analysis of patient data identified by the GEPIA network tool [53], PES1 was found to be highly expressed in pancreatic cancer. PES1 was observed to enhance the growth of pancreatic cancer cells in vitro and in vivo [19]. Western blots and IHC analysis results revealed that PES1 expression was upregulated in 35 pancreatic cancer tissues compared to 21 non-tumor pancreatic tissue samples [19]. After PES1 knockdown, the results of MTS, CCK8, BrdU cell proliferation and colony formation assays showed that the proliferation ability of pancreatic cancer cells decreased, while in mice, the Ki-67 protein level and the proliferative ability of pancreatic cancer cells were reduced. Moreover, knocking down PES1 increased the sensitivity of pancreatic cancer cells to BET inhibitors, AKT inhibitors and mTOR pathway inhibitors, and decreased the mRNA and protein levels of c-Myc, which is related to the sensitivity of pancreatic cancer cells to BET inhibitors. Finally, CDK5 increased the stability of PES1 protein by phosphorylation of PES1, but had no significant effect on PES1 mRNA levels.
Papillary thyroid cancer (PTC)
The results of IHC staining showed that the expression of PES1 and ERα protein was significantly increased in PTC, while the expression of ERβ protein was significantly reduced (P < 0.001). PES1 was found to promote the proliferation, invasion and migration of human PTC cells [20]. PES1, ERα and ERβ protein levels, respectively, were related to tumor size, extrathyroidal extension, lymph node metastasis, BRAFV600E mutation and TNM stage (all p <0.05) [20]. As discussed previously, knockdown of PES1 reduces the level of ERα protein and increases the level of ERβ protein. ERα promotes the proliferation, invasion and migration of PTC cells and normal thyroid cells, while ERβ has the opposite effect. Therefore, highly expressed PES1 promotes the proliferation, invasion and migration of PTC cells by adjusting the ratio of ERα/ERβ protein.
Gastric cancer (GC)
PES1 was found to be highly expressed in GC [26, 54]. IHC staining results showed that in the cancer tissues of 59 patients with GC, PES1 was highly expressed in 24 tissues, while no positive staining was detected in the corresponding normal tissues adjacent to the cancer. Silencing PES1 significantly reduced the expression of cyclin D1, HIF-1α and VEGF, increased the expression of p21WAF1, and caused cell cycle arrest in the G2 or G1 phase. Li et al. found that ablation of PES1 inhibited GC proliferation and growth both in vitro and in vivo [26].
Neuroblastoma
Nakaguro et al. reported that the PES1 expression level increased with the growth of neuroblastomas, and that PES1 expression in patients with advanced neuroblastoma was higher than that in patients with early stage [27]. The expression of PES1 was associated with poor prognosis. TUNEL assay results indicated that knockdown of PES1 inhibited the growth of neuroblastoma by inducing apoptosis. The results of quantitative PCR showed that after knocking down PES1, the expression of GAP43 and RET, two genes related to differentiation, was increased [55], but the morphology of the cells did not change significantly. Moreover, all-trans retinoic acid (ATRA)-induced differentiation may regulate PES1 expression in vivo and in vitro. The nucleolus is composed of three different sub-nucleolar compartments called the fibril center, the dense fibrillar component and granular composition, from inside to the outside of the nucleolus [56]. PES1 was localized to the dense fibrillar component. The nucleolus can act as a sensor of DNA damage [57, 58], and PES1 knockdown induced the expression of DNA damage marker phosphorylated H2AX (γH2AX) and delayed DNA repair [59]. PES1 and nucleolin were also found to be redistributed due to DNA damage [27].
Ovarian cancer
Similar to PTC, PES1 has been shown to be highly expressed in ovarian cancer. Li et al. found that PES1 ablation led to a decrease in the proliferation rate of ovarian cancer cells and a delay in the G2 phase of the cell cycle [25]. By differentially regulating the transcriptional activity of ERα and ERβ, PES1 increased ERα protein and reduced ERβ protein. It was found that cyclin D1 was regulated by E2 in breast cancer, where it was activated by ERα and inhibited by ERβ [60, 61]. Knockdown of PES1 downregulated cyclin D1 and upregulated cyclin-dependent kinase inhibitor p21WAF1 in ovarian cancer [25]. HIF-1α is the target of estrogen in ovarian cancer cells and regulates the expression of VEGF [62]. The expression levels of VEGF and HIF-1α decreased after PES1 was silenced [25]. The results of IHC staining of 20 normal human ovarian tissues and 54 ovarian cancer samples indicated that the expression of PES1 was positively correlated with the expression of ERα (p <0.05) and negatively correlated with the expression of ERβ (p <0.05) [25].
Colorectal cancer
Western blot analysis showed that PES1 is highly expressed in colorectal cancer [28]. It was reported that PES1 promotes the proliferation of colorectal cancer cells in vitro and in vivo [28]. IHC analysis showed that in the 265 colon cancer tissues, 89 tissues showed PES1 staining, while only seven of the matched adjacent control tissues showed such staining. The expression of PES1 was significantly different between colon cancer tissues and adjacent normal tissues (P <0.001) [28]. Silencing of PES1 in this study inhibited the proliferation and growth of colorectal cancer cells in vitro and in vivo.
Comparing RNA extracted from PES1-silenced cells and control cells, PES1 knockdown increased mRNA levels of RAD51, MSH6 and RAD51AP1, as shown by cDNA microarray analysis and quantitative PCR [59]. RAD51 protein is essential for homologous recombination repair of DNA breaks [63]. RAD51AP1 enhances the ATPase activity of RAD51 in DNA repair [64]. MSH6 plays an important role in DNA mismatch repair [65]. However, after PES1 ablation, the protein level of RAD51 did not change significantly, which indicates that the adaptive response to increased DNA damage may cause changes in the mRNA level of PES1 [66]. Immunofluorescence analysis showed that PES1 ablation reduced nuclear RAD51 levels induced by DNA damage, indicating that PES1 plays a role in the process of RAD51 entering the nucleus. Moreover, as a transcription factor responsible for inducing drug metabolizing enzymes and controlling DNA damage response [67], the aryl hydrocarbon receptor was downregulated after PES1 knockdown.
In addition, the ablation of PES1 impaired the ability of cells to repair DNA damage, reduced the rate of DNA repair and made cells more sensitive to chemotherapy drugs [59], which indicated that PES1 may be involved in regulating the sensitivity of colorectal cancer to chemotherapy. Furthermore, JNK increased the transcription of c-Jun by phosphorylating serine 63 and serine 73 in the NH2 terminal transactivation domain of c-Jun [68], and c-Jun increased the activity of the PES1 promoter. Therefore, the phosphorylation of c-Jun regulated by JNK was essential for activating the expression of PES1, and there was a positive correlation between c-Jun and PES1 (r = 0.580, P <0.0001) in human colon cancer tissue [28].
The role of PES1 in multiple signaling pathways
β-catenin/TCF signaling pathway
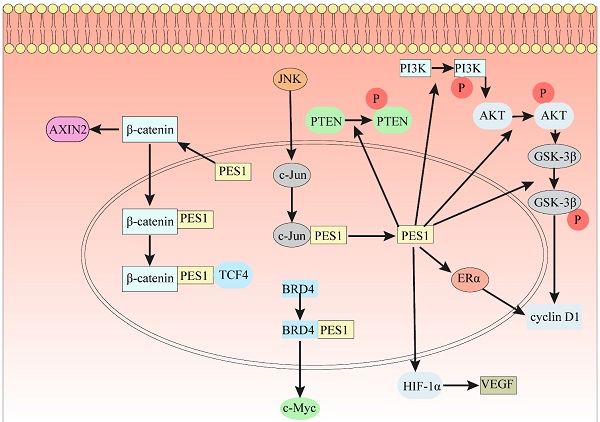
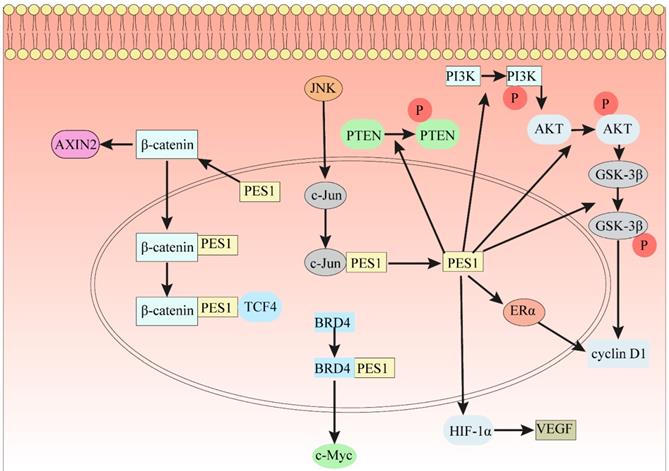
As is well known, β-Catenin affects gene transcription in many ways and plays an indispensable role in the occurrence and development of many cancers. In 7404 and Hep3B cells, ablation of PES1 decreased the protein expression levels of N-cadherin, Snail, vimentin and cyclin D1 [18], which is a downstream gene of the β-Catenin/TCF complex. The results of GST pulldown analysis indicated that PES1 interacts with β-Catenin [18]. Immunoprecipitation results in 7404 cell lysates showed that PES1 and β-Catenin formed a complex [18]. PES1 bridges β-Catenin and TCF4 together and enhances their interaction, thus activating β-Catenin/TCF signaling (Fig. 2). In addition, downregulation of β-Catenin rescued the function of PES1 in promoting HCC cell migration and colony formation [18].
Wnt/β-Catenin signaling pathway
The Wnt/β-Catenin signaling pathway regulates cell proliferation and apoptosis by inducing the expression of Wnt target genes [69], which indicates that the Wnt/β-Catenin signaling pathway plays an important role in the growth of cancer and stem cells [70]. After knocking down PES1, the level of stabilized β-Catenin increased, and the expression of AXIN2 increased significantly [71]. Knockdown of PES1 triggered nucleolar stress and further activated the Wnt/β-Catenin pathway [71]. This indicates that the loss of the key nucleolar factor PES1 can trigger nucleolar stress, further activate the Wnt/β-Catenin signaling pathway, and ultimately increase the expression of Wnt target protein AXIN2 (Fig. 2).
Interestingly, the overexpression of PES1 activates the β-Catenin/TCF signaling pathway, while the Wnt/β-Catenin signaling pathway is activated after PES1 is knocked down. Whether PES1 is involved in a negative feedback regulatory mechanism between the two pathways remains to be further explored.
The indispensable role of PES1 in different signaling pathways. (1) PES1 bridges β-Catenin and TCF4 to activate the β-Catenin/TCF signaling pathway. (2) Loss of PES1 increases the level of stabilized β-Catenin in the cytoplasm and the level of protein AXIN2. (3) The JNK signaling pathway directly binds to the PES1 promoter by c-Jun to promote the expression of PES1. (4) PES1 promotes the expression of cyclin D1 by upregulating the phosphorylation levels of PTEN, PI3K, AKT and GSK3β, and also promotes the expression of cyclin D1 by upregulating ERα. (5) PES1 promotes the expression of VEGF by upregulating HIF-1α, thereby promoting angiogenesis. (6) The bromodomain of BRD4 binds to the conserved lysine sequence in PES1 and acetylates the conserved lysine sequence, which promotes the expression of c-Myc.
PI3K/AKT/GSK3β/cyclin D1 signaling pathway
Changes in the PI3K-Akt signaling pathway have been reported in many human cancers [72]. The PI3K-Akt signaling pathway is an intracellular signaling pathway crucial in regulating the cell cycle. GSK-3β can phosphorylate cyclin D1 [73]. After knockdown of PES1 in HCC cells, phosphorylated phosphatase and tensin homolog (p-PTEN), p-PI3K, p-AKT, p-GSK-3β and cyclin D1 were all downregulated [16]. After knocking down PES1 in AGS and N87 cells, in addition to a significant decrease in the mRNA level of cyclin D1, it was also found that p21WAF1, one of the cyclin-dependent kinase inhibitors, increased significantly at the mRNA level [26]. Cyclin D1 is regulated by E2 in breast cancer, where it is activated by ERα and inhibited by ERβ [60, 61]. Highly expressed PES1 upregulates ERα and downregulates ERβ, so PES1 may activate cyclin D1 by upregulating ERα (Fig. 2). However, the specific molecular regulatory mechanism underlying the PES1 and PI3K/AKT signaling pathway needs to be further explored.
HIF-α Pathway
The HIF-1α pathway plays an important role in tumor metastasis and angiogenesis [74]. It was reported that inhibiting HIF-1 activity had a significant effect on tumor growth [75, 76]. In human squamous cell carcinoma, HIF-1α induced lymphangiogenesis and promoted tumor cell invasion by upregulating VEGF-C [77]. Li et al. found that after knocking down PES1 in AGS and N87 cells, the mRNA and protein levels of HIF-1α and VEGF decreased significantly [26]. The expression of VEGF was dependent on HIF-1α, so that when PES1 was knocked down, re-expression of PES1 increased the protein levels of HIF-1α and VEGF. In a word, PES1 can promote tumor invasion and lymphangiogenesis by regulating the expression of HIF-1α (Fig. 2).
C-Myc Pathway
Numerous reports have indicated that c-Myc regulates genes associated with the biogenesis of ribosomes and mitochondria and the metabolism of glucose and glutamine [78, 79]. Moreover, c-Myc induces genes involved in nucleotide metabolism and DNA replication [78]. Furthermore, c-Myc plays a crucial role in cancer cell energy metabolism and promotes cancer cell proliferation. Jin et al. found that the protein level of PES1 was positively correlated with the expression level of c-Myc [19]. The bromodomain-containing protein BRD4 can regulate c-Myc. Fan et al. found that knocking out BRD4 reduced PES1 expression in HepG2 and Huh7 cells [17]. The bromodomains (BD1 and BD2) of BRD4 can recognize and acetylate the conserved lysine sequence in PES1. PES1 binds to BRD4 exogenously in 293 T cells and binds to BRD4 endogenously in PANC-1 cells [19]. The interaction of PES1 and BRD4 is regulated by acetylation, and the deacetylation of PES1 leads to reduced binding to BRD4. Overexpression of PES1 upregulated the expression of c-Myc, but after overexpression of PES1 and knockdown of BRD4, c-Myc was not significantly upregulated [19]. Compared with PANS-1 cells in which PES1 or BRD4 was knocked down alone, the expression of c-Myc in PANS-1 cells in which both PES1 and BRD4 were knocked down did not change significantly [19]. In conclusion, PES1 interacts with BRD4 and promotes the expression of c-Myc (Fig. 2).
JNK signaling pathway
The JNK signal transduction pathway is an important branch of the MAPK pathway, which plays a crucial role in a variety of physiological and pathological processes such as cell cycle, reproduction, apoptosis and cell stress. JNK signaling promotes cancer cell survival [80, 81]. JNK increased the transcription of c-Jun by phosphorylating serine 63 and serine 73 in the NH2 terminal transactivation domain of c-Jun [68]. Chromatin immunoprecipitation analysis results indicated that c-Jun directly binds to the promoter of PES1 and interacts with it to regulate PES1 promoter activity [28] and increase expression of PES1. In short, the JNK signaling pathway increased the expression level of PES1 by regulating the activity of the PES1 promoter (Fig. 2).
Here, we hypothesize that with PES1 as an intermediate medium, the JNK signaling pathway is likely to form a closed feedback regulatory loop with the previously introduced β-Catenin/TCF and Wnt/β-Catenin signaling pathways. Whether this hypothesis is valid or not will require more research.
MicroRNAs, circRNA and PES1
MicroRNA (miRNA) is closely related to the occurrence, development and treatment of cancer [82]. Jiang et al. found that miR-1271 can specifically bind to PES1, which was regulated as a target gene of miR-1271 [12]. Wei et al. found that CD44 regulates the expression of miR-105-5p, and expression of miR-105-5p further targets and regulates PES1, thereby regulating the proliferation of liver cancer cells [15]. As a molecular inhibitor of PES1, circANRIL binds to the C-terminal domain of PES1, negatively regulates PeBoW activity and inhibits rRNA maturation [83].
In general, PES1 may also participate in many other signaling pathways which interact or share crosstalk with β-Catenin, PI3K/AKT, HIF-α and c-Myc, thus jointly regulating the pathways involved in the occurrence and development of tumors (Table 1).
Conclusion and perspectives
Tumorigenesis and proliferation of tumors is a very complex process, involving the formation of blood vessels and the replication of DNA. This process not only depends on changes in genetics, but also changes in epigenetics. A large number of studies have shown that PES1 is highly expressed in a variety of human cancers. PES1 is a nucleolar protein which is involved in the biogenesis of ribosomes and regulates DNA replication. PES1 can also regulate the glycolysis of tumor cells, which suggests that PES1 may be related to the “Warburg effect”, a common feature of solid tumors. PES1 is an important part of telomerase assembly and can regulate telomerase activity, maintain telomere length and regulate cellular senescence [8]. In the process of tumor cell development, the activity of telomerase is often higher, which indicates that PES1 may also regulate tumor progression by regulating the activity of telomerase. PES1 participates in the formation of the PeBoW complex and regulates the maturation of the 60S ribosomal subunit [10].
From its initial identification in zebrafish, PES1 has been studied for more than 20 years, and research on its function and related signal transduction pathways still provides surprising findings. PES1 is believed to be involved in a variety of physiological and pathological processes, including ribosome biogenesis, DNA replication and cancer [3-5]. PES1 is highly expressed in many tumors and promotes the progression of cancer. PES1 may also be a potential prognostic marker of cancer. Furthermore, given the emergence of tumor resistance to drugs including BET inhibitors, PES1 may serve as a potential target to increase tumor sensitivity to drugs.
PES1 is closely related to a variety of signaling pathways. PES1 is widely involved in regulating multiple locations and branches of the Wnt signaling pathway, which suggests an inseparable relationship between PES1 and Wnt. PES1 regulates different target proteins downstream in the Wnt signaling pathway, while playing different roles in tumor development. Knockdown of PES1 may activate the Wnt/β-Catenin signaling pathway, while the JNK signaling pathway can promote the expression of PES1, which implies that PES1 may form a regulatory loop in the Wnt signaling pathway. PES1 promotes the proliferation of breast cancer, ovarian cancer and PTC by differentially regulating ERα and ERβ. PES1 regulates the glycolysis of tumor cells by changing the gene expression of GLUT1, PKM2, ENO1, FBP1 and PCK1, which provides a new approach for the treatment of cancer. If a drug can target PES1 to inhibit the glycolysis of tumor cells, it may reduce the productivity of tumor cells and inhibit their growth. In addition, PES1 promotes tumor progression and angiogenesis by increasing factors such as cyclin D1, HIF-1α and VEGF. Therefore, drugs targeting PES1 may affect tumor progression through telomerase, glycolysis, angiogenesis, and other cellular functions.
Recent studies have also found that some proteins, miRNAs and circRNAs can target the expression of PES1. These data indicate that we can regulate PES1 at the epigenetic level to achieve tumor suppression. Various data indicate that PES1 participates in a very complex intracellular molecular regulation network, and this complex network is inextricable from the occurrence and development of tumors. Therefore, further research on PES1 will help to better evaluate cancer prognosis, develop new tumor-targeted therapeutic drugs and increase tumor sensitivity to drugs.
Abbreviations
PES1: Pescadillo ribosomal biogenesis factor 1; BOP1: block of proliferation 1; WDR12: WD repeat protein 12; miRNA: microRNA; circRNA: circular RNA; BRD4: bromodomain-containing protein 4; TERT: telomerase reverse transcriptase; TR: telomerase RNA; PTC: papillary thyroid cancer; GC: gastric cancer; PI3K: phosphatidylinositol 3'-kinase; GSK3β: glycogen synthase kinase 3β; TCF: transcription factor; JNK: c-Jun NH2 terminal kinase; HIF: hypoxia inducible factor; VEGF: vascular endothelial growth factor; BRCA1: breast cancer associated gene 1; ER: estrogen receptor; ERE: ER response element; IHC: Immunohistochemical; CCND1: cyclin D1; CTSD: cathepsin D; E2F1: E2F transcription factor 1; PCNA: proliferating cell nuclear antigen; CDK: cyclin-dependent kinase; MMP: matrix metalloproteinase; BET: bromodomain and extra-terminal motif; ATRA: all-trans retinoic acid; γH2AX: phosphorylated H2AX; p-PTEN: phosphorylated phosphatase and tensin homolog.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
2. Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453-8
3. Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141-55
4. Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835-48
5. Kinoshita Y, Jarell AD, Flaman JM, Foltz G, Schuster J, Sopher BL. et al. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. J Biol Chem. 2001;276:6656-65
6. Oeffinger M, Leung A, Lamond A, Tollervey D. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626-36
7. Zhang J, Yang Y, Wu J. B23 interacts with PES1 and is involved in nucleolar localization of PES1. Acta Biochim Biophys Sin (Shanghai). 2009;41:991-7
8. Cheng L, Yuan B, Ying S, Niu C, Mai H, Guan X. et al. PES1 is a critical component of telomerase assembly and regulates cellular senescence. Sci Adv. 2019;5:eaav1090
9. Lerch-Gaggl A, Haque J, Li J, Ning G, Traktman P, Duncan SA. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J Biol Chem. 2002;277:45347-55
10. Rohrmoser M, Holzel M, Grimm T, Malamoussi A, Harasim T, Orban M. et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27:3682-94
11. Tecza A, Bugner V, Kuhl M, Kuhl SJ. Pescadillo homologue 1 and Peter Pan function during Xenopus laevis pronephros development. Biol Cell. 2011;103:483-98
12. Jiang Z, Zhang Y, Chen X, Wang Y, Wu P, Wu C. et al. microRNA-1271 impedes the development of prostate cancer by downregulating PES1 and upregulating ERbeta. J Transl Med. 2020;18:209
13. Lazaro-Ibanez E, Lunavat TR, Jang SC, Escobedo-Lucea C, Oliver-De La Cruz J, Siljander P. et al. Distinct prostate cancer-related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer. 2017;17:92
14. Fu Z, Jiao Y, Li YQ, Ke JJ, Xu YH, Jia BX. et al. PES1 In Liver Cancer: A Prognostic Biomarker With Tumorigenic Roles. Cancer Manag Res. 2019;11:9641-53
15. Wei S, Liu K, He Q, Gao Y, Shen L. PES1 is regulated by CD44 in liver cancer stem cells via miR-105-5p. FEBS Lett. 2019;593:1777-86
16. Wang J, Sun J, Zhang N, Yang R, Li H, Zhang Y. et al. PES1 enhances proliferation and tumorigenesis in hepatocellular carcinoma via the PI3K/AKT pathway. Life Sci. 2019;219:182-9
17. Fan P, Wang B, Meng Z, Zhao J, Jin X. PES1 is transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2018;104:1-8
18. Wu N, Zhao J, Yuan Y, Lu C, Zhu W, Jiang Q. NOP7 interacts with beta-catenin and activates beta-catenin/TCF signaling in hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:6369-76
19. Jin X, Fang R, Fan P, Zeng L, Zhang B, Lu X. et al. PES1 promotes BET inhibitors resistance and cells proliferation through increasing c-Myc expression in pancreatic cancer. J Exp Clin Cancer Res. 2019;38:463
20. Qiu YB, Liao LY, Jiang R, Xu M, Xu LW, Chen GG. et al. PES1 promotes the occurrence and development of papillary thyroid cancer by upregulating the ERalpha/ERbeta protein ratio. Sci Rep. 2019;9:1032
21. Li S, Wang M, Qu X, Xu Z, Yang Y, Su Q. et al. SUMOylation of PES1 upregulates its stability and function via inhibiting its ubiquitination. Oncotarget. 2016;7:50522-34
22. Thomas C, Gustafsson JA. Targeting PES1 for restoring the ERalpha/ERbeta ratio in breast cancer. J Clin Invest. 2012;122:2771-3
23. Cheng L, Li J, Han Y, Lin J, Niu C, Zhou Z. et al. PES1 promotes breast cancer by differentially regulating ERalpha and ERbeta. J Clin Invest. 2012;122:2857-70
24. Li J, Yu L, Zhang H, Wu J, Yuan J, Li X. et al. Down-regulation of pescadillo inhibits proliferation and tumorigenicity of breast cancer cells. Cancer Sci. 2009;100:2255-60
25. Li J, Zhuang Q, Lan X, Zeng G, Jiang X, Huang Z. PES1 differentially regulates the expression of ERalpha and ERbeta in ovarian cancer. IUBMB Life. 2013;65:1017-25
26. Li J, Zhou X, Lan X, Zeng G, Jiang X, Huang Z. Repression of PES1 expression inhibits growth of gastric cancer. Tumour Biol. 2016;37:3043-9
27. Nakaguro M, Kiyonari S, Kishida S, Cao D, Murakami-Tonami Y, Ichikawa H. et al. Nucleolar protein PES1 is a marker of neuroblastoma outcome and is associated with neuroblastoma differentiation. Cancer Sci. 2015;106:237-43
28. Xie W, Feng Q, Su Y, Dong B, Wu J, Meng L. et al. Transcriptional regulation of PES1 expression by c-Jun in colon cancer. PLoS One. 2012;7:e42253
29. Prisco M, Maiorana A, Guerzoni C, Calin G, Calabretta B, Voit R. et al. Role of pescadillo and upstream binding factor in the proliferation and differentiation of murine myeloid cells. Mol Cell Biol. 2004;24:5421-33
30. Haque J, Boger S, Li J, Duncan SA. The murine Pes1 gene encodes a nuclear protein containing a BRCT domain. Genomics. 2000;70:201-10
31. Holzel M, Grimm T, Rohrmoser M, Malamoussi A, Harasim T, Gruber-Eber A. et al. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007;35:789-800
32. Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25-30
33. Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68-76
34. Koonin EV, Altschul SF, Bork P. BRCA1 protein products: Functional motifs. Nat Genet. 1996;13:266-8
35. Fox EM, Davis RJ, Shupnik MA. ERbeta in breast cancer-onlooker, passive player, or active protector? Steroids. 2008;73:1039-51
36. Shaaban AM, Green AR, Karthik S, Alizadeh Y, Hughes TA, Harkins L. et al. Nuclear and cytoplasmic expression of ERbeta1, ERbeta2, and ERbeta5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res. 2008;14:5228-35
37. Honma N, Horii R, Iwase T, Saji S, Younes M, Takubo K. et al. Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol. 2008;26:3727-34
38. Santen R, Cavalieri E, Rogan E, Russo J, Guttenplan J, Ingle J. et al. Estrogen mediation of breast tumor formation involves estrogen receptor-dependent, as well as independent, genotoxic effects. Ann N Y Acad Sci. 2009;1155:132-40
39. Foster JS, Henley DC, Ahamed S, Wimalasena J. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol Metab. 2001;12:320-7
40. Bretschneider N, Kangaspeska S, Seifert M, Reid G, Gannon F, Denger S. E2-mediated cathepsin D (CTSD) activation involves looping of distal enhancer elements. Mol Oncol. 2008;2:182-90
41. Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513-26
42. Sikorski EM, Uo T, Morrison RS, Agarwal A. Pescadillo interacts with the cadmium response element of the human heme oxygenase-1 promoter in renal epithelial cells. J Biol Chem. 2006;281:24423-30
43. Trudel D, Fradet Y, Meyer F, Harel F, Tetu B. Significance of MMP-2 expression in prostate cancer: an immunohistochemical study. Cancer Res. 2003;63:8511-5
44. London CA, Sekhon HS, Arora V, Stein DA, Iversen PL, Devi GR. A novel antisense inhibitor of MMP-9 attenuates angiogenesis, human prostate cancer cell invasion and tumorigenicity. Cancer Gene Ther. 2003;10:823-32
45. Mendez-Lucas A, Li X, Hu J, Che L, Song X, Jia J. et al. Glucose Catabolism in Liver Tumors Induced by c-MYC Can Be Sustained by Various PKM1/PKM2 Ratios and Pyruvate Kinase Activities. Cancer Res. 2017;77:4355-64
46. Hamaguchi T, Iizuka N, Tsunedomi R, Hamamoto Y, Miyamoto T, Iida M. et al. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008;33:725-31
47. Cho HS, Ahn JM, Han HJ, Cho JY. Glypican 3 binds to GLUT1 and decreases glucose transport activity in hepatocellular carcinoma cells. J Cell Biochem. 2010;111:1252-9
48. Liu MX, Jin L, Sun SJ, Liu P, Feng X, Cheng ZL. et al. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene. 2018;37:1637-53
49. Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK. et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251-5
50. Racker E. Warburg effect revisited. Science. 1981;213:1313
51. Vaupel P, Multhoff G. Revisiting the Warburg effect: historical dogma versus current understanding. J Physiol. 2021;599:1745-57
52. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-17
53. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
54. Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C. et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248-55
55. Matsushima H, Bogenmann E. Expression of trkA cDNA in neuroblastomas mediates differentiation in vitro and in vivo. Mol Cell Biol. 1993;13:7447-56
56. Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325-34
57. Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068-77
58. Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665-72
59. Xie W, Qu L, Meng L, Liu C, Wu J, Shou C. PES1 regulates sensitivity of colorectal cancer cells to anticancer drugs. Biochem Biophys Res Commun. 2013;431:460-5
60. Welboren WJ, Sweep FC, Span PN, Stunnenberg HG. Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated? Endocr Relat Cancer. 2009;16:1073-89
61. Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008;27:1019-32
62. Ho QT, Kuo CJ. Vascular endothelial growth factor: biology and therapeutic applications. Int J Biochem Cell Biol. 2007;39:1349-57
63. Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247-51
64. Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai MS, Leung SG. et al. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem. 2012;287:12343-7
65. Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407-20
66. Smith JJ, Cole ES, Romero DP. Transcriptional control of RAD51 expression in the ciliate Tetrahymena thermophila. Nucleic Acids Res. 2004;32:4313-21
67. Puga A, Marlowe J, Barnes S, Chang CY, Maier A, Tan Z. et al. Role of the aryl hydrocarbon receptor in cell cycle regulation. Toxicology. 2002;181-182:171-7
68. Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354:494-6
69. MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9-26
70. Tran FH, Zheng JJ. Modulating the wnt signaling pathway with small molecules. Protein Sci. 2017;26:650-61
71. Dannheisig DP, Bachle J, Tasic J, Keil M, Pfister AS. The Wnt/beta-Catenin Pathway is Activated as a Novel Nucleolar Stress Response. J Mol Biol. 2021;433:166719
72. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin Cancer Biol. 2019;59:147-60
73. Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102-14
74. Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378-89
75. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721-32
76. Ma Z, Xiang X, Li S, Xie P, Gong Q, Goh BC. et al. Targeting hypoxia-inducible factor-1, for cancer treatment: Recent advances in developing small-molecule inhibitors from natural compounds. Semin Cancer Biol. 2020 S1044-579X(20)30202-9
77. Schito L, Rey S, Tafani M, Zhang H, Wong CC, Russo A. et al. Hypoxia-inducible factor 1-dependent expression of platelet-derived growth factor B promotes lymphatic metastasis of hypoxic breast cancer cells. Proc Natl Acad Sci U S A. 2012;109:E2707-16
78. Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479-83
79. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024-39
80. Wu Q, Wu W, Fu B, Shi L, Wang X, Kuca K. JNK signaling in cancer cell survival. Med Res Rev. 2019;39:2082-104
81. Abdelrahman KS, Hassan HA, Abdel-Aziz SA, Marzouk AA, Narumi A, Konno H. et al. JNK signaling as a target for anticancer therapy. Pharmacol Rep. 2021;73:405-34
82. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
83. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429
Author contact
![]() Corresponding author: Dong-Qiu Dai, MD, PhD, Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, No. 4 Chongshan East Road, Shenyang 110032, China. Telephone: +86-24-62043110, E-mail: daidq63com.
Corresponding author: Dong-Qiu Dai, MD, PhD, Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, No. 4 Chongshan East Road, Shenyang 110032, China. Telephone: +86-24-62043110, E-mail: daidq63com.

 Global reach, higher impact
Global reach, higher impact