Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(2):481-495. doi:10.7150/jca.65012 This issue Cite
Review
Biomarkers for predicting the efficacy of immune checkpoint inhibitors
1. Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China.
2. Department of Hematology, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China.
3. Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University & Capital Medical University, Beijing Tongren Hospital, Beijing 100730, China.
#These authors contributed equally to this work.
Received 2021-7-16; Accepted 2021-12-5; Published 2022-1-1
Abstract
Immune checkpoint blockade has vastly changed the landscape of cancer treatment and showed a promising prognosis for cancer patients. However, there is still a large portion of patients who have no response to this therapy. Therefore, it's essential to investigate biomarkers to predict the efficacy of immune checkpoint inhibitors. This article summarizes the predictive value of established biomarkers, including programmed cell death ligand 1(PD-L1) expression level, tumor mutational burden, tumor-infiltrating lymphocytes, and mismatch repair deficiency. It also addresses the predictive value of tumorous mutations, circulation factors, immune-related factors, and gut microbiome with immunotherapy treatment. Furthermore, some of the emerging novel biomarkers, and potential markers for hyper progressive disease are discussed, which should be validated in clinical trials in the future.
Keywords: Predictive biomarkers, PD-L1 expression, circulation biomarkers, immunotherapy, hyper progressive disease
Introduction
Programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) is believed to be a pathway closely related to tumor growth and immune evasion [1]. PD-1/PD-L1 axis is considered to inhibit proliferation and activation of T cells, breaking the balance of immune surveillance and immune resistance [2]. PD-1 and PD-L1 blockade has greatly improved the prognosis of cancer patients and favorable outcomes have been observed since Food and Drug Administration (FDA) approved the first immune checkpoint inhibitor (pembrolizumab) in 2014. Some biomarkers are believed to have power in predicting the effect of PD-1/PD-L1 antibodies, such as PD-L1 expression levels and tumor mutation burden (TMB) [3,4].
However, not all patients could obtain clinical benefits from anti-PD-1/PD-L1 therapy, and still a large number of patients, with shorter progression-free survival (PFS), have no response to PD-1 inhibitors, which limited the clinical applications of anti-PD-1/PD-L1 therapy [5]. Predictive biomarkers can be used to select patients most likely to be benefited from immunotherapy, reducing costs and treatment-related adverse reactions. This article reviewed current potential biomarkers by categories of tumor-related factors, immune-related factors, and factors detected in tumor tissue and peripheral blood. The effect of biomarkers for hyper progressive disease (HPD) was also discussed, but the role of these biomarkers is still controversial with the current available clinical data.
Established biomarkers
PD-L1 expression
PD-L1 expression has remained to be the focus of various studies since the early stage of immunotherapy. Based on the results of the phase III KEYNOTE-024 trial, patients treated with pembrolizumab showed much longer progression-free survival (PFS) and overall survival (OS) than those treated with platinum-based chemotherapy, and patients of both groups had at least 50% PD-L1 expression on their tumor cells [3]. In the KEYNOTE-052 trial, a phase II study of pembrolizumab for patients with urothelial carcinoma, the objective response rate (ORR) in the group with PD-L1 expression higher than 10% was significantly better than that in the group with PD-L1 expression lower than 1% (39% vs 11%) [6].
The efficacy of immune checkpoint inhibitors (ICIs) varied with tumor types, but it is generally believed that high PD-L1 expression level is associated with favorable clinical outcomes to anti PD-1/anti PD-L1 therapy (Table 1). Schmid et al. [7] observed that PD-L1 expression on tumor-infiltrating immune cells as a percentage of tumor area is predictive in advanced triple-negative breast cancer patients treated with atezolizumab and nab-paclitaxel. Patients in the PD-L1 positive group (> 1%) had a longer median overall survival (25 months vs 21.3 months) than all patients [7]. But in the OAK study of non-small cell lung cancer (NSCLC) patients treated with atezolizumab, patients with low or negative PD-L1 (PD-L1 expression < 1%) also had a longer median OS than those treated with docetaxel (12.6 months vs 8.9 months), while clinical benefit was more significant in patients with positive PD-L1 expression [8]. This finding suggests PD-L1 expression level is not the predictive determinant for efficacy of ICIs.
Recently, PD-L1 structural rearrangements (PD-L1MUT) was reported as a potential biomarker for response to immunotherapy in patients with relapse/refractory natural-killer/T cell lymphoma. This frequent somatic mutation led to the disruption of the 3'-UTR of PD-L1 gene, was found to be significantly enriched in the responders to pembrolizumab (4 of 9) compared to the non-responders (0 of 10) [9].
Besides PD-L1 expression levels on T cells and tumor cells, Liu et al. [10] found that high-level of PD-L1 expression on CD68+ macrophages correlated with longer OS in NSCLC patients treated with immunotherapy. PD-L1 levels on macrophages were significantly associated with PD-L1 expression on tumor cells and CD8+ T-cell infiltration [10]. However, defective and controversial aspects about the predictive role of PD-L1 could not be ignored. Not all predictions by PD-L1 expression for immunotherapy were positively correlated. Motzer et al. [11] reported that PD-L1 negative group (<1%) showed a longer median PFS (27.4 months vs 21.8 months) than PD-L1 (>1%) group in advanced clear-cell renal-cell carcinoma patients treated with nivolumab. Based on the results of CheckMate 040, a study of advanced hepatocellular carcinoma treated with nivolumab, PD-L1 predictive effect was not that significant: objective response in PD-L1 positive (cut-off value was 1%) group was 26%(9/34), while in PD-L1 negative group, that was 19%(26/140) [12]. Remarkable intra-tumoral heterogeneity of PD-L1 expression was observed across breast cancer sub-types and stages [13]. Ilie et al. [14] discovered that PD-L1 expression was often inconsistent between removed tissues and matched biopsy specimens (the overall discordance rate was 48%), which was measured by highly sensitive SP142 immunohistochemical (IHC) assay. Hong et al. [15] found that the heterogeneity of PD-L1 expression was influenced by anatomic locations of distant metastases and treatment stages in NSCLC patients. The course of immunotherapy also changed PD-L1 levels [15]. These factors would result in inaccurate PD-L1 expression measurements from biopsies or surgically removed specimens.
IHC is widely used to assess the level of PD-L1 expression, which includes 4 main IHC assays: 22C3, 28-8, SP142, and SP263. Hirsch et al. [16] demonstrated that 28-8, 22C3 and SP263 were highly consistent in evaluating the PD-L1 expression on tumor cells, while the positive rate of SP142 antibody was lower compared with the other three assays. The discordance of PD-L1 expression with two rabbit monoclonal antibodies, E1L3N and SP142, was also observed in NSCLC patients [17]. And no consensus of the cut-off value of PD-L1 expression had been reached across various tumor types.
Besides IHC, novel measurement of PD-L1 expression is being investigated. Conroy et al. [18] demonstrated that the PD-L1 mRNA expression, which was measured by next generation RNA sequencing, was associated with PD-L1 expression levels in a study of 209 patients. Furthermore, non-invasive positron emission tomography (PET) for evaluating PD-L1 expression levels was proved to be valid in immunotherapy effect prediction [19,20]. Bensch et al. [20] claimed PET for PD-L1 status evaluation was more relevant to clinical response than IHC or RNA sequencing for PD-L1 levels. However, challenges like how to interpret and integrate PET data into medical practice are remained to be solved [21].
Predictive effect of PD-L1 expression for PD-1/PD-L1 inhibitors
| Immunological checkpoint inhibitor | Study | Tumors | Participants | Cut-off | Efficacy (95% CI) | Ref |
|---|---|---|---|---|---|---|
| Pembrolizumab | NCT01295827 | Advanced NSCLC | n=101 | <1% 1%-49% ≥50% | Median PFS, months 3.5 (2.1 to 19.0) Median PFS, months 4.2 (3.1 to 6.4) Median PFS, months 12.5 (6.2 to NR) | [33] |
| NCT01295827 | Melanoma | n=451 | <10% ≥10% | Median PFS, months 5.6 (4.4 to 8.1) Median PFS, months 2.8 (2.7 to 2.8) | [34] | |
| NCT02335424 | Urothelial carcinoma | n=370 | All patients ≥10% | ORR: 24% (20% to 29%) ORR: 38% (29% to 48%) | [6] | |
| NCT02255097 | Squamous cell carcinoma | n=171 | <50% ≥50% | ORR: 13% (7% to 20%) ORR: 27% (15% to 42%) | [35] | |
| Nivolumab | NCT01721772 | Metastatic melanoma | n=418 | <5% ≥5% | ORR 33.1% (25.2% to 41.7%) ORR 52.7% (40.8% to 64.3%) | [36] |
| NCT02387996 | Urothelial carcinoma | n=265 | <1% ≥1% | ORR: 16.1% (10.5% to 23.1%) ORR: 23.8% (16.5% to 32.3%) | [37] | |
| Nivolumab & Ipilimumab | NCT02477826 | Acute myeloid leukemia | n=70 | <1% ≥1% | Median OS, months: 17.2 (12.8 to 22.0) Median OS, months: 17.1 (15.8 to 20.1) | [38] |
| Atezolizumab | NCT02008227 | NSCLC | n=425 | <1% ≥1% | Median PFS, months 12.6 (9.6 to 15.2) Median PFS, months 20.5 (17.5 to NR) | [8] |
| NCT02108652 | Urothelial carcinoma | n=119 | <5% ≥5% | ORR: 21% (11% to 35%) ORR: 28% (14% to 47%) | [39] |
Tumor microenvironment immune types (TMIT)
| Classification | TIL and PD-L1 expression | Characteristics |
|---|---|---|
| TMIT 1 | TIL+ PD-L1+ | Sensitive to immunotherapy |
| TMIT 2 | TIL- PD-L1- | Low response rate to ICIs |
| TMIT 3 | TIL- PD-L1+ | Dysfunction of T cells |
| TMIT 4 | TIL+ PD-L1- | Lack of target |
Although detection of PD-L1 expression levels is the major way to predict the efficacy, its predictive value might get improved when combined with other biomarkers [22], which would be further explained in this article.
Tumor infiltrating lymphocytes (TILs)
TILs are crucial components of tumor microenvironment, which is related to tumor growth and regression. Tumeh et al. [23] found that proliferation of intra-tumoral CD8+ T cells was associated with tumor regression in metastatic melanoma patients treated with pembrolizumab. Furthermore, more T-cell antigen receptors (TCRs) and PD-1+ CD8+ T cells were observed inside and on the edge of the invasive tumors of the responders. Thommen et al. [24] reported that high baseline levels of PD-1+ CD8+ T cells (>1%, assessed by multiple immunohistochemistry) was correlated with longer OS in advanced NSCLC patients treated with nivolumab. CD45RO+ T cells were reported as independent predictive factors for favorable outcomes of immunotherapy across various cancers, while FOXP3+ Treg cells were considered to be related to poor prognosis [25-27].
Chen et al. [28] classified tumor microenvironment into four immune types by TILs (presence/absence) and PD-L1 expression (positive/negative) to evaluate clinical benefits of immunotherapy (Table 2). The cut-off values of TILs and PD-L1 expression were adjusted for various tumor types (Figure 1).
In metastatic melanoma and NSCLC patients, TILs and high PD-L1 expression levels were observed to be correlated with significantly longer OS and better prognosis [29,30]. But resistance to immunotherapy in tumor microenvironment immune type 1(TMIT1) patients with urothelial carcinoma was also observed [31]. Kim et al. [32] summarized and concluded that about 45% to 50% of patients had primary resistance to immunotherapy, but this data was influenced by the detection methods of PD-L1 expression and TILs, for example, the initial sampling of PD-L1 expression evaluation, the effectiveness of research drugs, and the accuracy of IHC methods.
Tumor Mutational Burden
Tumor Mutational Burden (TMB) is defined as the density of nonsynonymous mutations in the coding region of the tumor genome and calculated as mutations per DNA Megabase (Mb) [40]. In NSCLC patients with high TMB, the subgroup treated with nivolumab plus ipilimumab had higher 1-year PFS rate (42.6% vs 13.2%), median FPS (7.2 months vs 5.5 months) and better ORR (45.3% vs 26.9%) than those treated with chemotherapy, where TMB cut-off value was 10 mutations per Mb [41]. Higher TMB increased the likelihood of neoantigen production, promoting immune recognition and tumor cell killing [42]. A statistic correlation between TMB and response rate to anti-PD-1 therapy was observed across different types of tumor [43,44]. However, TMB cut-off value varies remarkably across tumor types because of different histological distribution of TMB, it is unrealistic to predict clinical benefits by a universal TMB cut-off value [44]. PD-L1 expression and TMB are not related, but increased and higher PD-L1 levels predicts better clinical benefits, according to a study of 240 advanced NSCLC patients treated with PD-1 blockades [45].
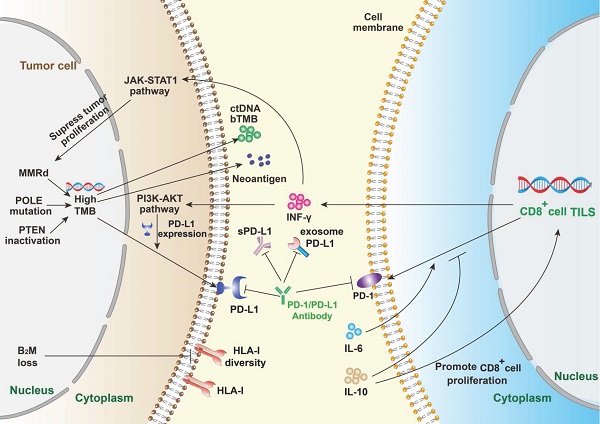
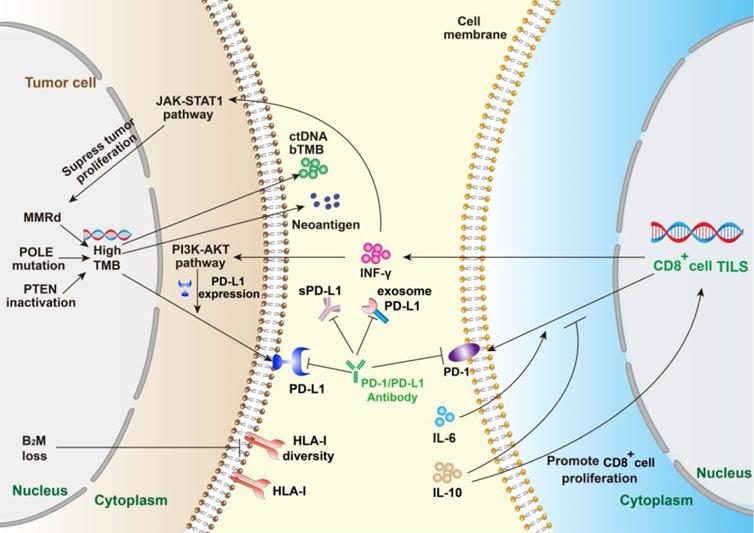
Summary of mechanism of PD-1/PD-L1 and anti PD-1/PD-L1 immunotherapy. The efficacy of PD-1/PD-L1 antibodies therapy is mainly predicted by PD-L1 expression, tumor infiltrating lymphocytes, tumor mutational burden and peripheral cytokines. PD-L1 expression reflects immune resistance, which is the target of PD-1/PD-L1 inhibitors. Mismatch repair deficiency (MMRd) and mutations of related genes will contribute to high TMB. Meanwhile, TMB enhances immunogenicity, which can be detected in the periphery. B2M loss suppresses the expression of HLA-I and leads to ICIs escape. IFN-γ, mainly derived from TILs, can enhance immune activity and inhibit tumor proliferation, but can also up-regulate the expression level of PD-L1 on tumor cells. IL-6 and IL-10 mainly play a regulatory role by affecting the PD-1 expression on immune cells.
Whole exome sequencing (WES) is the gold standard for TMB assessment, but it is only accessible to a few patients due to high cost and complex testing process. Therefore, next-generation sequencing (NGS) for blood TMB (bTMB) is growing popular these days. Rizvi et al. [46] found that in metastatic NSCLC patients with higher bTMB (bTMB ≥20 mutations per Mb), the subgroup treated with durvalumab plus tremelimumab had a significantly better median OS (21.9 months vs 10.0 months) than those treated with chemotherapy. No correlation was found between PD-L1 expression and bTMB [46]. Gandara et al. [47] observed that higher bTMB (cut-off value: 16 mutations per Mb) was correlated with remarkable longer OS (13.0 months with atezolizumab vs 7.4 months with docetaxel) in NSCLC patients. In general, good consistency between tissue TMB and blood TMB has been observed, suggesting bTMB as a predictive biomarker for efficacy of ICIs, but more data and evidence are needed [48].
Mismatch repair deficiency (MMRd)
Mismatch repair (MMR) system takes part in modifying mismatch, insertion and deletion of DNA replication. Le et al. [49] found that metastatic carcinoma patients with MMRd showed significantly higher ORR (40% vs 0%) and PFS rate (78% vs 11%) than those with mismatch repair-proficient, both groups treated with pembrolizumab. Higher ORR and better clinical benefits were observed in patients with ICIs and MMRd across 12 tumor types [50]. In the Phase II KEYNOTE-158 Study, Pembrolizumab showed clinical benefits in MMRd/MSI-H (high microsatellite instability) patients with 27 advanced cancers, and outcomes were promising: ORR was 34.3%, the median PFS was 4.1 months and the median OS was 23.5 months [51].
Mutations resulting from MMRd are most frequently located in monomorphic microsatellites, which is called high microsatellite instability (MSI-H). The MMR system usually counts on four vital genes: mutL homologue 1 (MLH1), post meiotic segregation increased 2 (PMS2), mutS homologue 2 (MSH2), and mutS 6 (MSH6), which are detected by related proteins through IHC. If two or more genes altered or inactivated, the tumor is called MSI-H. If one genetic abnormality was observed, it is usually called low microsatellite instability (MSI-L). Or it is called microsatellite stability (MSS) [52]. Great consistency (>90%) between MMRd and MSI-H has been observed across cancer types [53]. Furthermore, MSH2 expression was found to be associated with high TMB, increased PD-L1 expression and CD8+ T cells infiltration in lung adenocarcinoma (LUAD) patients with ICIs, suggesting MSH2 as a potential predictive biomarker [54].
The positive rate of PD-L1 expression was much higher (38.9% vs 16.5%) in solid tumors with MLH1/MSH2 loss than overall samples [55]. The significant correlation between positive PD-L1 and MMRd was also described in endometrial carcinomas and colorectal cancer [56,57]. Howitt et al. [58] reported that MSI-H was relevant to high TMB and TILs density. It is generally believed that high TMB are associated with MSI-H and MMRd, which is correlated with high level of PD-L1 expression.
Tumorous factors
PTEN inactivation
Phosphatase and tension homology deleted on chromosome 10 (PTEN) is a vital tumor suppressor gene controlling the activation of the PI3K-AKT pathway, which participates in the process of cell proliferation. PTEN inactivation is believed to correlate with DNA repair and plays a role in MMRd/MSI-H [59]. Loss of PTEN inhibits T-cell mediated tumor killing and lowers the density of TILs in tumor microenvironment, resulting in poor efficacy of immunotherapy [60]. Zhao et al. [61] found that PTEN inactivation was enriched in non-responders with glioblastomas, compared with patients who responded to ICIs. Barroso-Sousa et al. [62] demonstrated that PTEN mutations were significantly associated with lower ORR (6% vs 48%), shorter PFS (2.3 months vs 6.1 months) and OS (9.7 months vs 20.5 months) in triple-negative breast cancer patients treated with anti PD-1/PD-L1 therapy. George et al. [63] found that loss of PTEN was correlated with reduced tumor neoantigens which activated T cells in vitro, suggesting PTEN inactivation as a predictive biomarker for immunotherapy.
POLE Mutations
The DNA polymerase epsilon catalytic subunit A (POLE) participates in DNA replication and DNA repair pathways, playing a crucial role in tumor mutations. The loss of POLE raises the tumor mutation rate, resulting in a significant increase of TMB and tumor growth [64]. Though uncommon, it was reported that POLE mutations were associated with higher TMB, PD-L1 expression and CD8+ TILs density in NSCLC patients treated with immunotherapy, compared with patients with wild-type POLE [65]. Increased neoantigen loads and high density of TILs were also observed in patients with POLE mutation across multiple tumor types, suggesting better response to immunotherapy [66,67]. Wang et al. [66] reported that the POLE mutation rate was 2.79% across 47721 patients with various cancers, and the mutation rate in non-melanoma skin cancer patients was the highest (16.59%). Under immunotherapy, POLE mutations were correlated with increased OS (34 months vs 18 months with POLE wild-type) [66]. In conclusion, current data and studies suggest that POLE mutations predict better clinical benefits for PD-1 blockade therapy, but larger scale of studies are needed for validation.
Co-Mutation of KRAS and STK11
The correlation between KRAS and STK11 mutations was found based on a large analysis of NGS of 1343 NSCLC tumor samples [68]. Skoulidis et al. [69] demonstrated KRAS and STK11/LKB1 co-mutation was relevant to PD-L1 negativity in intermediate or high TMB lung adenocarcinoma (LUAC). Furthermore, patients treated with nivolumab with KRAS and STK11 co-mutations showed a much lower ORR (7.4%), compared with those with KRAS and p53 co-mutations (35.7%) and KRAS mutations alone (28.6%) [69]. In patients with NSCLC, alteration of KRAS/STK11 was found to have influence on tumor microenvironment and decrease PD-L1 expression levels, suggesting a lower response rate to ICIs and poorer prognosis [70,71]. Deficiency of STK11/LKB1 was correlated with T cell exhaustion and accumulation of neutrophils with T-cell suppressive effects [71]. These studies indicate that KRAS and STK11 mutations predict poorer clinical outcomes for immunotherapy.
EGFR mutation
EGFR signaling modulates the immunoregulatory effects of MHC I/II and PD-L1 expression on activity of tumor cells and T cells. EGFR mutation (mEGFR) is associated with decreased PD-L1 expression level, lower TMB and decreased density of CD8+ T cells infiltration, but the detection of mEGFR is mainly used in NSCLC [72]. In advanced NSCLC patients with EGFR mutations (including sensitizing and non-sensitizing mutations) and positive PD-L1 expression (>1%), only 1 in 11 patients (9%) responded to pembrolizumab [73]. EGFR sensitizing mutations are the most vital driving gene mutations in lung cancer, and exon19 deletion or L858R point mutation are the most common. Sensitizing mutations tend to induce tyrosine kinase inhibitors resistance and co-mutations, which predict a poor prognosis [74]. Oxnard et al. [75] reported that combination of durvalumab and osimertinib (a third generation of EGFR tyrosine kinase inhibitor) would increase the morbidity of interstitial lung disease (38%) in NSCLC patients with mEGFR (L858R or exon19 insertion/deletion), while significant clinical benefits were not observed. However, not all patients with mEGFR could not have clinical benefits from the immunotherapy. Based on the results of the IMpower150 trial, the atezolizumab, bevacizumab, carboplatin, and paclitaxel (ABCP) subgroup had significantly longer OS and PFS than the bevacizumab, carboplatin, and paclitaxel (BCP) subgroup in patients with NSCLC who harbored EGFR sensitizing mutations [76]. The clinical efficacy of PD-1/PD-L1 inhibitors combined with tyrosine kinase inhibitors (TKI) was limited in mEGFR patients, but immunotherapy combined with chemotherapy or anti-vascular therapy suggested favorable outcomes. In cancer types of stomach, brain, breast, endometrium and colon, increased EGFR expression levels were also detected, suggesting possibility of EGFR mutations and poor prognosis [77,78]. However, studies on EGFR mutations as predictors of immunotherapy have been widely carried out in NSCLC. EGFR-mutated tumors have significantly higher richness and lower T-cell clonality than EGFR-wild-type tumors, although no difference is noted in T cell repertoire, which could be a possible explanation of the lower activity of ICIs.
TET
DNA hydroxymethylated enzyme Ten-Eleven-Translocation (TET) is capable of demethylation of 5-methylcytosine (5mC) and plays a crucial role in genomic epigenetic regulation. The TET family contains TET1, TET2 and TET3, where mutations of TET2 are thought to be related with hematopoietic malignancy [79]. Xu et al. [80] showed that TET2 controlled chemokine and PD-L1 expression, and lymphocyte infiltration through IFNγ-JAK-STAT signaling pathway. Lower activity of TET was associated with decreased TILs and tumor progression and immune evasion. High activity of TET suggested favorable outcomes in patients treated with immunotherapy, which could be enhanced by vitamin C or ascorbate [80]. Moreover, Chen et al. [81] found de-ubiquitination enzyme 15 (USP15) could suppress tumor immunity and promote tumor growth through inactivation of TET2. Wu et al. [82] found that TET1-mutant was more common in patients treated with ICIs, and patients with TET1-mutant showed significantly higher ORR (60.9% vs 28.8%) and durable clinical benefits (71.4% vs 31.6%) than patients with TET1-wildtype.
Biomarkers in peripheral blood
Neutrophil-to-lymphocyte ratio
Peripheral blood cells are associated with prognosis in patients receiving immunotherapy, especially lymphocyte counts and neutrophil-to-lymphocyte ratio (NLR). Diem et al. [83] found that higher NLR was associated with lower ORR, shorter OS and PFS in NSCLC patients treated with nivolumab. Ren et al. [84] reported that in the advanced NSCLC patients treated with immunotherapy, in the group with TMB>10 mutations per Mb, an association between lower NLR (NLR<2.5) and longer OS and PFS was observed, compared with group with TMB<10 mutation per Mb. Moreover, low absolute lymphocyte count (ALC) was considered to be a biomarker for poorer prognosis. Decreased clinical benefits from immunotherapy and shorter median PFS (60 days vs 141 days) have been observed in patients with lower ALC (ALC < 600 cells/μl) [85]. Absolute neutrophil counts (ANC) alone was also relevant to the efficacy of anti PD-1 therapy. Patients with high baseline of ANC (ANC>7500) and elevated NLR (NLR>3) had very low 1- and 2- year survival rates, only 2% and 0% respectively, compared with 43% and 24% for those with lower ANC and NLR [86].
Combined with other established biomarkers, NLR also has the potential for prognosis prediction of patients receiving ICIs. In advanced non-small cell lung cancer (aNSCLC), patients with NLR>5, combined with high PD-L1 expression levels (>80%), showed favorable outcome following first-line pembrolizumab monotherapy [87]. But no association between PD-L1 expression and NLR was found [88]. Patients with a lower NLR (NLR<2.5) combined with a high TMB (TMB>10) showed favorable OS and PFS than those with a higher TMB and NLR (NLR>2.5). Furthermore, Platelet-to-Lymphocyte ratio (PLR) level below 200 was correlated with longer OS and FPS, which might also have the potential to serve as a potential biomarker for prognosis [89].
Lactate dehydrogenase
High level of lactate dehydrogenase (LDH) is considered to be a marker for poorer prognosis in various tumors. Kelderman et al. [90] observed that the efficacy of ipilimumab treatment was limited in metastatic melanoma patients with baseline serum LDH greater than twice the upper limit of normal (ULM). Mezquita et al. [91] demonstrated that LDH greater than ULM, and derived neutrophils/(leukocytes minus neutrophils) ratio (dNLR) greater than 3, were considered to be markers for poor treatment outcomes of immunotherapy. However, the level of LDH elevation lacks specificity and may be caused by liver or muscle lesions, which should be excluded from clinical studies.
Peripheral immune cells
Peripheral immune cells reflect the functions and subtypes of TILs. Gros et al. [92] found that neoantigen-specific CD8+ T cells, which could recognize autologous tumors, existed in the circulating PD-1+ CD8+T-cell population. This study suggested that peripheral blood PD-1+CD8+ T cells might reflect the change of neoantigen-specific CD8+ T cells and predict the efficacy of immunotherapy. Kamphorst et al. [93] reported that 70% of patients with disease progression showed a delayed or absent circulating PD-1+CD8+ T cells response, while 80% of patients with clinical benefits had a PD-1+CD8+ T cells response in four weeks. The proliferation of circulating Ki-67+PD-1+CD8+ T cells was also observed in the first or second treatment cycle, associated with a promising treatment efficacy [93]. Krieg et al. [94] demonstrated that high frequency of peripheral CD14+ CD16- HLA-DRhi monocytes was correlated with increased ORR to anti-PD-1/PD-L1 therapy, and decreased number of peripheral T cells was also observed in responding patients. Higher abundance of CD45RO+ T memory cells was observed in responders, suggesting favorable outcomes of immunotherapy [94,95].
Predictive effect of some peripheral immune cells
| Predictive factors | Cancer types | Predictive effect | Ref |
|---|---|---|---|
| Ki-67+ PD-1+ CD8+ T cells | NSCLC | Favorable objective response | [93] |
| CD14+CD16-HLA-DRhi monocytes | Stage IV melanoma | Favorable objective response | [94] |
| CD45RO+ T memory cells | NSCLC | Favorable objective response | [25] |
| FOXP3+ Treg cells | metastatic gastric cancer | Poor objective response | [26] |
Circulating tumor DNA
Circulating tumor DNA (ctDNA) is generally believed to reflect the mutation load of tumor tissues, and it is more accessible than TMB examinations. Bratman et al. [96] reported that ctDNA baseline level was a sensitive predictor for the efficacy and prognosis in solid tumor patients treated with pembrolizumab, and patients with ctDNA below median baseline levels had a longer OS. The dynamic changes of ctDNA can further predict the clinical benefits. In a cohort study of 73 patients, the ctDNA levels of 33 patients dropped from their baselines, and 14 (42%) of them had objective remission. While among 40 patients with an elevated ctDNA levels from the baselines, only 1 (2%) reached objective remission. Decreased levels of ctDNA after treatment of immunotherapy predict that the patients will have a longer OS [97,98]. Gandara et al. [47] reported a positive correlation between ctDNA, obtained by next- generation sequencing method, and tissue TMB. They retrospectively analyzed the outcomes of randomized phase II study POPLAR and phase III OAK trial, defining ctDNA ≥ 16 mutations per megabase (Mut/Mb) as a cut-off point in NSCLC for predicting atezolizumab benefits. But this cut-off value is controversial and may have trouble in applying to other tumor types [99].
Soluble PD-L1
Like membrane-binding PD-L1, soluble PD-L1 (sPD-L1) inhibits proliferation and activation of T cells, promoting tumor growth and escape. A positive correlation of soluble PD-1 (sPD-1) and favorable clinical outcomes was observed in advanced pancreatic cancer patients. The group with high sPD-L1 levels (sPD-L1 > 0.012 ng/ml) showed a shorter median OS (9.53 months vs 11.92 months) than patients with low sPD-L1 levels. Furthermore, sPD-1 and sPD-L1 levels are closely correlated [100]. High baseline levels of sPD-L1 are also associated with increased possibility of tumor progression in patients with melanoma and NSCLC [101,102]. However, sPD-L1 levels were not relevant to PD-L1 expression levels of tumor cells [102]. The concentration of sPD-L1 is easier to measure than tumor PD-L1 expression levels, but available clinical data is limited and more studied are needed to further unravel the predictive role of sPD-L1.
Besides sPD-L1, exosome PD-L1 levels are also investigated as a predictive biomarker. Theodoraki et al. [103] found that PD-L1 levels on exosomes, instead of levels of sPD-L1, were associated with tumor progression of head and neck squamous cell carcinomas. Fan et al. [104] reported that higher exosome PD-L1 levels, which were an independent prognostic factor, were correlated with lower OS compared with lower level of exosome PD-L1 group in gastric cancer patients. Chen et al. [105] demonstrated that stimulation with interferon-γ (IFN-γ) increased the amount of exosome PD-L1, thereby inhibiting the functions of CD8+ T cells and promoting tumor growth.
Peripheral blood T-cell receptor (TCR)
TILs are not necessarily tumor-recognizing lymphocytes, and the immune response cannot be judged simply by the numbers of TILs. The detection of TCR can reflect T-cell functions of recognizing tumor cells and predict the efficacy of immunotherapy more accurately [106,107]. Scheper et al. [107] found that only about 10% of tumor infiltrating T cells could recognize autologous tumor cells by analyzing intra-tumoral TCR repertoire of CD8+ T cells in ovarian and colorectal cancer. Postow et al. [108] found that higher evenness (similarities between the rearrangement frequencies of specific V and J genes) of peripheral TCR was associated with better PFS in 12 patients with metastatic melanoma, but no significant difference in OS was observed. Hogan et al. [109] observed that low baseline level of diversity evenness of the TCR repertoire before treatment was related to better FPS and promising response to anti-PD-1 therapy. Moreover, peripheral blood TCR repertoire was correlated to Immune-related Adverse Events (IrAEs) in early stages of treatment. Lack of diversity of CD4+ and CD8+ TCR was observed in patients with severe IrAEs [110].
Peripheral cytokines
The alteration of peripheral cytokines can reflect the conditions of tumor microenvironment and T cells response [111]. TGF-β is considered as a factor of tumor evasion and immune suppression. High baseline levels of peripheral TGF-β (≥200 pg/mL) are associated with poor prognosis in patients with hepatocellular carcinoma who were treated with pembrolizumab [112]. TGF-β, originating from peri-tumor fibroblasts, prevents T cells from infiltrating into the tumor parenchyma, which makes tumor-specific T cells more likely to be distributed in the peri-tumor stroma rather than the intratuminal parenchyma [113]. TGF-β inhibitors can increase the sensitivity of anti PD-1/PD-L1 therapy in the mouse models of progressive liver metastatic diseases, which may be due to TGF-β inhibition resulting in an effective and long-lasting cytotoxic T-cell response to tumor cells [114].
IFN-γ, mainly derived from TILs, could promote immune activity and inhibit tumor proliferation, but it also regulates up the expression levels of PD-L1 on tumor cells [115]. IFN-γ could activate the JAK2-STAT1 pathway, which suppressed the tumor cell proliferation [116]. However, IFN-γ also induces the activation of PI3K-AKT pathway, which could increase PD-L1 expression levels. Blockade of PI3K-AKT pathway would maximize the effect of IFN-γ in anti-tumor therapy [117]. The results of phase II POPLAR trial showed increased expression of IFN-γ induced by T-cell effector was correlated to favorable OS in patients with NSCLC who received atezolizumab [118]. Furthermore, IFN-γ positive mRNA signature suggested clinical benefits with anti-PD-1/PD-L1 treatment [119]. But more clinical data are needed to support this conclusion.
IL-6 is relevant to poor prognosis in patients with NSCLC [120]. High levels of IL-6 is associated with significant increase of Treg cells and high expression level of PD-1 on CD4+ and CD8+ T cells [121]. Keegan et al. [122] observed that decreased levels of IL-6 were associated with better FPS in patients with NSCLC who received immunotherapy than those with high or normal levels of IL-6 (median PFS: 11 vs 4 months). These evidences indicate that IL-6 levels might be a potential biomarker for prognosis.
IL-10 is considered as an immunoregulatory cytokine, but it also plays a role in proliferation of CD8+ T cells. Li et al. [123] found that IL-10 improved the expression of IFN-γ and inhibited the expression of PD-1 on CD8+ T cells in peripheral blood and tumor tissues. Giunta et al. [124] reported higher IFN-γ/IL-10 ratio was observed in responding patients treated with immunotherapy. Boutsikou et al. [125] observed that increased levels of cytokines (TNF-α, IL-1β, IL-2, IL-4 and IL-8) were related to favorable outcomes of anti PD-1/anti PD-L1 therapy, and these cytokines were independent from PD-L1 expression. Additionally, Sanmamed et al. [126] reported a strong correlation between decreased levels of IL-8 and better response to immunotherapy and longer OS in NSCLC and melanoma patients.
Vascular endothelial growth factor (VEGF) plays a crucial role in angiogenesis, which results in tumor growth and metastasis [127]. VEGF can inhibit the function of T cells and increase the recruitment of myeleloids-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), and the differentiation and activation of dendritic cells (DCs) are obstructed [128]. Wallin et al. [129] demonstrated that anti PD-1 and anti VEGF combined would lead to proliferation of CD8+ T cells in tumor microenvironment and improving tumor-specific T-cell migration. Atkins et al. [130] claimed that pembrolizumab combined VEGF inhibitors showed better clinical benefits and appeared to be tolerable in advanced renal cell carcinoma patients. Shibaki et al. [131] observed that high levels of peripheral VEGF were related to worse outcomes and poorer efficacy of immunotherapy in NSCLC patients older than 75 years (Figure 2).
Immune-related factors
Beta-2-microglobulin
Beta-2-microglobulin (B2M) is a vital component of human leukocyte antigen class I (HLA-I) molecules, whose mutation inhibits antigen presentation and tumor evasion. Sade-Feldman et al. [132] found that 5 of 17 patients with metastatic melanoma and immunotherapy had point mutations, deletions or loss of heterozygosity (LOH) in B2M. Non-responders had three times more B2M LOH (30% vs 10%) than responders and were associated with poorer OS. Gettinger et al. [133] found that deletion and decreased expression of B2M on tumor cells were correlated with resistance to PD-1 or PD-L1 inhibitors. Class I HLA disruption mediated by B2M deletion leads to ICIs escape in lung cancer. Immunocompetent lung cancer mouse, whose B2M genes were knocked out, also showed the resistance to ICIs. Janikovits et al. [134] found that increased density of PD-1+ T cells was associated to B2M mutations, suggesting that deficiency of HLA-I induced by B2M mutations mainly occurred in PD-1+ T-cell infiltrating microenvironment.
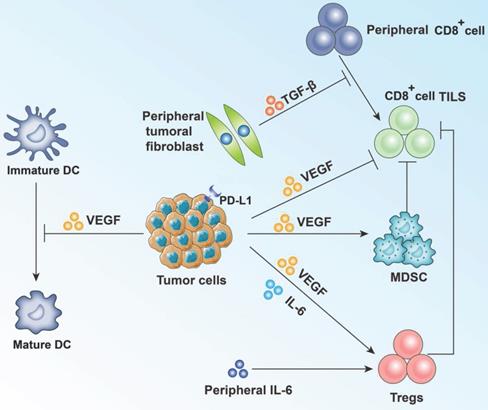
Summary of effect of VEGF and other main cytokines on tumor cells and TILs. VEGF, mainly secreted by tumor cells, shows immunosuppressive effect by activating Treg cells and bone marrow-derived suppressor cells (MDSC). VEGF can also inhibit T-cell function directly. Besides, maturation of dendritic cells (DC) is blocked. IL-6 and TGF-β also participate in the immune response and tumor evasion.
Pereira et al. [135] reported that 5% lung cancer patients carried B2M mutations, and most of them impair the correct formation of HLA-I complex. Other genetic mutations involved in HLA-I complex maturation had also been observed. The levels of B2M and HLA-I proteins were associated with lower cytotoxic CD8+ lymphocyte infiltration and PD-L1 expression. Chowell et al. [136] found that maximal heterozygosity (different alleles at HLA-I locus) at HLA-I was related to better OS with immunotherapy treatment, compared to those patients who were homozygous for at least one HLA locus. In advanced melanoma patients, the group with the most (>50% cells) and complete loss of major histocompatibility complex(MHC) I expression on tumor cells showed primary resistance to anti cytotoxic T lymphocyte antigen 4 (CTLA-4) therapy, while MHC II expression positive (>1%) on tumor cells suggested favorable response to anti PD-1 therapy [137].
B7-H4
B7-H4, which belongs to the B7 immunoglobulin superfamily, inhibits tumor immune response by inhibiting T-cell proliferation and cytokines production. And B7-H4 is highly expressed in tumor tissues, but low or no expression in normal tissues [138]. B7-H4 also participates in tumor growth and immune escape through suppressing functions and proliferation of T cells [139]. Genova et al. [140] found that B7-H4 expression was correlated with the decrease of PFS (1.7 vs 2.0 months) and OS (4.4 vs 9.8 months) in NSCLC patients treated with nivolumab. And no significant association was observed between B7-H4 expression and outcomes of patients receiving platinum-based chemotherapy. Shrestha et al. [141] observed that high expression level of B7-H4 suggested poor prognosis in hepatocellular carcinoma patients treated with immunotherapy. And B7-H4 expression was independently associated with worse prognosis.
TOX
Scott et al. [142] reported high expression of thymocyte selection-associated high mobility group box gene (TOX) in tumor specific T cells and exhausted T cells during chronic viral infection. The loss of TOX in tumor-specific T cells removes the T-cell failure procedure, but these non-exhausted T cells remains dysfunctional [142]. Guo L et al. [143] found that increased TOX expression was positively correlated with higher density of TILs, and high expression of TOX indicated favorable prognosis for various tumor types, especially for lung adenocarcinoma (LUAD). However, Kim et al. [144] found that TOX promoted the exhaustion of CD8+ T cells in tumor microenvironment by up-regulating intercellular adhesion molecules, and inhibition of TOX expression was associated with increased efficacy of anti PD-1 therapy. Although TOX showed predictive value for immunotherapy effect, TOX measurement was hard to be applied in daily clinical practice.
Gut microbiota
Gut microbiota could influence host immune response and have an impact on the efficacy of PD-1 based immunotherapy (Table 4). Sivan et al. [145] found that oral administration of bifidobacteria alone was associated with enhanced function of dendritic cells (DC), which resulted in increased density of CD8+ T cells in tumor microenvironment and better effect of ICIs. But this phenomenon disappeared after fecal transfer. Routy et al. [146] found that oral supplemented or fecal microbiota transplanted Akkermansia muciniphila would enrich the CRCX3+ CD4+ and CD8+ T cells in tumor bed, and enhance cytotoxic CD8+ lymphocyte infiltration and T-cell immune functions, which was correlated with improvement of the efficacy of immunotherapy. Enterococcus hirae was observed to help increase the ratio of CD8+ T-cell/Treg after transferring to secondary lymphoid organs [147]. Enrichment of Barnesiella intestinihominis in the colon was related to the increase of tumor infiltrating T cells [147]. Furthermore, Matson et al. [148] found that Bifidobacterium longum, Colinthia, and Enterococcus faecalis were more abundant in the responders, which could be repeated in germ-free mouse by fecal microbiota transplantation from the responders. Gopalakrishnan et al. [149] observed much higher alpha diversity and relative abundance of bacteria of the Ruminococcaceae family in melanoma patients responding to PD-1 based immunotherapy. Therefore, the dominant presence of different species of microbe in different cancers conferred sensitivity to immunotherapy, and analysis of gut microbiota composition can be helpful to predict the efficacy of anti-PD-1/PD-L1 treatment.
Favorable predictive effect of gut microbiota
| Favorable predictive factors | Cancer types | Model | Ref |
|---|---|---|---|
| Bifidobacterium | Melanoma | Mouse | [145] |
| Faecalibacterium | Metastatic melanoma | Human | [158] |
| Akkermansia muciniphila | Metastatic melanoma | Mouse/human | [149] |
| Bacteroides | Melanoma | Human/mouse | [159] |
| Higher diversity of gut microbiota | Metastatic melanoma | Mouse/human | [149] |
Biomarkers for hyper progressive disease
Hyper progressive disease (HPD) is referred to as abnormal tumor growth related to very poor prognosis (Table 5). No consensus has been reached on the definition of HPD, but it is acknowledged that if the tumor growth kinetics and/or tumor growth rate increased by at least two times during treatment, it can fall into this category [150,151]. Ferrar et al. [152] found that HPD was more common in NSCLC patients who received immunotherapy (13.8%, 56 in 406 patients) than those treated with chemotherapy (5.1%, 3 in 59 patients). Kim et al. [153] also observed this phenomenon, and they found that low frequency of CD8+ T cells, CCR7-CD45RA- T memory cells and high frequency of exhausted T cells in peripheral blood were associated with higher occurrence of HPD and worse outcome in NSCLC patients treated with ICIs.
Predictive biomarkers for HPD
| Biomarkers | Cancer types | Predictive effect on HPD | Ref |
|---|---|---|---|
| MDM2 family amplification | Not mentioned | Positive (4 in 6 patients, 67%) | [160] |
| EGFR mutation | Not mentioned | Positive (2 in 10 patients, 20%) | [160] |
| Elevated NLR | NSCLC | Positive | [153] |
| Elevated levels of LDH, ANC and CRP | Advanced gastric cancer | Positive | [161] |
| Ki67+ PD1+ Treg cells | Gastric cancer | Positive | [162] |
| CCR7- CD45RA-CD8+T memory cells | NSCLC | Negative | [153] |
| TIGIT+PD1+CD8+ T cells | NSCLC | Positive | [153] |
Advanced age is a risk factor for HPD. The incidence rate of HPD is significantly higher in patients over the age of 65 than those under 65 (37% vs 19%) [154]. Lo Russo et al. [155] found that increased infiltration of M2 tumor-associated macrophages (TAMs), which participated in immunotherapy, was related to HPD. Increased infiltration of M2 TAMs in tumor microenvironment activated Treg cells and accelerated apoptosis of effector T cells to suppress immune response [156]. Furthermore, Chen et al. [157] demonstrated that high concentrations of ctDNA and MIKI67 mutations were correlated with HPD in 22 advanced NSCLC patients treated with immunotherapy.
Conclusion
It is generally believed that PD-L1 expression is the main predictive biomarker for anti PD-1 therapy. But PD-L1 expression is not an independent factor, which could be affected by TMB, MMRd and other predictive factors. PD-L1 expression combined with other established biomarkers, such as TMB and TILs, could better predict the immunotherapy effects in certain tumor types. Mutations in tumor genes are considered to be the initiating factors for tumor growth and progression. Some specific mutations, such as POLE and PTEN, are greatly associated with efficacy of ICIs. However, whole exome-sequencing (WES) or next-generation sequencing (NGS) has not been widely applied in clinical practice due to high cost and technical factors. Proliferation and activation of T cells plays a crucial role in immunotherapy. T-cell related factors can reflect immunotherapy effects more intuitively. But these factors have not been universally tested in clinical practice. Peripheral biomarkers are more accessible than established biomarkers mentioned above. However, these factors are more susceptible to interference. Some biomarkers, like high levels of LDH, have low specificity. The examinations of major biomarkers are limited to laboratory studies and there is still a long way to go between laboratory researches and clinical applications.
There are a large number of novel biomarkers emerging correlated with tumor progression or regression in the process of immunotherapy treatment, which are not fully covered in this article. Additionally, some of these factors may have values in prognosis prediction for the patients who are not treated with PD-1 inhibitors. Overall, more data and evidence are needed to support the predictive values of these emerging biomarkers.
Abbreviations
PD-1: programmed cell death protein 1; PD-L1: programmed cell death ligand 1; TMB: tumor mutation burden; ICIs: immune checkpoint inhibitors; NSCLC: non-small cell lung cancer; TILs: Tumor infiltrating lymphocytes; TCRs: T-cell antigen receptors; TMIT: tumor microenvironment immune types; Mb: Megabase; MMRd: Mismatch repair deficiency; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability; MLH-1: mutL homologue 1; PMS2: post meiotic segregation increased 2; MSH2: mutS homologue 2; MSH6: mutS 6; MSS: microsatellite stability; LUAD: lung adenocarcinoma; EGFR: epidermal growth factor receptor; TKI: tyrosine kinase inhibitors; TET: hydroxymethylated enzyme Ten-Eleven-Translocation; 5mC: demethylation of 5-methylcytosine; USP15: de-ubiquitination enzyme 15; NLR: neutrophil-to-lymphocyte ratio; ALC: absolute lymphocyte count; ANC: absolute neutrophil counts; PLR: platelet-to-lymphocyte ratio; LDH: lactate dehydrogenase; ULM: upper limit of normal; dNLR: derived neutrophils/(leukocytes minus neutrophils) ratio; ctDNA: circulating tumor DNA; sPD-L1: soluble PD-L1; IFNγ: interferon-γ; IrAEs: Immune-related Adverse Events; TGF-β: transforming growth factor-β; VEGF: vascular endothelial growth factor; IL-6: interleukin- 6; IL-8: interleukin- 8; IL-10: interleukin- 10; MDSCs: myeleloids-derived suppressor cells; Tregs: regulatory T cells; DCs: dendritic cells; B2M: beta-2-microglobulin; HLA-I: human leukocyte antigen class I; LOH: loss of heterozygosity; MHC: major histocompatibility complex; CTLA-4: cytotoxic T lymphocyte antigen 4; TOX: thymocyte selection-associated high mobility group box gene; TAMs: tumor-associated macrophages; CRP: C-reactive protein; IHC: immunohistochemical; PET: positron emission tomography; WES: Whole exome sequencing; NGS: next-generation sequencing; bTMB: blood tumor mutation burden; PFS: progression-free survival; OS: overall survival; ORR: objective response rate; HPD: hyper progressive disease.
Acknowledgements
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant No. 81873450, 82170181), the Open Research Fund from Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beijing Tongren Hospital, Beihang University, and Capital Medical University (grant No. BHTR-KFJJ-202009), and Beijing Municipal Administration of Hospitals' Youth Programme (code: QMS20200201) to Liang Wang.
Author Contributions
LW designed the study; CJW and HNW collected data, and wrote the manuscript; all authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kythreotou A, Siddique A, Mauri FA, Bower M, Pinato DJ. PD-L1. J Clin Pathol. 2018;71:189-94
2. Ribas A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 2015;5:915-9
3. Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A. et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375:1823-33
4. Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A. et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843-52
5. Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9-16
6. Balar AV, Castellano D, O'Donnell PH, Grivas P, Vuky J, Powles T. et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18:1483-92
7. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H. et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108-21
8. Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255-65
9. Lim JQ, Huang D, Tang T, Tan D, Laurensia Y, Peng R. et al. Whole-genome sequencing identifies responders to Pembrolizumab in relapse/refractory natural-killer/T cell lymphoma. Leukemia. 2020;34:3413-9
10. Liu Y, Zugazagoitia J, Ahmed FS, Henick BS, Gettinger SN, Herbst RS. et al. Immune Cell PD-L1 Colocalizes with Macrophages and Is Associated with Outcome in PD-1 Pathway Blockade Therapy. Clin Cancer Res. 2020;26:970-7
11. Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S. et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373:1803-13
12. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. The Lancet. 2017;389:2492-502
13. Dill EA, Gru AA, Atkins KA, Friedman LA, Moore ME, Bullock TN. et al. PD-L1 Expression and Intratumoral Heterogeneity Across Breast Cancer Subtypes and Stages: An Assessment of 245 Primary and 40 Metastatic Tumors. Am J Surg Pathol. 2017;41:334-42
14. Ilie M, Long-Mira E, Bence C, Butori C, Lassalle S, Bouhlel L. et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol. 2016;27:147-53
15. Hong L, Negrao MV, Dibaj SS, Chen R, Reuben A, Bohac JM. et al. Programmed Death-Ligand 1 Heterogeneity and Its Impact on Benefit From Immune Checkpoint Inhibitors in NSCLC. J Thorac Oncol. 2020;15:1449-59
16. Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K. et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12:208-22
17. McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J. et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. Jama Oncol. 2016;2:46-54
18. Conroy JM, Pabla S, Nesline MK, Glenn ST, Papanicolau-Sengos A, Burgher B. et al. Next generation sequencing of PD-L1 for predicting response to immune checkpoint inhibitors. J Immunother Cancer. 2019;7:18
19. Lv G, Sun X, Qiu L, Sun Y, Li K, Liu Q. et al. PET Imaging of Tumor PD-L1 Expression with a Highly Specific Nonblocking Single-Domain Antibody. J Nucl Med. 2020;61:117-22
20. Bensch F, van der Veen EL, Lub-de HM, Jorritsma-Smit A, Boellaard R, Kok IC. et al. (89)Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat Med. 2018;24:1852-8
21. Nimmagadda S. Quantifying PD-L1 Expression to Monitor Immune Checkpoint Therapy: Opportunities and Challenges. Cancers (Basel). 2020 12
22. Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB. et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. Jama Oncol. 2019;5:1195-204
23. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568-71
24. Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S. et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994-1004
25. Paulsen EE, Kilvaer T, Khanehkenari MR, Maurseth RJ, Al-Saad S, Hald SM. et al. CD45RO(+) Memory T Lymphocytes-a Candidate Marker for TNM-Immunoscore in Squamous Non-Small Cell Lung Cancer. Neoplasia. 2015;17:839-48
26. Park HS, Kwon WS, Park S, Jo E, Lim SJ, Lee CK. et al. Comprehensive immune profiling and immune-monitoring using body fluid of patients with metastatic gastric cancer. J Immunother Cancer. 2019;7:268
27. Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y. et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci U S A. 2019;116:9999-10008
28. Chen YP, Zhang Y, Lv JW, Li YQ, Wang YQ, He QM. et al. Genomic Analysis of Tumor Microenvironment Immune Types across 14 Solid Cancer Types: Immunotherapeutic Implications. Theranostics. 2017;7:3585-94
29. Althammer S, Tan TH, Spitzmüller A, Rognoni L, Wiestler T, Herz T. et al. Automated image analysis of NSCLC biopsies to predict response to anti-PD-L1 therapy. J Immunother Cancer. 2019;7:121
30. Bence C, Hofman V, Chamorey E, Long-Mira E, Lassalle S, Albertini AF. et al. Association of combined PD-L1 expression and tumour-infiltrating lymphocyte features with survival and treatment outcomes in patients with metastatic melanoma. J Eur Acad Dermatol Venereol. 2020;34:984-94
31. Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L. et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med. 2017;376:1015-26
32. Kim TK, Herbst RS, Chen L. Defining and Understanding Adaptive Resistance in Cancer Immunotherapy. Trends Immunol. 2018;39:624-31
33. Hui R, Garon EB, Goldman JW, Leighl NB, Hellmann MD, Patnaik A. et al. Pembrolizumab as first-line therapy for patients with PD-L1-positive advanced non-small cell lung cancer: a phase 1 trial. Ann Oncol. 2017;28:874-81
34. Daud AI, Wolchok JD, Robert C, Hwu WJ, Weber JS, Ribas A. et al. Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J Clin Oncol. 2016;34:4102-9
35. Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV, Gilbert J. et al. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results From a Single-Arm, Phase II Study. J Clin Oncol. 2017;35:1542-9
36. Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L. et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320-30
37. Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J. et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312-22
38. Hellmann MD, Paz-Ares L, Bernabe CR, Zurawski B, Kim SW, Carcereny CE. et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2019;381:2020-31
39. Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J. et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67-76
40. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214-8
41. Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C. et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018;378:2093-104
42. Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM. et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271-81
43. Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med. 2017;377:2500-1
44. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202-6
45. Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D. et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol. 2018;36:633-41
46. Rizvi NA, Cho BC, Reinmuth N, Lee KH, Luft A, Ahn MJ. et al. Durvalumab With or Without Tremelimumab vs Standard Chemotherapy in First-line Treatment of Metastatic Non-Small Cell Lung Cancer: The MYSTIC Phase 3 Randomized Clinical Trial. Jama Oncol. 2020;6:661-74
47. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y. et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24:1441-8
48. Sholl LM, Hirsch FR, Hwang D, Botling J, Lopez-Rios F, Bubendorf L. et al. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2020;15:1409-24
49. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD. et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-20
50. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-13
51. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP. et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol. 2020;38:1-10
52. Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol. 2019;12:54
53. Cicek MS, Lindor NM, Gallinger S, Bapat B, Hopper JL, Jenkins MA. et al. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumors results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13:271-81
54. Jia M, Yao L, Yang Q, Chi T. Association of MSH2 Expression With Tumor Mutational Burden and the Immune Microenvironment in Lung Adenocarcinoma. Front Oncol. 2020;10:168
55. Kim ST, Klempner SJ, Park SH, Park JO, Park YS, Lim HY. et al. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget. 2017;8:77415-23
56. Sloan EA, Ring KL, Willis BC, Modesitt SC, Mills AM. PD-L1 Expression in Mismatch Repair-deficient Endometrial Carcinomas, Including Lynch Syndrome-associated and MLH1 Promoter Hypermethylated Tumors. Am J Surg Pathol. 2017;41:326-33
57. Rosenbaum MW, Bledsoe JR, Morales-Oyarvide V, Huynh TG, Mino-Kenudson M. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod Pathol. 2016;29:1104-12
58. Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S. et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. Jama Oncol. 2015;1:1319-23
59. Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP. et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157-70
60. Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT. et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202-16
61. Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T. et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462-9
62. Barroso-Sousa R, Keenan TE, Pernas S, Exman P, Jain E, Garrido-Castro AC. et al. Tumor Mutational Burden and PTEN Alterations as Molecular Correlates of Response to PD-1/L1 Blockade in Metastatic Triple-Negative Breast Cancer. Clin Cancer Res. 2020;26:2565-72
63. George S, Miao D, Demetri GD, Adeegbe D, Rodig SJ, Shukla S. et al. Loss of PTEN Is Associated with Resistance to Anti-PD-1 Checkpoint Blockade Therapy in Metastatic Uterine Leiomyosarcoma. Immunity. 2017;46:197-204
64. Agbor AA, Göksenin AY, LeCompte KG, Hans SH, Pursell ZF. Human Pol ε-dependent replication errors and the influence of mismatch repair on their correction. DNA Repair (Amst). 2013;12:954-63
65. Song Z, Cheng G, Xu C, Wang W, Shao Y, Zhang Y. Clinicopathological characteristics of POLE mutation in patients with non-small-cell lung cancer. Lung Cancer. 2018;118:57-61
66. Wang F, Zhao Q, Wang YN, Jin Y, He MM, Liu ZX. et al. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. Jama Oncol. 2019;5:1504-6
67. Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K. et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126:2334-40
68. Legras A, Barritault M, Tallet A, Fabre E, Guyard A, Rance B. et al. Validity of Targeted Next-Generation Sequencing in Routine Care for Identifying Clinically Relevant Molecular Profiles in Non-Small-Cell Lung Cancer: Results of a 2-Year Experience on 1343 Samples. J Mol Diagn. 2018;20:550-64
69. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF. et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822-35
70. Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860-77
71. Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS. et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res. 2016;76:999-1008
72. Li X, Lian Z, Wang S, Xing L, Yu J. Interactions between EGFR and PD-1/PD-L1 pathway: Implications for treatment of NSCLC. Cancer Lett. 2018;418:1-9
73. Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T. et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients With Advanced NSCLC. J Thorac Oncol. 2018;13:1138-45
74. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169-81
75. Oxnard GR, Yang JC, Yu H, Kim SW, Saka H, Horn L. et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol. 2020;31:507-16
76. Reck M, Mok T, Nishio M, Jotte RM, Cappuzzo F, Orlandi F. et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir Med. 2019;7:387-401
77. Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172-87
78. Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9-15
79. Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C. et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11-24
80. Xu YP, Lv L, Liu Y, Smith MD, Li WC, Tan XM. et al. Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J Clin Invest. 2019;129:4316-31
81. Chen LL, Smith MD, Lv L, Nakagawa T, Li Z, Sun SC. et al. USP15 suppresses tumor immunity via deubiquitylation and inactivation of TET2. Sci Adv. 2020 6
82. Wu HX, Chen YX, Wang ZX, Zhao Q, He MM, Wang YN. et al. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J Immunother Cancer. 2019;7:264
83. Diem S, Schmid S, Krapf M, Flatz L, Born D, Jochum W. et al. Neutrophil-to-Lymphocyte ratio (NLR) and Platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lung Cancer. 2017;111:176-81
84. Ren F, Zhao T, Liu B, Pan L. Neutrophil-lymphocyte ratio (NLR) predicted prognosis for advanced non-small-cell lung cancer (NSCLC) patients who received immune checkpoint blockade (ICB). Onco Targets Ther. 2019;12:4235-44
85. Ho WJ, Yarchoan M, Hopkins A, Mehra R, Grossman S, Kang H. Association between pretreatment lymphocyte count and response to PD1 inhibitors in head and neck squamous cell carcinomas. J Immunother Cancer. 2018;6:84
86. Ferrucci PF, Ascierto PA, Pigozzo J, Del VM, Maio M, Antonini CG. et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann Oncol. 2016;27:732-8
87. Banna GL, Signorelli D, Metro G, Galetta D, De Toma A, Cantale O. et al. Neutrophil-to-lymphocyte ratio in combination with PD-L1 or lactate dehydrogenase as biomarkers for high PD-L1 non-small cell lung cancer treated with first-line pembrolizumab. Transl Lung Cancer Res. 2020;9:1533-42
88. Wang X, Cao L, Li S, Wang F, Huang D, Jiang R. Combination of PD-L1 expression and NLR as prognostic marker in patients with surgically resected non-small cell lung cancer. J Cancer. 2019;10:6703-10
89. Russo A, Russano M, Franchina T, Migliorino MR, Aprile G, Mansueto G. et al. Neutrophil-to-Lymphocyte Ratio (NLR), Platelet-to-Lymphocyte Ratio (PLR), and Outcomes with Nivolumab in Pretreated Non-Small Cell Lung Cancer (NSCLC): A Large Retrospective Multicenter Study. Adv Ther. 2020;37:1145-55
90. Kelderman S, Heemskerk B, van Tinteren H, van den Brom RR, Hospers GA, van den Eertwegh AJ. et al. Lactate dehydrogenase as a selection criterion for ipilimumab treatment in metastatic melanoma. Cancer Immunol Immunother. 2014;63:449-58
91. Mezquita L, Auclin E, Ferrara R, Charrier M, Remon J, Planchard D. et al. Association of the Lung Immune Prognostic Index With Immune Checkpoint Inhibitor Outcomes in Patients With Advanced Non-Small Cell Lung Cancer. Jama Oncol. 2018;4:351-7
92. Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433-8
93. Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A. et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A. 2017;114:4993-8
94. Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM. et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144-53
95. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J. et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell. 2019;35:238-55
96. Lee JH, Long GV, Menzies AM, Lo S, Guminski A, Whitbourne K. et al. Association Between Circulating Tumor DNA and Pseudoprogression in Patients With Metastatic Melanoma Treated With Anti-Programmed Cell Death 1 Antibodies. Jama Oncol. 2018;4:717-21
97. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449-58
98. Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A. et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017;28:1996-2001
99. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J. et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. Jama Oncol. 2019;5:696-702
100. Kruger S, Legenstein ML, Rösgen V, Haas M, Modest DP, Westphalen CB. et al. Serum levels of soluble programmed death protein 1 (sPD-1) and soluble programmed death ligand 1 (sPD-L1) in advanced pancreatic cancer. Oncoimmunology. 2017;6:e1310358
101. Zhou J, Mahoney KM, Giobbie-Hurder A, Zhao F, Lee S, Liao X. et al. Soluble PD-L1 as a Biomarker in Malignant Melanoma Treated with Checkpoint Blockade. Cancer Immunol Res. 2017;5:480-92
102. Costantini A, Julie C, Dumenil C, Hélias-Rodzewicz Z, Tisserand J, Dumoulin J. et al. Predictive role of plasmatic biomarkers in advanced non-small cell lung cancer treated by nivolumab. Oncoimmunology. 2018;7:e1452581
103. Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients. Clin Cancer Res. 2018;24:896-905
104. Fan Y, Che X, Qu J, Hou K, Wen T, Li Z. et al. Exosomal PD-L1 Retains Immunosuppressive Activity and is Associated with Gastric Cancer Prognosis. Ann Surg Oncol. 2019;26:3745-55
105. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382-6
106. Reuben A, Zhang J, Chiou SH, Gittelman RM, Li J, Lee WC. et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun. 2020;11:603
107. Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij M. et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25:89-94
108. Postow MA, Manuel M, Wong P, Yuan J, Dong Z, Liu C. et al. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J Immunother Cancer. 2015;3:23
109. Hogan SA, Courtier A, Cheng PF, Jaberg-Bentele NF, Goldinger SM, Manuel M. et al. Peripheral Blood TCR Repertoire Profiling May Facilitate Patient Stratification for Immunotherapy against Melanoma. Cancer Immunol Res. 2019;7:77-85
110. Arakawa A, Vollmer S, Tietze J, Galinski A, Heppt MV, Bürdek M. et al. Clonality of CD4(+) Blood T Cells Predicts Longer Survival With CTLA4 or PD-1 Checkpoint Inhibition in Advanced Melanoma. Front Immunol. 2019;10:1336
111. Mitsuhashi A, Okuma Y. Perspective on immune oncology with liquid biopsy, peripheral blood mononuclear cells, and microbiome with non-invasive biomarkers in cancer patients. Clin Transl Oncol. 2018;20:966-74
112. Feun LG, Li YY, Wu C, Wangpaichitr M, Jones PD, Richman SP. et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer-Am Cancer Soc. 2019;125:3603-14
113. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544-8
114. Tauriello D, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M. et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538-43
115. Ni L, Lu J. Interferon gamma in cancer immunotherapy. Cancer Med. 2018;7:4509-16
116. Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18:545-58
117. Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X. et al. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931-43
118. Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J. et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837-46
119. Higgs BW, Morehouse CA, Streicher K, Brohawn PZ, Pilataxi F, Gupta A. et al. Interferon Gamma Messenger RNA Signature in Tumor Biopsies Predicts Outcomes in Patients with Non-Small Cell Lung Carcinoma or Urothelial Cancer Treated with Durvalumab. Clin Cancer Res. 2018;24:3857-66
120. Silva EM, Mariano VS, Pastrez P, Pinto MC, Castro AG, Syrjanen KJ. et al. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. Plos One. 2017;12:e181125
121. Rice SJ, Liu X, Zhang J, Jia B, Zheng H, Belani CP. Advanced NSCLC patients with high IL-6 levels have altered peripheral T cell population and signaling. Lung Cancer. 2019;131:58-61
122. Keegan A, Ricciuti B, Garden P, Cohen L, Nishihara R, Adeni A. et al. Plasma IL-6 changes correlate to PD-1 inhibitor responses in NSCLC. J Immunother Cancer. 2020 8
123. Li C, Zuo W. IL-10 in vitro could enhance IFNγ expression and suppress PD-1 expression in CD8 T cells from esophageal cancer patients. Exp Cell Res. 2019;379:159-65
124. Giunta EF, Barra G, De Falco V, Argenziano G, Napolitano S, Vitale P. et al. Baseline IFN-γ and IL-10 expression in PBMCs could predict response to PD-1 checkpoint inhibitors in advanced melanoma patients. Sci Rep. 2020;10:17626
125. Boutsikou E, Domvri K, Hardavella G, Tsiouda D, Zarogoulidis K, Kontakiotis T. Tumour necrosis factor, interferon-gamma and interleukins as predictive markers of antiprogrammed cell-death protein-1 treatment in advanced non-small cell lung cancer: a pragmatic approach in clinical practice. Ther Adv Med Oncol. 2018;10:433584018
126. Sanmamed MF, Perez-Gracia JL, Schalper KA, Fusco JP, Gonzalez A, Rodriguez-Ruiz ME. et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann Oncol. 2017;28:1988-95
127. Hegde PS, Wallin JJ, Mancao C. Predictive markers of anti-VEGF and emerging role of angiogenesis inhibitors as immunotherapeutics. Semin Cancer Biol. 2018;52:117-24
128. Yang J, Yan J, Liu B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front Immunol. 2018 9
129. Wallin JJ, Bendell JC, Funke R, Sznol M, Korski K, Jones S. et al. Atezolizumab in combination with bevacizumab enhances antigen-specific T-cell migration in metastatic renal cell carcinoma. Nat Commun. 2016;7:12624
130. Atkins MB, Plimack ER, Puzanov I, Fishman MN, McDermott DF, Cho DC. et al. Axitinib in combination with pembrolizumab in patients with advanced renal cell cancer: a non-randomised, open-label, dose-finding, and dose-expansion phase 1b trial. Lancet Oncol. 2018;19:405-15
131. Shibaki R, Murakami S, Shinno Y, Matsumoto Y, Yoshida T, Goto Y. et al. Predictive value of serum VEGF levels for elderly patients or for patients with poor performance status receiving anti-PD-1 antibody therapy for advanced non-small-cell lung cancer. Cancer Immunol Immunother. 2020;69:1229-36
132. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP. et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136
133. Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R. et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov. 2017;7:1420-35
134. Janikovits J, Müller M, Krzykalla J, Körner S, Echterdiek F, Lahrmann B. et al. High numbers of PDCD1 (PD-1)-positive T cells and B2M mutations in microsatellite-unstable colorectal cancer. Oncoimmunology. 2018;7:e1390640
135. Pereira C, Gimenez-Xavier P, Pros E, Pajares MJ, Moro M, Gomez A. et al. Genomic Profiling of Patient-Derived Xenografts for Lung Cancer Identifies B2M Inactivation Impairing Immunorecognition. Clin Cancer Res. 2017;23:3203-13
136. Chowell D, Morris L, Grigg CM, Weber JK, Samstein RM, Makarov V. et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582-7
137. Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C. et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. 2018 10
138. MacGregor HL, Ohashi PS. Molecular Pathways: Evaluating the Potential for B7-H4 as an Immunoregulatory Target. Clin Cancer Res. 2017;23:2934-41
139. Wang JY, Wang WP. B7-H4, a promising target for immunotherapy. Cell Immunol. 2020;347:104008
140. Genova C, Boccardo S, Mora M, Rijavec E, Biello F, Rossi G. et al. Correlation between B7-H4 and Survival of Non-Small-Cell Lung Cancer Patients Treated with Nivolumab. J Clin Med. 2019 8
141. Shrestha R, Prithviraj P, Anaka M, Bridle KR, Crawford D, Dhungel B. et al. Monitoring Immune Checkpoint Regulators as Predictive Biomarkers in Hepatocellular Carcinoma. Front Oncol. 2018;8:269
142. Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571:270-4
143. Guo L, Li X, Liu R, Chen Y, Ren C, Du S. TOX correlates with prognosis, immune infiltration, and T cells exhaustion in lung adenocarcinoma. Cancer Med. 2020;9:6694-709
144. Kim K, Park S, Park SY, Kim G, Park SM, Cho JW. et al. Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer. Genome Med. 2020;12:22
145. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-9
146. Routy B, Le Chatelier E, Derosa L, Duong C, Alou MT, Daillère R. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-7
147. Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V. et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity. 2016;45:931-43
148. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-8
149. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103
150. Saâda-Bouzid E, Defaucheux C, Karabajakian A, Coloma VP, Servois V, Paoletti X. et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2017;28:1605-11
151. Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel-Vinay S. et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin Cancer Res. 2017;23:1920-8
152. Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier-Valette C, Tessonnier L. et al. Hyperprogressive Disease in Patients With Advanced Non-Small Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or With Single-Agent Chemotherapy. Jama Oncol. 2018;4:1543-52
153. Kim CG, Kim KH, Pyo KH, Xin CF, Hong MH, Ahn BC. et al. Hyperprogressive disease during PD-1/PD-L1 blockade in patients with non-small-cell lung cancer. Ann Oncol. 2019;30:1104-13
154. Brower V. Hyperprogressive disease with anti-PD-1 and anti-PD-L1. Lancet Oncol. 2016;17:e527
155. Lo RG, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G. et al. Antibody-Fc/FcR Interaction on Macrophages as a Mechanism for Hyperprogressive Disease in Non-small Cell Lung Cancer Subsequent to PD-1/PD-L1 Blockade. Clin Cancer Res. 2019;25:989-99
156. Wang X, Wang F, Zhong M, Yarden Y, Fu L. The biomarkers of hyperprogressive disease in PD-1/PD-L1 blockage therapy. Mol Cancer. 2020;19:81
157. Chen Y, Li X, Liu G, Chen S, Xu M, Song L. et al. ctDNA Concentration, MIKI67 Mutations and Hyper-Progressive Disease Related Gene Mutations Are Prognostic Markers for Camrelizumab and Apatinib Combined Multiline Treatment in Advanced NSCLC. Front Oncol. 2020;10:1706
158. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368-79
159. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-84
160. Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res. 2017;23:4242-50
161. Sasaki A, Nakamura Y, Mishima S, Kawazoe A, Kuboki Y, Bando H. et al. Predictive factors for hyperprogressive disease during nivolumab as anti-PD1 treatment in patients with advanced gastric cancer. Gastric Cancer. 2019;22:793-802
162. Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y. et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci U S A. 2019;116:9999-10008
Author contact
![]() Corresponding author: Liang Wang, MD, Department of Hematology, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China; Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University & Capital Medical University, Beijing Tongren Hospital, Beijing 100730, China. Office Phone: +861058268442; E-mail: wangliangtrhoscom.
Corresponding author: Liang Wang, MD, Department of Hematology, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China; Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University & Capital Medical University, Beijing Tongren Hospital, Beijing 100730, China. Office Phone: +861058268442; E-mail: wangliangtrhoscom.

 Global reach, higher impact
Global reach, higher impact