Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(2):681-690. doi:10.7150/jca.62671 This issue Cite
Research Paper
Long Noncoding RNA PCAT6 Regulates Cell Proliferation and Migration in Human Esophageal Squamous Cell Carcinoma
1. Department of Thoracic Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu Province, China.
2. Department of Thoracic Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui Province, China.
3. Department of Thoracic Surgery, The First People's Hospital of Lianyungang, Lianyungang, Jiangsu Province, China.
4. State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, Jiangsu Province, China.
*Weibing Wu, Haixing Wei and Shuo Hu had equal contribution to the work and they should be identified as co-first authors.
Received 2021-5-12; Accepted 2021-12-11; Published 2022-1-1
Abstract
Esophageal squamous cell carcinoma (ESCC) is the sixth most common cancer type in East Asian countries. Mounting evidences illustrated that long noncoding RNAs (lncRNAs) play important roles in a variety of human cancers, including ESCC. LncRNA PCAT6 has been identified as a tumor promoter in multiple cancers. However, the roles and underlying mechanism of PCAT6 in ESCC remain largely unclear. In the current study, we discovered that lncRNA PCAT6, which was aberrantly upregulated in ESCC tumor tissues, significantly promoted cell proliferation and migration in ESCC cell lines Eca-109 and Kyse-30 cells. Flow cytometry assays showed that PCAT6 knockdown promoted the apoptosis of ESCC cells. Mechanistically, RNA-seq and Gene Ontology analyses indicated that PCAT6 knockdown influenced the expression of genes that participated in cell proliferation and migration. Furthermore, real-time PCR and western blot assays validated that knockdown of PCAT6 could increase the levels of GDF15 and DUSP4 in Eca-109 and Kyse-30 cells. In brief, our findings reveal that lncRNA PCAT6 plays an oncogenic role in the progression of ESCC by inhibiting the expression of genes related to cell proliferation and migration.
Keywords: LncRNA, PCAT6, cell proliferation, cell migration, ESCC.
Introduction
Esophageal cancer is a common malignant disease of the digestive tract and one of the six most common malignant tumors in the world. There are two types of esophageal carcinoma, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). EAC is highly prevalent in western countries, while ESCC is the most primary subtype in China. In 2018, it was estimated that there were 307 thousand newly diagnosed ESCC cases and 283 thousand ESCC related deaths in China [1, 2]. Although the diagnosis and treatment techniques of ESCC are developing rapidly, the prognosis is still poor. The 5 year overall survival of ESCC patients is less than 30% in most countries [3]. Therefore, it is very important to further explore the pathogenesis of esophageal cancer to identify more crucial biomarkers and therapeutic targets.
In recent years, owing to the development of Encyclopedia of DNA Elements project, long non-coding RNA (lncRNA) has attracted more and more attention [4, 5]. LncRNA is one of the RNA transcripts with approximately 200 to 100000 nucleotides in length [6, 7]. With in-depth research, the biological functions of lncRNAs have been initially revealed. LncRNAs participate in a series of cellular progressions, such as cellular development, apoptosis, transcriptional regulation, intracellular material transport and chromosome remodeling, etc. [8-11]. Moreover, the abnormal expression of lncRNAs has been established to play significant roles in regulating tumorigenesis including ESCC [12-19].
Prostate cancer associated transcript-6 (PCAT6), which has been reported in previous studies, was found upregulated in tumor tissues and could promote the tumor progression in multiple cancer types [20-23]. For example, Wan et al observed that PCAT6 was significantly elevated in lung tumor tissues and patients with a high expression of PCAT6 showed poorer prognosis. Functionally, they discovered that knockdown of PCAT6 significantly repressed the proliferation and invasion of lung cancer cells [20]. Similarly, the expression of PCAT6 was aberrantly elevated in colorectal cancer (CRC) tissues and knockdown of PCAT6 attenuated CRC chemoresistance to 5‐FU [21]. Nevertheless, the specific function of PCAT6 in the development of ESCC is still unclear.
In this study, we found that PCAT6 was significantly upregulated in ESCC tissues, and we confirmed that PCAT6 could promote cell proliferation and migration by the loss and gain of function assays. Flow cytometry assays showed that PCAT6 knockdown promoted the apoptosis of ESCC cells. In addition, RNA-seq and Gene Ontology (GO) analysis suggested that PCAT6 associated genes were mainly participated in pathways such as cell proliferation and cell migration. Prior to this, the downstream global gene expression profile of PCAT6 had not been studied, and we determined global gene expression profiling regulated by PCAT6 for the first time. The real-time PCR and western blot assays further proved that knockdown of PCAT6 could upregulate the expression of GDF15 and DUSP4, both of which have been reported associated with cell proliferation. In conclusion, our study reveals that lncRNA PCAT6 functioned as a tumor promoter in the progression of ESCC by regulating the expression of genes related to cell proliferation and migration.
Materials and Methods
Human samples and cell lines
Tissues used in this study were collected from 44 patients with ESCC underwent esophagectomy at the First Affiliated Hospital of Nanjing Medical University. All collected tissue specimens were immersed in RNA Later stabilization solution (Qiagen) and were immediately frozen in liquid nitrogen and conserved at - 80 °C until RNA extraction. This study was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, and written informed consent was acquired from all the patients.
ESCC cell lines (Eca-109, Kyse-30, TE-1, Kyse-70, Kyse-150) and human esophageal mucosal epithelial cell line (Het-1A) were purchased from Shanghai Cell Bank (Shanghai, China). These cells were cultivated in RPMI-1640 or DMEM medium (GIBCO‐BRL) with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were incubated at 37 °C 5% CO2.
Transfection of cell lines
We transfected Eca-109 and Kyse-30 cell lines with the siRNAs targeting PCAT6 or si-NC by utilizing Lipofectamine-2000 (Invitrogen) based on the protocol. The plasmid was transfected into Eca-109 and Kyse-30 cells with the X-tremeGENE™ HP DNA Transfection Reagent (Roche) following the manufacturer's protocol. After transfection for 48h, we harvested the cells for qRT-PCR to quantify the expression of related genes. All sequences of siRNAs used in this study were shown in Supplementary Table S1.
RNA extraction, reverse transcription and qRT-PCR
Total RNA was isolated from ESCC cells using TRIzol reagent (Invitrogen, Carlsbad, CA) following the instructions. 1μg of RNA from each specimen was reverse transcribed into complementary DNA (cDNA) for qRT‐PCR by utilizing the Reverse Transcription Kit (Takara, Dalian, China). We used SYBR PCR Master Mix reagent kit (Takara, Dalian China) for qRT‐PCR assays based on the publisher's protocol. The result was standardized with the endogenous expression of GAPDH. All the relative primer sequences were presented in Supplementary Table S1.
Cell proliferation assay
Cell proliferation assay was examined by using Cell Proliferation Reagent Kit I (MTT) (Sigma) following the standard procedures. After transfection for 24h, Eca-109 or Kyse-30 cell lines were placed in 96-well plates. We tested the cell viability every 24h based on the protocol. After cells transfection with si-PCAT6 or si-NC for 24h, a certain amount of cell suspension was grown in six-well plates for colony formation experiments. They were cultured in the cell incubator with 5% CO2 at 37 °C for ten days, replacing the fresh RPMI-1640 medium every 5 days. Then cells were treated with methanol for 30 mins and dyed by using crystal violet for 15 mins. We counted the number of clones and compared the results. All experiments were performed three times independently.
Ethynyldeoxyuridine (EdU) analysis
According to the instruction, we utilized an EdU labeling/detection kit (Ribobio, China) to valuate proliferating cells. Eca-109 or Kyse-30 cells were placed in 24-well plates at 3*10^4 cells per well. After transfection for 48h, the cell culture was replaced with 50 μM EdU labeling medium and they were still incubated for 4h in a cell incubator Then, the cells were fixed with 4% paraformaldehyde (pH 7.4) for 30 mins and then treated with 0.5% Triton X-100 for 15 mins at room temperature. Next, we stained the cells by using anti-EdU working solution for 15 mins. Finally, the cells were incubated with 300 μl Hoechst 33342 (5 μg/ml). The ratio of EdU-positive cells was calculated and compared. The assay was repeated three times independently.
Cell migration assays
After transfection for 24 h, 6×104 Eca-109 or 3×104 Kyse-30 cells in medium containing 1% FBS were seeded in the upper chambers of inserts (Millipore, Billerica, USA). The medium including 10% FBS was added to the lower chamber. The cells were treated with methanol and 0.1% crystal violet 48 hours after incubation. Then we wiped away the cells that remained on the upper chamber with the cotton swab. The cell which had migrated through the membrane was evaluated through the inverted microscope. The assay was replaced in triplicate independently.
Flow cytometric analysis of apoptosis
Following the instructions, ESCC cells were harvested 48 hours after transfection and performed Annexin V-FITC and Propidium iodide for 10 mins with the Annexin V-FITC Apoptosis Detection Kit (Vazyme BioTech, Nanjing, China). Next, the cells were conducted by flow cytometry (FACScan®; BD Biosciences) for analysis of apoptosis.
RNA-seq bioinformatic analysis
After Eca-109 cell lines treated with si-PCAT6-1# or si-NC for 48 hours, total RNA was extracted and quantified separately. The concentration of each specimen was measured with using NanoDrop 2000. The Agilent2200 (Agilent, USA) was conducted for assessing the quality. The sequencing library of every RNA specimen was established with using Ion Proton Total RNA-Seq Kit v2 based on the protocol provided by manufacturer (Life technologies, USA).
Gene ontology analysis
In order to explore the biological implications of representative profiles of the target gene of the differentially expressed mRNA in the assay, GO analysis was further performed by using GO annotations from three public-available databases, including NCBI, UniProt and the Gene Ontology.
Western blot
ESCC cells transfected with the specific siRNAs were lysed by utilizing RIPA buffer supplemented with Proteinase inhibitor. Total protein was extracted and separated with 10% SDS-PAGE gel, then transferred to a 0.45 μm polyvinylidene fluoride (PVDF) membrane. Then the PVDF membrane was incubated with specific antibodies. The protein was visualized through the autoradiography. The bodies (anti-GDF15 and anti-DUSP4) were purchased from Abcam.
Statistical analysis
The expression of PCAT6 in ESCC tumor tissues and adjacent normal tissues was obtained from TCGA database. Association between PCAT6 expression and the prognosis of ESCC patients was evaluated using Log-rank test. Student's t test was adopted to examine the differences between NC cells and PCAT6 knockdown cells. All statistical analyses were performed based on R software v3.6.0 or GraphPad Prism v.7.00. In this study, P values less than 0.05 were regarded as statistically significant.
Results
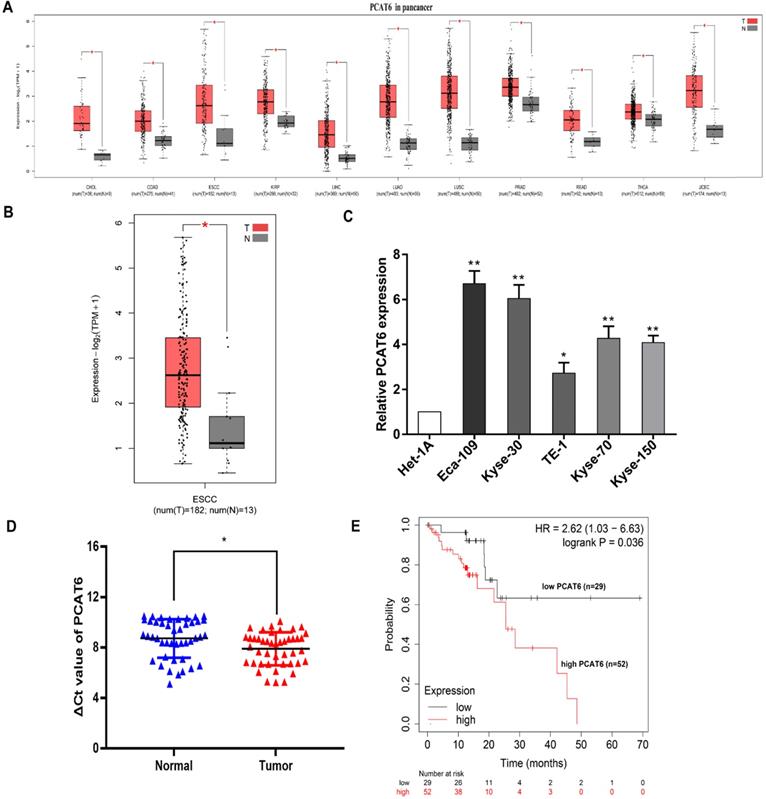
PCAT6 is upregulated in ESCC tumor tissues
To identify the clinical significance of PCAT6, we initially analyzed The Cancer Genome Atlas (TCGA) database (Figure 1A). Higher expression levels of PCAT6 were identified in a variety of cancers through pan-cancer analysis. And differential expression of PCAT6 was observed in the database which containing 182 ESCC tumor tissues and 13 normal tissues were included. From the results, we observed that PCAT6 was significantly elevated in ESCC tumor tissues (Figure 1B). After that, we examined the expression of PCAT6 in five ESCC cells and control cells. PCAT6 was noticeably upregulated in ESCC cells than in control cells (Figure 1C). Eca-109 and Kyse-30 with higher levels of PCAT6 were chosen for the following assays. To confirm the results, we collected 44 pairs of ESCC tissues and adjacent normal tissues from our center. qRT-PCR analysis demonstrated that PCAT6 was significantly upregulated in ESCC patients (Figure 1D). Further, we explored the relationship between PCAT6 expression levels and prognosis of patients with ESCC by Kaplan-Meier Plotter analysis [24]. As shown in Figure 1E, the results demonstrated that higher expression of PCAT6 were correlated with significantly shorter overall survival. From the above, PCAT6 may play a vital role in the tumorigenesis of ESCC.
Identification of the expression of PCAT6 in ESCC. (A) Data from TCGA database showed the relative expression of lncRNA PCAT6 in several cancers and normal tissues. (B) The relative expression of PCAT6 in in ESCC (n = 182) and nontumorous tissues (n = 13) from TCGA database. (C) PCAT6 expression evaluated using qRT-PCR in ESCC cell lines (Eca-109, Kyse-30, TE-1, Kyse-70, Kyse-150) and normal esophageal mucosal epithelial cell line (Het-1A). (D) qRT-PCR analysis of the expression of PCAT6 expression in 44 pairs of ESCC tissues and adjacent normal tissues collected from our center. The ΔCt value was determined by subtracting the GAPDH Ct value from the PCAT6 Ct value. A smaller ΔCt value indicates higher expression. (E) Kaplan-Meier survival plots revealed that higher PCAT6 expression correlated with poor overall survival in ESCC patients. (*P < 0.05 and **P < 0.01)
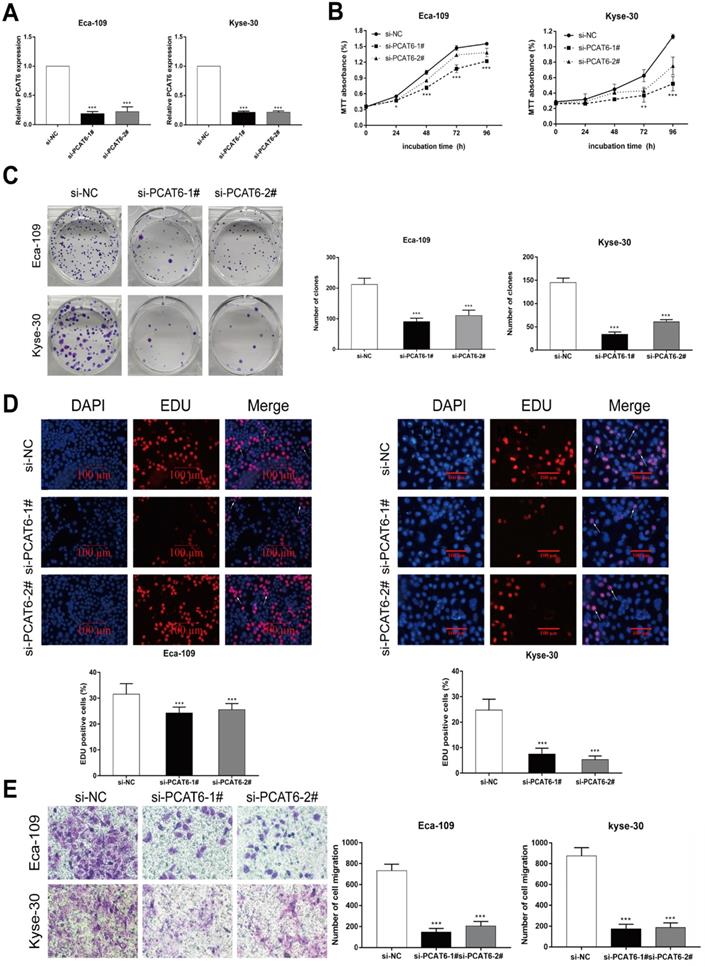
Knockdown of PCAT6 inhibits ESCC cells proliferation and migration
To investigate the biological role of PCAT6 in ESCC tumorigenesis, we first knocked down the expression level of PCAT6 using siRNAs targeting PCAT6. As shown in Figure 2A, the expression of PCAT6 in Eca-109 and Kyse-30 cells was significantly decreased after transfection with PCAT6 siRNAs. MTT assays indicated that cell viability was significantly inhibited when PCAT6 levels were decreased in Eca-109 and Kyse-30 cell lines (Figure 2B, P < 0.001). Similarly, the colony formation experiment showed that the growth ability of ESCC cells with PCAT6 knockdown was inhibited in both Eca-109 and Kyse-30 cells (Figure 2C, P < 0.001). Moreover, EdU staining assays also revealed that PCAT6 had a significant influence on the proliferation of ESCC cells (Figure 2D, P < 0.001). In addition, down-regulation of PCAT6 significantly suppressed cell migration ability in both Eca-109 and Kyse-30 cells (Figure 2E, P < 0.001). Our findings indicated that PCAT6 could promote the proliferation and migration of ESCC cells.
The downregulation of PCAT6 regulates ESCC cells proliferation and migration. (A) The expression of PCAT6 was tested using qRT-PCR after transfection with siRNAs in Eca-109 and Kyse-30 cells. (B) MTT assays were performed to confirm the viability of cell treated with PCAT6 siRNAs. PCAT6 knockdown significantly inhibited the viability of Eca-109 and Kyse-30 cells. (C) Colony formation assays exhibited that PCAT6 knockdown significantly inhibited the proliferation of Eca-109 and Kyse-30 cells. (D) EDU staining assays were conducted to determine cells proliferation following knockdown of PCAT6. The arrow points to the positive cell. Scale bars = 100 μm. (E) The migration abilities of Eca-109 and Kyse-30 cells were examined by transwell assays after transfection, respectively. (Data are mean ± SD. **P < 0.01).
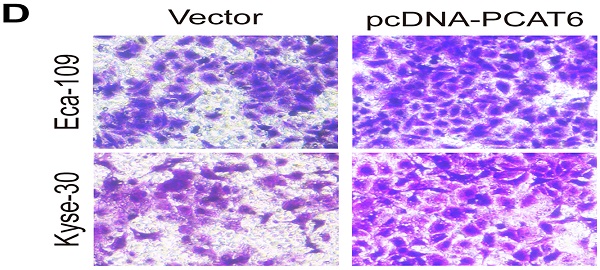
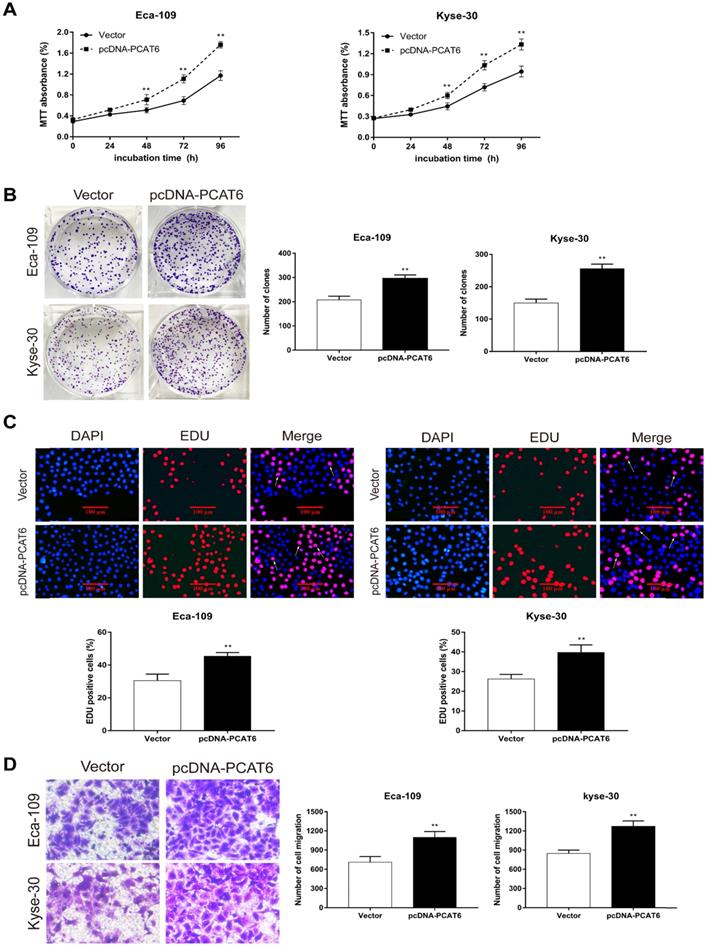
PCAT6 promotes cell proliferation and migration in ESCC
To conform the oncogenic role of PCAT6 investigate the effect of overexpression of PCAT6 on ESCC cells proliferation and migration, we conducted corresponding cell functional assays. As shown in Figure 3A, MTT analysis revealed that PCAT6 significantly increased cell activity in Eca-109 and Kyse-30 cells (P < 0.01). Then, colony formation assays were also performed, and the results indicated that the clonogenic ability of ESCC cells with PCAT6 upregulation was improved (Figure 3B, P < 0.01). Furthermore, Edu staining experiments also showed that overexpression of PCAT6 promoted cell proliferation (Figure 3C, P < 0.01). Additionally, transwell experiments demonstrated that overexpressed PCAT6 significantly increased cell migration ability in ESCC cells (Figure 3D, P < 0.01). Together, these results demonstrate that PCAT6 exerts an oncogenic role in ESCC through affecting cell proliferation and migration.
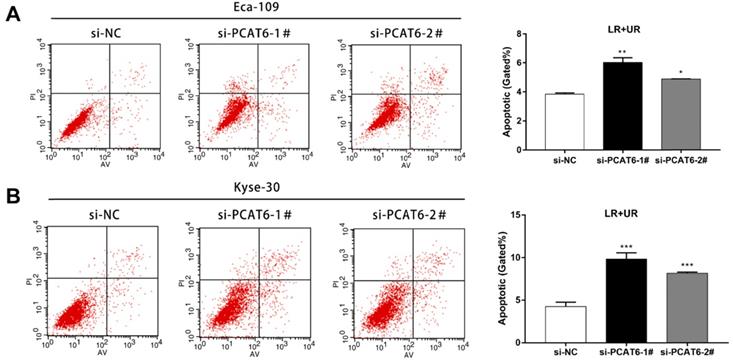
PCAT6 knockdown promotes ESCC cells apoptosis
Furthermore, flow cytometry was applied to confirm whether knockdown of PCAT6 affected ESCC cells apoptosis. As presented in Figure 4A, the rate of both early apoptotic and late apoptotic cells significantly increased when PCAT6 was down-regulated in Eca-109 cells compared to NC cells (P < 0.001). Consistently, PCAT6 inhibition in Kyse-30 cells resulted in a similar inducement of apoptotic cells (Figure 4B, P < 0.001). Accordingly, our results demonstrated that PCAT6 facilitates proliferation ability by regulating apoptosis in ESCC cells.
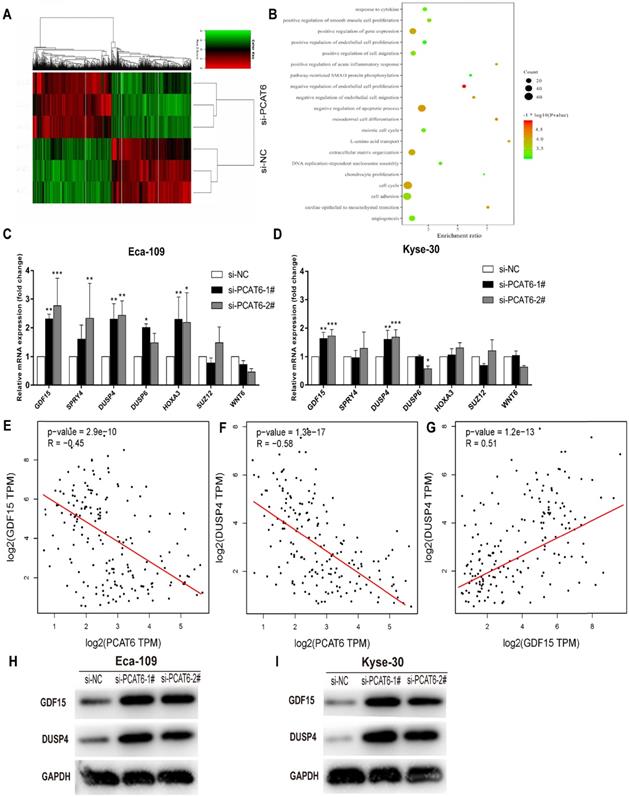
The global gene expression profile regulated by PCAT6 in ESCC Cells
Although PCAT6 has been reported to promote cell proliferation in a variety of cancers, the global gene expression profile regulated by PCAT6 has not been clarified. In order to probe the molecular mechanisms underlying the increased proliferation of PCAT6 in ESCC, RNA-Seq of PCAT6 knockdown Eca-109 cells and control cells was carried out. As a consequence, the transcript levels of 775 mRNAs displayed ≤ 2-fold down-regulation in abundance in Eca-109 cells following PCAT6 knockdown, while 503 protein-coding genes showed ≥ 2-fold up-regulation (Figure 5A; data are available in Supplementary Table S2). Furthermore, GO enrichment analysis demonstrated that these genes were mainly enriched in cell proliferation, cell migration pathways (Figure 5B).
As described above, PCAT6 might participate in pathways associated with cell growth, migration and apoptosis in ESCC. As expected, we found that many well-known genes that associated with cell proliferation and migration (e.g. WNT6, DUSP6, HOXA3, SUZ12, DUSP4, SPRY4, GDF15, et al.) were involved in these pathways. To determine the potential target genes of PCAT6, we used qRT-PCR to verify the expression changes of the 7 genes. GDF15, SPRY4, DUSP4 and HOXA3 were observed upregulated in Eca-109 cells while GDF15 and DUSP4 upregulated in Kyse-30 cells with the suppression of PCAT6 (Figures 5C and D). The consistent changes of GDF15 and DUSP4 levels in Eca-109 and Kyse-30 cells implies the potential regulatory roles of PCAT6. Previous studies have reported that GDF15 had anti-tumorigenic and pro-apoptotic activity, which was first identified in colorectal cancer [25]. In addition, overexpression of DUSP4 in glioblastoma cells could result in a significant decrease of cellular proliferation ability specifically via dephosphorylating and inactivating MAPKs [26]. Moreover, we analyzed the respective relationships between the expression levels of PCAT6 and GDF15, DUSP4 in TCGA ESCC samples. As shown in Figure 5E and F, the expression of PCAT6 was negatively associated with the expression of GDF15 and DUSP4 in ESCC. Further , through exploring the correlation between GDF15 and DUSP4 in ESCC samples from TCGA, we found that there is a positive relationship between GDF15 and DUSP4 (Figure 5G).Western blot assays were further carried out to verify the expression changes of GDF15 and DUSP4 following PCAT6 knockdown in Eca-109 and Kyse-30 cells. The protein levels were reduced by PCAT6 knockdown (Figure 5H and I). These evidences suggested that the dysregulated genes GDF15 and DUSP4 might be the underlying downstream mediators of PCAT6.
Discussion
Increasing evidences showed that lncRNAs played vital roles in cell growth and human disease development, especially in cancer [27, 28]. The biological functions and roles of lncRNA PCAT6 in multiple cancers have been revealed in previous studies. For example, high PCAT6 expression was significantly associated with TNM stage and indicated poor overall survival of patients with lung cancer [20]. Shi et al. found that PCAT6 exerted the oncogenic activity by suppressing LATS2 through binding to the epigenetic repressor EZH2 in non-small-cell lung cancer [29]. Moreover, PCAT6 promoted cell proliferation and suppressed cell apoptosis via sponging miR-143-3p to upregulate PRDX5 in gastrointestinal stromal tumor [30]. However, the possible role of PCAT6 in ESCC is still unclear.
Effect of overexpressed PCAT6 on cell proliferation and migration in ESCC. (A) MTT analysis of Eca-109 and Kyse-30 cells transfected with an overexpression plasmid of PCAT6. (B) Colony formation assays were conducted to determine cell proliferation of Eca-109 and Kyse-30 cells treated with the overexpression plasmid of PCAT6. (C) Edu assays of Eca-109 and Kyse-30 cells transfected with an overexpression plasmid of PCAT6. The arrow points to the positive cell. Scale bars = 100 μm. (D) Transwell assays were performed to detect the cell migration ability of Eca-109 and Kyse-30 cells following overexpressed PCAT6. (Data are mean ± SD. **P < 0.01).
In the current study, we found that PCAT6 was significantly upregulated in ESCC tumor tissues, suggesting that PCAT6 might function as a tumor promoter in the progression of ESCC. As expected, knockdown of PCAT6 in Eca-109 and Kyse-30 cells significantly inhibited the proliferation of ESCC cells. Consistently, the migration ability of ESCC cells was apparently suppressed in PCAT6 knockdown cells compared to control cells. Furthermore, the cell apoptosis assay showed that down-regulation of PCAT6 significantly promoted the apoptosis of ESCC cells in both Eca-109 and Kyse-30 cells. On the contrary, overexpression of PCAT6 significantly promoted ESCC cells proliferation through the MTT, clone formation and Edu analysis. And the ability of ESCC cells migration was improved following overexpressed PCAT6. Taken together, all these findings revealed that PCAT6 played an oncogenic role in ESCC tumorigenesis.
To further investigate the potential mechanisms of PCAT6 in ESCC, RNA transcriptome sequencing was conducted. We obtained the downstream global gene expression profile of PCAT6 in Eca-109 cells. GO enrichment analysis indicated that PCAT6 associated genes were enriched in pathways related to cell proliferation and migration, which suggested that PCAT6 could regulate the expression of genes involved in cell proliferation or migration. Further, the results of qRT-PCR and western blot proved that inhibition of PCAT6 significantly increased the expression of GDF15 and DUSP4, suggesting GDF15 and DUSP4 could be the target genes of PCAT6. Except for GDF15 and DUSP4, HOXA3 was also observed to be upregulated in Eca-109 cells, it indicates that PCAT6 might be involved in other regulatory pathways which needs further exploration.
GDF15, belonging to the TGF-β superfamily, has been reported to serve as a tumor suppressor in several cancers, including esophageal carcinoma [31-34]. Lu X et al observed that GDF15 was apparently downregulated in non-small-cell lung cancer tissues. Functional experiments indicated that overexpression of GDF15 significantly reduced cell proliferation and induced apoptosis [33]. In our study, GDF15 was upregulated along with the knockdown of PCAT6. It indicated that PCAT6 may promotes development of ESCC via suppressing GDF15. With regard to DUSP4, previous research reported that downregulation of DUSP4 enhanced cell proliferation in colorectal carcinomas (CRC). Recovery of DUSP4 led to inactivation of ERKs, inhibiting the proliferation and invasiveness of CRC cells [35]. Zhang et al had illuminated that down-regulation of DUSP4 expression in gastric cancer patients was associated with clinicopathological features, including tumor size and distant metastasis. Overexpression of DUSP4 effectively suppressed the proliferation of gastric cancer cells and induced apoptosis [36]. However, the roles and functions of DUSP4 in ESCC have not been revealed in previous studies. Our results confirmed that DUSP4 was upregulated following PCAT6 knockdown. PCAT6 may enhance ESCC cell proliferation by down-regulating DUSP4. Highlight these evidences, we speculated that PCAT6 could promote the progression of ESCC by regulating the expression of GDF15 and DUSP4. In addition, positive relationship between GDF15 and DUSP4 indicated their synergy effect to cell proliferation and migration.
To conclude, our findings revealed that lncRNA PCAT6 was aberrantly elevated in ESCC tissues, and elucidated that PCAT6 played an oncogenic role in ESCC cells. We also determined the global gene expression profile regulated by PCAT6 by RNA-seq assays, and GDF15 and DUSP4 were identified as potential targets of PCAT6. Though further explorations were needed, our results provide a better understanding of the role of PCAT6 in tumor progression and a potential therapeutic target and prognostic predictor against this ESCC.
Effect of PCAT6 on cell apoptosis in ESCC cells. Flow cytometry assays were performed to detected the cell apoptosis in (A) Eca-109 and (B) Kyse-30 cells. PCAT6 knockdown resulted in increased cell apoptotic rate. LR, early apoptotic cells. UR, terminal apoptotic cells. (Data are mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001).
The global gene expression profile regulated by PCAT6 and underlying target genes. (A) Mean-centered, hierarchical clustering of 1,278 gene transcripts altered (≥ 2-fold change) in the control and si-PCAT6-1#-treated Eca-109 cells, with three repeats. Red represents the high expression value, while green represents the low. (B) Gene ontology analysis for all genes with altered expressions. qRT-PCR was used to determine the changes of the altered genes expression which related to the cell proliferation after PCAT6 knockdown in (C) Eca-109 and (D) Kyse-30 cells. (E and F) The respective negative correlations were found between the expression levels of PCAT6 and GDF15, DUSP4 in TCGA ESCC samples. (G) Correlation between GDF15 and DUSP4 mRNA from TCGA database. Western blot assays were conducted to verify the change in the protein levels of related genes following knockdown of PCAT6 in (H) Eca-109 and (I) Kyse-30 cells. (*P < 0.05, **P < 0.01 and ***P < 0.001).
Abbreviations
lncRNAs: long non-coding RNAs; ESCC: esophageal squamous cell carcinoma; EAC: esophageal adenocarcinoma; PCAT6: Prostate cancer associated transcript-6; GO: Gene Ontology.
Supplementary Material
Supplementary table 1.
Supplementary table 2.
Acknowledgements
Thanks to TCGA database for using the data of ESCC tissues.
Ethics approval and consent to participate
The study was approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University. Written informed consent was acquired from all the patients.
Funding
This work was supported by the Program of Jiangsu Medical Innovation Team (No. CXTDA2017006) and Jiangsu Province 333 Talents Project (No. BRA2017545).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Schweigert M, Dubecz A, Stein HJ. Oesophageal cancer—an overview. Nature Reviews Gastroenterology & Hepatology. 2013;10:230-44
3. Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A. et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. The Lancet Oncology. 2011;12:681-92
4. Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20:994-1000
5. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756-70
6. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178-81
7. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208
8. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298-307
9. Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harbor perspectives in biology. 2014;6:a018614
10. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407
11. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46
12. Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC, An O. et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology. 2018;154:2137-51 e1
13. Xiao G, Yao J, Kong D, Ye C, Chen R, Li L. et al. The Long Noncoding RNA TTTY15, Which Is Located on the Y Chromosome, Promotes Prostate Cancer Progression by Sponging let-7. Eur Urol. 2018;76:315-26
14. Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J. et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612-27
15. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P. et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65:1494-504
16. Zhang XD, Huang GW, Xie YH, He JZ, Guo JC, Xu XE. et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46:1793-809
17. Lin C, Zhang S, Wang Y, Wang Y, Nice E, Guo C. et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res. 2018;24:486-98
18. Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X. et al. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25:1989-2000
19. Zhang E, He X, Zhang C, Su J, Lu X, Si X. et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154
20. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of Long Noncoding RNA PCAT6 Inhibits Proliferation and Invasion in Lung Cancer Cells. Oncol Res. 2016;24:161-70
21. Wu H, Zou Q, He H, Liang Y, Lei M, Zhou Q. et al. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8:2484-95
22. Huang W, Su G, Huang X, Zou A, Wu J, Yang Y. et al. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18:69-83
23. Chen S, Chen Y, Qian Q, Wang X, Chang Y, Ju S. et al. Gene amplification derived a cancer-testis long noncoding RNA PCAT6 regulates cell proliferation and migration in hepatocellular carcinoma. Cancer Med. 2019;8:3017-25
24. Gyorffy B, Molnar B, Lage H, Szallasi Z, Eklund AC. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS One. 2009;4:e5645
25. Wang X, Baek SJ, Eling TE. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol. 2013;85:597-606
26. Waha A, Felsberg J, Hartmann W, von dem Knesebeck A, Mikeska T, Joos S. et al. Epigenetic downregulation of mitogen-activated protein kinase phosphatase MKP-2 relieves its growth suppressive activity in glioma cells. Cancer Res. 2010;70:1689-99
27. Yin D, Lu X, Su J, He X, De W, Yang J. et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17:92
28. Xu Y, Ge Z, Zhang E, Zuo Q, Huang S, Yang N. et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8:e3104
29. Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X. et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177-87
30. Bai F, Zhang N, Fang W, He X, Zheng Y, Gu D. PCAT6 mediates cellular biological functions in gastrointestinal stromal tumor via upregulation of PRDX5 and activation of Wnt pathway. Mol Carcinog. 2020;59:661-9
31. Baek SJ, Okazaki R, Lee SH, Martinez J, Kim JS, Yamaguchi K. et al. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology. 2006;131:1553-60
32. Kim KS, Baek SJ, Flake GP, Loftin CD, Calvo BF, Eling TE. Expression and regulation of nonsteroidal anti-inflammatory drug-activated gene (NAG-1) in human and mouse tissue. Gastroenterology. 2002;122:1388-98
33. Lu X, He X, Su J, Wang J, Liu X, Xu K. et al. EZH2-Mediated Epigenetic Suppression of GDF15 Predicts a Poor Prognosis and Regulates Cell Proliferation in Non-Small-Cell Lung Cancer. Mol Ther Nucleic Acids. 2018;12:309-18
34. Leung AC, Wong VC, Yang LC, Chan PL, Daigo Y, Nakamura Y. et al. Frequent decreased expression of candidate tumor suppressor gene, DEC1, and its anchorage-independent growth properties and impact on global gene expression in esophageal carcinoma. Int J Cancer. 2008;122:587-94
35. Ichimanda M, Hijiya N, Tsukamoto Y, Uchida T, Nakada C, Akagi T. et al. Downregulation of dual-specificity phosphatase 4 enhances cell proliferation and invasiveness in colorectal carcinomas. Cancer Sci. 2018;109:250-8
36. Zhang R, Wang G, Zhang PF, Zhang J, Huang YX, Lu YM. et al. Sanguinarine inhibits growth and invasion of gastric cancer cells via regulation of the DUSP4/ERK pathway. J Cell Mol Med. 2017;21:1117-27
Author contact
![]() Corresponding author: Liang Chen, The First Affiliated Hospital of Nanjing Medical University, No. 300, Guangzhou Road, Nanjing 210029, Jiangsu Province, China. Tel: +86-13913872727. Fax: +86-25-83718836. E-mail: clbright0909edu.cn
Corresponding author: Liang Chen, The First Affiliated Hospital of Nanjing Medical University, No. 300, Guangzhou Road, Nanjing 210029, Jiangsu Province, China. Tel: +86-13913872727. Fax: +86-25-83718836. E-mail: clbright0909edu.cn

 Global reach, higher impact
Global reach, higher impact