Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(5):1630-1639. doi:10.7150/jca.59286 This issue Cite
Research Paper
Identification of a Metastasis-Related Protein IFI16 in Esophageal Cancer using a Proteomic Approach
1. Guizhou University School of Medicine, Guizhou, 550025, China.
2. State Key Lab of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China.
Received 2021-2-10; Accepted 2021-11-30; Published 2022-3-6
Abstract
Background: Metastasis is the leading cause of the high morality of esophageal squamous cell carcinoma (ESCC), so early monitoring metastasis of esophageal cancer is the key to improve the survival rate of ESCC patients. However, there have not been effective biomarkers for predicting metastasis of ESCC patients,it is an urgent need to identify ESCC metastasis-related proteins.
Methods: iTRAQ-based proteomic method was performed in highly metastatic 30M cell established in our previous study and the corresponding parental cells KYSE30.The expression of IFI16 was verified using western blotting and immunohistochemistry (IHC). Then, cck8, transwell assay,mouse metastasis experiments were performed to determine the functional role of IFI16 in esophageal cancer. Finally, RAN-Seq, qpcr, transwell assay were used to investigate the underlying mechanism of IFI16 in esophageal cancer metastasis.
Results: The data showed that IFI16 was upregulated in 30M cell compared with KYSE30 cell. IFI16 also increased in ESCC tumor compared with non-tumor tissue. Kaplan-Meier survival curve analysis showed that the relapse-free survival (RFS) of patients with high IFI16 level was worse than that of patients with low IFI16 level (P=0.0449). In addition, IFI16 knockdown did not affect the cell growth, but inhibited ESCC cell migration and invasion in ESCC cells. Moreover, IFI16 knockdown suppressed the lung metastasis of 30M cells in mouse models. Finally, we performed an RNA-Seq assay in IFI16-knocking down 30M cells and identified that knocking down IFI16 downregulated the expression of fibroblast growth factor proteins (FGF1, FGF2 etc.). Furthermore, overexpressing FGF1 and FGF2 rescued the lost of migration and invasion ability of 30M mediated by IFI16 knockdown.
Conclusion: Our results demonstrated that IFI16 was a key ESCC metastasis-related protein and played a role in ESCC metastasis through promoting the FGF proteins expression.
Keywords: esophageal squamous cell carcinoma (ESCC), metastasis, proteomics, IFI16, FGF1, FGF2
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the common types of the gastrointestinal tumors with high mortality rate in China [1]. The prognosis of patients with esophageal cancer is very poor, and their 5-year survival rate is less than 20% [2], one important reason is the high invasion and metastasis characteristics of esophageal cancer. However, the biological mechanism of esophageal cancer metastasis is still unclear. To develop better diagnosis and treatment approaches, it is important to identify proteins related to ESCC metastasis and understand the molecular basis of ESCC metastasis.
With the exploration of tumor invasion and metastasis, it is found that tumor invasion and metastasis is a complex biological process [3], which can be divided into: reduced adhesion between tumor cells, degradation of extracellular matrix [4], escape of tumor cells from basement membrane, migration into lymphatic vessels or microvessels, immune escape of tumor cells [5], formation of micrometastases and metastatic microenvironment [6]. Studies have shown that this complex biological behavior in cancer is an abnormal process regulated by multiple molecules [3, 7]. Therefore, finding key molecules for invasion and metastasis is a great significance for further finding of the targets of invasion and metastasis.
The current research on the invasion and metastasis of esophageal cancer mainly focuses on esophageal cancer cell lines with different metastatic capabilities, especially cell lines with the same genetic background but different metastatic capabilities, it's a ideal cell model for studying esophageal cancer metastasis and performing genes differential analysis [8]. Here, we directly used the esophageal cancer cell line 30-D (hereinafter referred to as 30M) with high metastasis ability obtained from the KYSE30 cell sieve in our laboratory through the screening model as the research object [9].
The proteome is the functional translation of the genome and the ultimate embodiment of life activities [10]. Therefore, quantitative proteomics is one of the hot research fields in the post-genomic era, and it has been widely used in oncology to identify biomarkers with diagnostic and therapeutic potential. In this project, we used iTRAQ (isobaric tags for relative and absolute quantification) combined with liquid chromatography and tandem mass spectrometry to analyze the protein expression of cell 30M with high metastasis capabilities and its parent cell KYSE30 and identify IFI16 as a key molecule involved in cancer metastasis. Our in vivo and in vitro experiments both demonstrated that IFI16 knockdown inhibited ESCC cell migration and invasion. In addition, ESCC patients with high IFI16 had shorter relapse-free survival (RFS) compared with low IFI16 patients. Finally, we revealed that IFI16 promoted metastasis mainly through increasing FGF proteins expression. Collectively, our results indicated that IFI16 is a key ESCC metastasis-related protein and provided us a possible opportunity for the therapy against ESCC metastasis.
Materials and Methods
Cell lines and cell culture
The human ESCC cell lines KYSE30, KYSE180, KYSE450 and other KYSE series were generously provided by Dr. Shimada Y. (Kyoto University, Kyoto, Japan) and cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS).The esophageal cancer high metastasis model 30M cells was derived from KYSE30 parental cells in our lab before. The HEK293T cell line was purchased from the American Type Culture Collection (ATCC), preserved in our laboratory, and cultured at 37 °C in 5% CO2 in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% FBS (HyClone, South Logan, UT, USA).
Protein extraction, digestion and labeling with iTRAQ reagents
Cells were sonicated three times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail), and then centrifuged at 12,000 rpm for 10 min at 4 °C. Finally, the supernatant was collected, and the protein concentration was determined with BCA kit according to the manufacturer's instructions. The protein preparation and LC-MS/MS detection using the iTRAQ technique were performed in Jingjie PTM Biolab Co. Ltd (Hangzhou, China).
Antibodies and reagents
The anti-IFI16 antibody (ab55328) was purchased from Abcam (Cambridge, UK); the anti-FGF1 antibody (ab179455) was purchased from Abcam (Cambridge, UK); the anti-β-actin antibody (A5316) was purchased from Sigma (St. Louis, MO, USA).
Plasmid construction and lentivirus infection
Short hairpin RNA (shRNA) sequences were cloned into the pSIH1-Puro vector (#26597, Addgene). The shRNA sequences for IFI16 were as follows: sh1, GGAAACTCTGAAGATTGATA, sh2, GATCATTGCCATAGCAAATTA. The full-length cDNA of human IFI16, FGF1 and FGF2 was cloned into the pLVX-IRES-Neo vector (632181, Clontech, CA, USA). Lentivirus were produced by HEK293-T cells and harvested 48 h after transfection, cells in which IFI16 was knocked down or overexpression were selected with 1 μg/ml puromycin or 200 μg/ml G418 for two weeks.
Transwell assays
In vitro, cell invasion and migration assays were evaluated in 24-well Boyden chambers (Corning Incorporated, Corning, NY, USA). The wells were coated with or without Matrigel (BD Biosciences) and then incubated at 37 °C for 1 hour. Then, medium containing 20% FBS (680 μL) was added to each lower chamber, and the cells were placed in the upper chamber.
Cell proliferation
Cells were seeded into a 96-well plate at a density of 1×103 cells per well. Cell Counting Kit-8 (CCK-8) reagents (Dojindo, Tokyo, Japan) were added at a dilution of 1:10 to each well and incubated for 1h. The absorbance values were then measured at a wavelength of 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent and reverse transcribed using a QuantScript RT Kit (Tiangen, Beijing, China) in accordance with a standard protocol. QRT-PCR was performed on an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using PowerUp SYBR Green Master Mix (Applied Biosystems). The following primers were used in the qRT-PCR assay: IFI16 (5′-TGAGCCCAAAGAGCAGAA-3′; 5′-GGTCAGCATTCACATCAGC-3′), IVL (5′-ACTGGCTCCACTTATTTCGGG-3′; 5′-GAGGTTGGGATTGGGGTCAT-3′), DHX58 (5′-GGTATCATCTTCACCCGCAC-3′, 5′-TCACTTCTTGCTGGTCCCTC-3′), ICAM1 (5′-AGCGGCTGACGTGTGCAGTAAT-3′, 5′-TCTGAGACCTCTGGCTTCGTCA-3′), BCAT1 (5′-TGTATCGCTCTGCTGTGAGG-3′, 5′-GGCTTTGGTAGGCTTCTTGAC-3′), ABCC2 (5′-CTCCTGTCTTCACCATCATCG-3′, 5′-AAAGGCACGGATAACTGGCAA-3′), FGF1 (5′-ACGGGCTTTTATACGGCTCA-3′, 5′-CCAACAAACCAATTCTTCTCT-3′), FGF2 (5′-GCTGTACTGCAAAAACGGGG-3′, 5′-AGCCAGGTAACGGTTAGCAC-3′), GAPDH (5′-CCGGGAAACTGTGGCGTGATGG-3′, 5′-AGGTGGAGGAGTGGGTGTCGCTGTT-3′).
Animal experiments
Animal experiments were performed according to protocols approved by the Chinese Academy of Medical Sciences Cancer Hospital Animal Care and Use Committee. Male severe combined immune deficiency beige (SCID/Beige) mice (6 weeks old) purchased from Vital River (Beijing, China) were bred under specific pathogen-free conditions. SCID/Beige mice were used for the lung metastasis experiments.
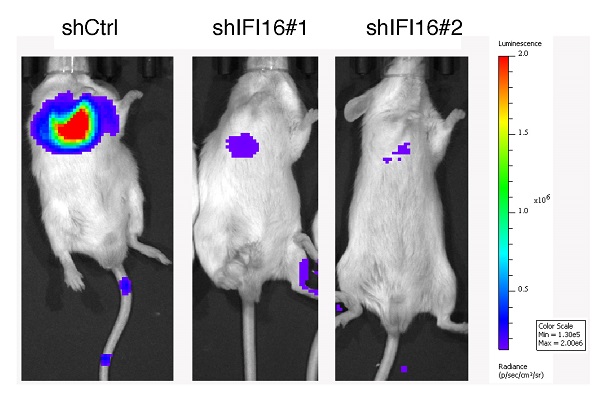
For the lung metastasis model, equal number of control and IFI16-knockdown 30M cells were injected into the tail vein of SCID/Beige mice. After 45 days, these mice were injected with D-luciferin (15 mg/ml), and observed with the IVIS Lumina XRMS in vivo Imaging System (PerkinElmer, Waltham, MA, USA). Then, the mice were sacrificed and the lungs were harvested for bouin staining.
Bioinformatics Methods in Proteomic Data
A total of 6,634 proteins (<1% FDR) were identified in the 30M/KYSE30 comparison group, of which 5,893 proteins contained quantitative information. The proteins which met the following criteria were confidently considered as differentially expressed proteins (DEPs): proteins showed an average ratio-fold change ≥1.5 or ≤1/1.5 and the same change trend in the three times experiments between the two cells (t-test; P < 0.05). In total, 383 DEPs were identified, including 295 up-regulated proteins and 88 down-regulated proteins (Table S1). The identified DEPs were then subjected to functional enrichment analysis, and the statistical significance of the functional enrichment was determined by Fisher's exact test.
Bioinformatics Methods in RNA-Seq Data
The RNA-Seq assay was performed in IFI16-knocking down 30M cells by two shRNA (#1 and #2) and control cells to determine regulated genes by IFI16. The genes which met the following criteria were confidently considered as differentially expressed genes (DEGs): RNAs showed an average ratio-fold change ≥2 or ≤1/2 and the same change trend in the three times experiments between the two cells (t-test; p-adjusted < 0.05). We found that 98 genes were upregulated and 94 genes were downregulated in both shRNA #1 and #2 (Table S2). These DEGs were then subjected to functional enrichment analysis, and the statistical significance of the functional enrichment was determined by Fisher's exact test.
Results
The proteomic characteristics of 30M with high metastasis capacity
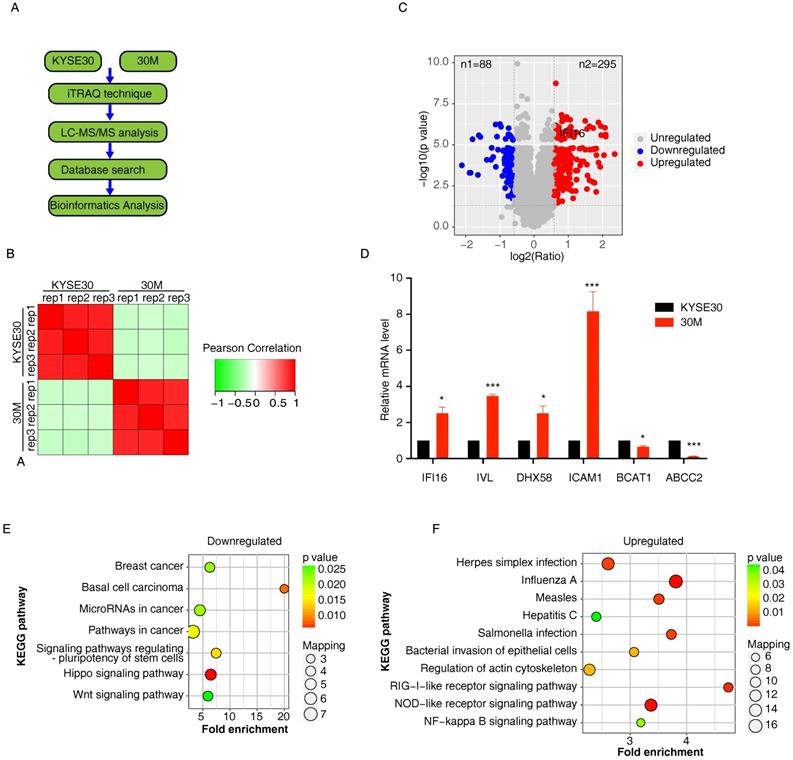
A summary of the workflow for iTRAQ-based quantification of differentially expressed proteins (DEPs) is shown in Figure 1A. We selected a high metastasis capacity cell 30M and its parent KYSE30 control cells to perform proteomic analysis by iTRAQ technique in three replicate samples per group with consistent quality control (Figure 1B). A total of 6,634 proteins (<1% FDR) were identified in the 30M/KYSE30 comparison group, of which 5,893 proteins contained quantitative information. In total, 383 DEPs with statistical significance (p < 0.05 and fold change (FC) of T/N >1.5 or <1/1.5) were identified, including 295 up-regulated proteins and 88 down-regulated proteins (Figure 1C and Table S1). Quantitative real-time PCR (qRT-PCR) was performed to confirm the expression of 6 random selected DEPs in 30M/KYSE30 cells (Figure 1D).
The down-regulated DEPs were mainly localized in the plasma membrane (25%) and extracellularm (23%), and the up-regulated DEPs in 30M/KYSE30 was found that they were mainly localized in the cytoplasm (41%) and nucleus (29%). The KEGG pathway enrichment analysis demonstrated that the top 3 pathways enriched by downregulated DEPs in 30M/KYSE30 were Hippo signaling pathway, Basal cell carcinoma and signaling pathways regulating pluripotency of stem cells (Figure 1E). And top 3 pathways enriched by upregulated DEPs in 30M/KYSE30 were Influenza A, NOD-like receptor signaling pathway and RIG-I-like receptor signaling pathway (Figure 1F).
IFI16 played a key role in ESCC metastasis
Our proteomic data showed that IFI16, a molecule of NOD-like receptor signaling pathway was up-regulated in 30M/KYSE30 (FC=1.85, p=4.15×10-6) (Figure 2A), and the qRT-PCR and western blot results also confirmed this result (Figure 1D and Figure 2B). To evaluate the significance of IFI16 in ESCC development, we also detected the IFI16 proteins levels in different ESCC cell lines and ESCC tumor tissues by western blot. The result showed that IFI16 also increased in most of ESCC cells lines and ESCC tumor compared with immortalized esophageal cells (HET1A and NE2) and non-tumor tissue (Figure 2B and Figure 2C).
iTRAQ analysis of highly metastatic 30M cell and the parental cells KYSE30. A. Experimental design for the quantitative proteomics analysis. The proteins from KYSE30 and 30 M cells treated with iTRAQ technique and LC-MS/MS analysis. The differentially expressed proteins (DEPs) through database searches and bioinformatics analysis. B. Repeatability test between KYSE30 and 30M samples. C. Volcano plot showed the distribution of DEPs in the highly metastatic 30M cell compared with the parental cells KYSE30. The horizontal axis is the logarithmic value of the relative quantiative value of the protein after log2 conversion, and the vertical axis is the p-value after the -log10 conversion. The red dots indicate significantly up-regulated DEPs, and the blue dots indicate significantly down-regulated DEPs. D. Six random selected DEPs (upregulated DEPs: ICAM1, DH58, IVL, IFI16; downregulated DEPs: BCAT1 and ABCC2) in 30M/KYSE30 cells were confirmed by quantitative real-time PCR (qRT-PCR) (*p<0.05, **p<0.01, *** p<0.001). E. KEGG pathway enrichment analysis of downregulated DEPs that are significantly altered in 30M/KYSE30. F. KEGG pathway enrichment analysis of upregulated DEPs that are significantly altered in 30M/KYSE30.
IFI16 was overexpressed in ESCC tumor and predicted poor prognosis. A. Heatmap of protein abundance of 14 DEPs in NOD-like receptor signaling pathway proteins. The colour represents the proteins abundance. B. The IFI16 protein levels were checked by western blot in ESCC cell lines. Among them, HET1A and NE2 are immortalized esophageal cells. C. A representative result of western blot shows the expression of IFI16 in the esophageal cancer tissues and normal tissues. D. Immunohistochemical staining for IFI16 expression in ESCC tissues and non-tumor tissue. Negative and weakly positive IFI16 were considered to represent low protein expression, while positive and strong IFI16 staining were considered to represent high protein expression. E. Kaplan-Meier analysis of the relationship between the expression of IFI16 and the survival time of relapse-free survival (RFS) in patients with esophageal cancer. The log-rank test p-values are shown.
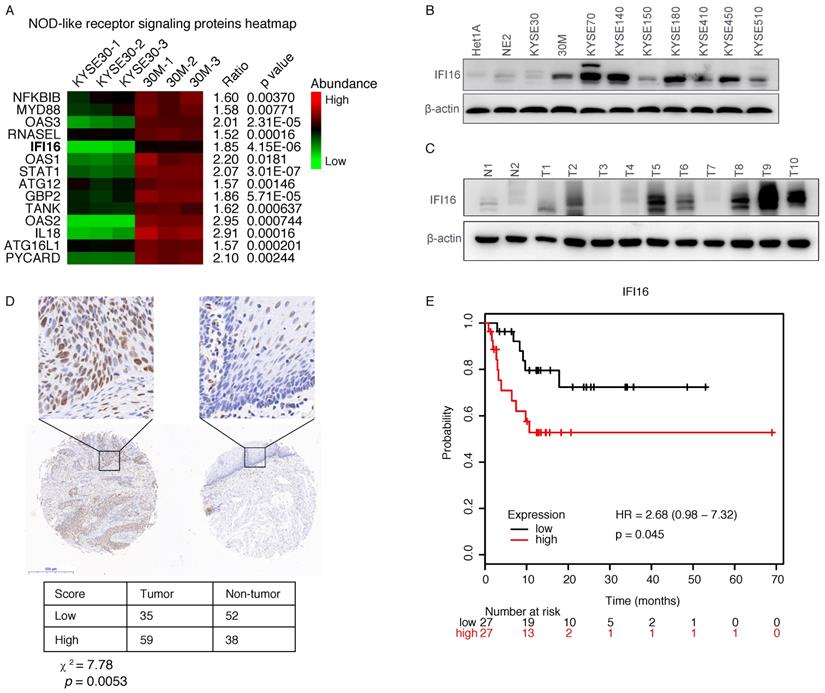
Next, to evaluate the clinical relevance of IFI16, we conducted further research on IFI16 through immunohistochemistry (IHC) assays, which produced similar results, with an increase expression of IFI16 in esophageal cancer tissues as compared with paracancerous tissue (Figure 2D). In addition, we also analyzed the relationship between IFI16 expression and the survival prognosis of esophageal cancer patients from the Kaplan-Meier (http://kmplot.com/analysis/) database. Compared with patients with esophageal cancer with low IFI16 expression, patients with high IFI16 expression had poorer relapse-free survival (RFS) (P=0.045) (Figure 2E).
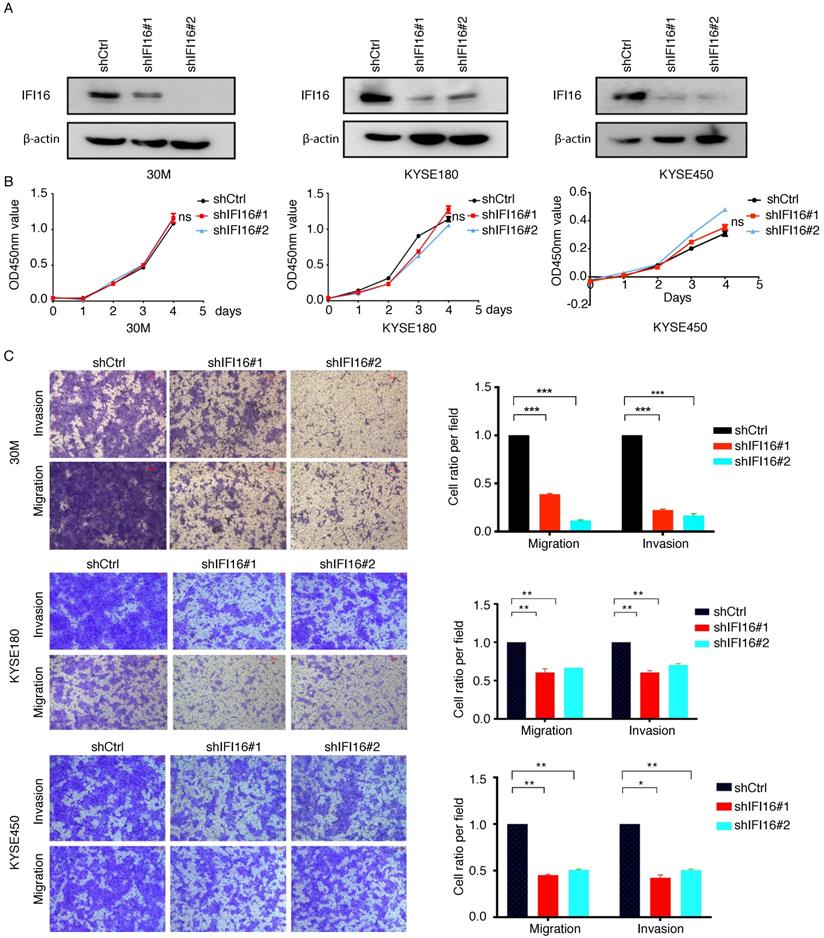
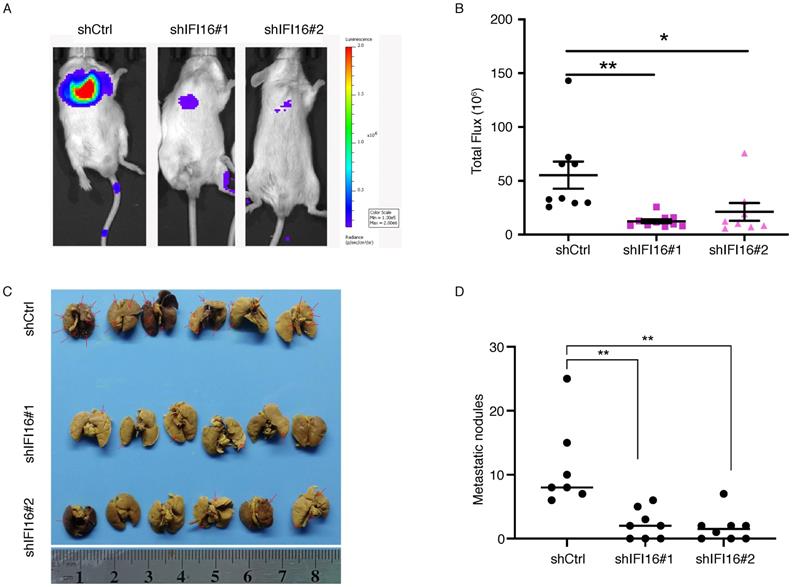
The significant increase of IFI16 expression in the high metastatic potentials 30M cell and tumor tissues hinted us to investigate its role in metastasis regulation both in vitro and in vivo. We generated 30M, KYSE180 and KYSE450 cell lines with knockdown of IFI16 (Figure 3A). CCK8 assay showed that proliferation of 30M, KYSE180, and KYSE450 cells were not affected by IFI16 knockdown (Figure 3B). However, knockdown IFI16 markedly inhibited migration and invasion of 30M, KYSE180 and KYSE450 cells (Figure 3C). The results suggested that IFI16 plays a key role in increasing esophageal cancer cell migration and invasion in vitro. To explore whether IFI16 influences ESCC metastasis in vivo, we used a tail vein injection mouse models to evaluate the effect of IFI16. The number of lung nodules was significantly decreased by IFI16 knockdown in 30M cells (Figure 4A-D).
Knocking down IFI16 inhibited ESCC cell migration and invasion in vitro. A-B. The proliferation of IFI16 knockdown (shIFI16#1 and shIFI16#2) and control (shCtrl) cells were measured by CCK8 in 30M, KYSE180 and KYSE450 cells. ns: not significant. C. Representative images (left) and quantitative analysis (right) of migration and invasion assays in 30M, KYSE180 and KYSE450 cells. Scale bars=100 µm.
IFI16 promotes ESCC metastasis through increasing FGF proteins expression
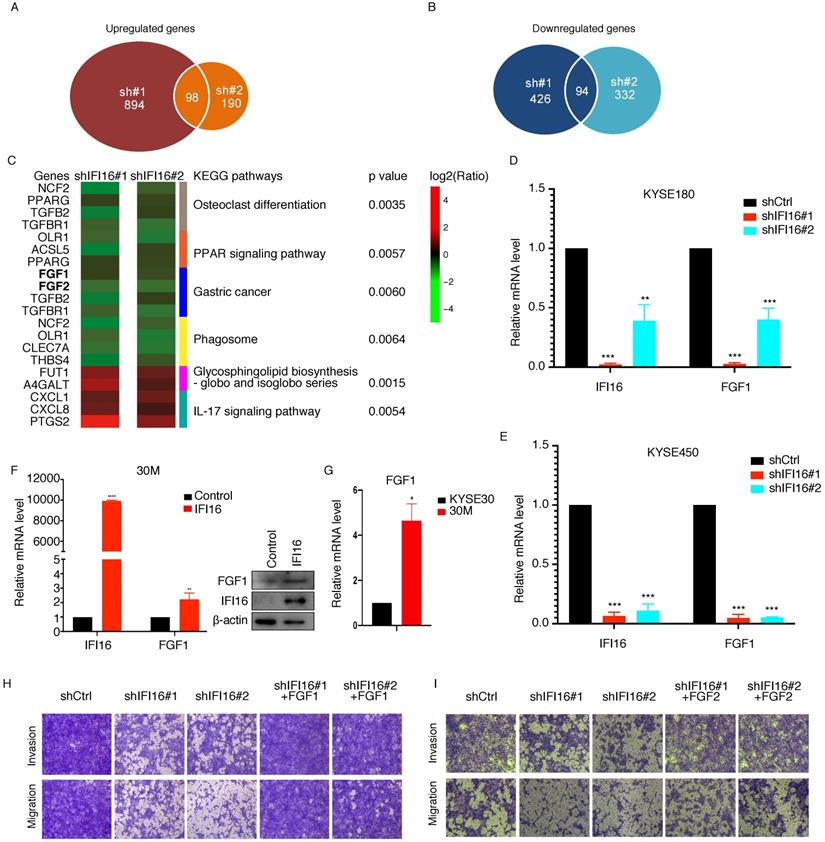
IFI16 located in the nucleoplasm and nucleolus is a member of the IFN-inducible HIN-200 gene family and as a regulatory factor that mediates the genes expression [11]. To determine the underlying mechanism by which IFI16 promotes metastasis, we performed an RNA-Seq assay in IFI16-knocking down 30M cells by two shRNA (#1 and #2) and control cells, and the differentially expressed genes (DEGs) were further analysed (Table S2). After knocking down of IFI16 expression in both groups of sh#1 and sh#2, we identified 98 DEGs with upregulated expression and 94 DEGs with downregulated expression (Figure 5A-B). KEGG pathway enrichment analysis revealed that top 3 KEGG pathways enriched by upregulated DEGs were Glycosphingolipid biosynthesis, IL-17 signaling pathway and Bladder cancer and top 3 KEGG pathways enriched by downregulated DEGs were Osteoclast differentiation, PPAR signaling pathway and Gastric cancer pathway (Figure 5C). In addition, NOD-like receptor signaling pathway was also enriched by downregulated DEGs after knocking down IFI16.
IFI16 promotes lung metastasis in vivo. A. Control cells and IFI16-knockdown 30M cells were injected into the tail vein of SCID/Beige mice and the lungs were harvested after 45 days for fluorescence. 30M cells labeled with luciferase in live imaging of small animals. The color scale describes the photon flux emitted by tumor cells. B. Results of quantitative analysis of fluorescence values (p<0.05). C. The lungs were harvested for bouin staining. D. Tumor nodules in lung tissues were counted (p<0.01).
Moreover, our RNA-Seq results showed that FGF proteins (FGF1, FGF2) were decreased in both IFI16-shRNA#1 and IFI16-shRNA#2 30M cells (Figure 5C). To further confirm that IFI16 can increase FGF1 expression in ESCC cells, we also knocked down IFI16 in KYSE180 and KYSE450 cells, and qRT-PCR results had showed that FGF1 also decreased in both two cells (Figure 5D-E). And overexpression of IFI16 could increase the FGF1 mRNA and protein level in 30M cell (Figure 5F). Moreover, FGF1 expression level was higher in 30M than in KYSE30 parent cell (Figure 5G). To further identify IFI16 promoted ESCC migration and invasion through increasing FGF gene expression, we overexpressed FGF1 and FGF2 in 30M cell after knocking down IFI16, and then measured the migration and invasion ability of cells. Our rescue experiment results showed that overexpression of FGF1 and FGF2 could rescue the inhibited migration and invasion ability by IFI16-depletion in 30M cells (Figure 5H & I).
Discussion
In this study, we applied used iTRAQ combined with LC-MS/MS technology to compare the proteome of two esophageal cancer cells KYSE30 and 30M with the same genetic background but different metastatic potential, and further combined in vivo and in vitro experiments to identify the IFI16 protein related to esophageal cancer metastasis. Increasing evidence has extended the concept that chronic inflammation meditated by abberant NOD-like receptor signaling is a powerful driver of carcinogenesis [12], and served as the potential targets in cancer therapy [13]. It was reported that chronic inflammation is associated with increased risk of ESCC development [14-16]. IFI16, a component of NOD-like receptor signaling pathway, provides a broad surveillance role against viral DNA by promoting restriction of gene expression from the exogenous DNA and inducing innate immune responses [17] and upregulated in 30M cell with high metastasis capability. Our data had showed that IFI16 was also upregulated in ESCC tumors. Kaplan-Meier survival curve analysis showed that the relapse-free survival (RFS) of patients with high IFI16 level was significantly worse than that of patients with low IFI16 level. These results demonstrated that IFI16 played a key role in progress of ESCC tumors. In vitro transwell model and in vivo mouse model both showed that knockdown IFI16 remarkably decreased the metastasis ability of ESCC cells.
IFI16 promoted the invasion and migration of ESCC cells through increasing FGF proteins expression. A. Number of upregulated genes in both IFI16-shRNA#1 and IFI16-shRNA#2 cells. B. Number of downregulated genes in both IFI16-shRNA#1 and IFI16-shRNA#2 cells. C. Heatmap showing the genes expressing level ratio of shIFI16/shCtrl in top significant enriched KEGG pathway (p<0.01). D-E. IFI16 knockdown decreased the FGF1 gene expression in KYSE180 (D) and KYSE450 (E) cells. QRT-PCR analysis of FGF1 expression in IFI16-knockdown KYSE180 and KYSE450 cells. (*p<0.05, **p<0.01,*** p<0.001) F. IFI16 overexpression increased the FGF1 gene expression in 30M measured by realtime-PCR (left) and western blotting (right). G. FGF1 mRNA level was measured in KYSE30 and 30M cells by realtime-PCR. H-I. FGF1 (H) and FGF2 (I) overexpression rescued the inhibited migration and invasion ability by IFI16-depletion. The migration and invasion ability of FGF1 and FGF2 overexpression 30M-shIFI16 cells was analyzed by the transwell assay.
IFI16, also known as PYHIN, a transcriptional regulator, member of the PYHIN protein family that contains a pyrin domain and two DNA binding HIN domains [11], is involved in transcriptional regulation. MTA1 complex composed of metastasis-associated protein 1 (MTA1) and IFN-γ-inducible protein 16 (IFI16) contribute to the epigenetic regulation of genes expression [18]. In addition, IFI16 regulates both self-renewal and differentiation gene expression during hESC trilineage specification through interaction with p53 [19]. To further clarify the mechanism of how IFI16 promoted ESCC metastasis, the RNA-seq analysis was performed in 30M cells by knockdown IFI16 by two shRNAs. The results showed that knocking down IFI16 could downregulate the expression of FGF proteins (FGF1, FGF2) expression. Similarly, we got the same result in KYSE180 and KYSE450 cells. The protein encoded by this gene is a member of the fibroblast growth factor (FGF) family. FGF family members possess broad mitogenic and cell survival activities, and are involved in a variety of biological processes, including cell proliferation, survival, migration, invasion, differentiation, and even angiogenesis [20]. It was reported that FGF was also aberrantly expressed in a series of cancer, including pancreatic cancer [21], glioblastoma [22], gastrointestinal cancer [23] and ESCC [24]. Deregulation of FGF signaling is implicated in both tumorigenesis and tumor invasion and metastasis [25]. Moreover, overexpression FGF1 and FGF2 could rescue the migration and invasion ability of ESCC cells after IFI16 knockdown. Therefore, our results demonstrated that IFI16 promoted ESCC metastasis through increasing FGF proteins expressing.
In conclusion, our quantitative proteomics data in this study demonstrated that the IFI16 upregulated in highly metastatic ESCC cell played a key role in promoting the invasion and migration of ESCC cells through increasing FGF proteins expression.
Supplementary Material
Supplementary table 1.
Supplementary table 2.
Acknowledgements
This project was funded by National Natural Science Foundation of China (81972634, 81773134), the CAMS Innovation Fund for Medical Sciences (2021-1-I2M-018), the fund of “San-ming” Project of Medicine in Shenzhen (SZSM201812062) and the Guangdong Basic and Applied Basic Research Foundation (2019B030302012).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Lin DC, Wang MR, Koeffler HP. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology. 2018;154:374-89
3. Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19:1450-64
4. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161-74
5. Nash GF, Turner LF, Scully MF, Kakkar AK. Platelets and cancer. Lancet Oncol. 2002;3:425-30
6. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
7. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913-7
8. Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467-73
9. Ma G, Jing C, Li L, Huang F, Ding F, Wang B. et al. MicroRNA-92b represses invasion-metastasis cascade of esophageal squamous cell carcinoma. Oncotarget. 2016;7:20209-22
10. Ma S, Sun Y, Zhao X, Xu P. [Recent advance in high accuracy iTRAQ for quantitative proteomics]. Sheng Wu Gong Cheng Xue Bao. 2014;30:1073-82
11. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997-1004
12. Saxena M, Yeretssian G. NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Front Immunol. 2014;5:327
13. Liu P, Lu Z, Liu L, Li R, Liang Z, Shen M. et al. NOD-like receptor signaling in inflammation-associated cancers: From functions to targeted therapies. Phytomedicine. 2019;64:152925
14. Lin R, Zhang C, Zheng J, Tian D, Lei Z, Chen D. et al. Chronic inflammation-associated genomic instability paves the way for human esophageal carcinogenesis. Oncotarget. 2016;7:24564-71
15. Maghsudlu M, Farashahi Yazd E. Heat-induced inflammation and its role in esophageal cancer. J Dig Dis. 2017;18:431-44
16. Goertzen C, Mahdi H, Laliberte C, Meirson T, Eymael D, Gil-Henn H. et al. Oral inflammation promotes oral squamous cell carcinoma invasion. Oncotarget. 2018;9:29047-63
17. Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A. 2013;110:E4492-501
18. Kang HJ, Lee MH, Kang HL, Kim SH, Ahn JR, Na H. et al. Differential regulation of estrogen receptor alpha expression in breast cancer cells by metastasis-associated protein 1. Cancer Res. 2014;74:1484-94
19. He Q, Wu Z, Yang W, Jiang D, Hu C, Yang X. et al. IFI16 promotes human embryonic stem cell trilineage specification through interaction with p53. NPJ Regen Med. 2020;5:18
20. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116-29
21. Yamanaka Y, Friess H, Buchler M, Beger HG, Uchida E, Onda M. et al. Overexpression of acidic and basic fibroblast growth factors in human pancreatic cancer correlates with advanced tumor stage. Cancer Res. 1993;53:5289-96
22. Stefanik DF, Rizkalla LR, Soi A, Goldblatt SA, Rizkalla WM. Acidic and basic fibroblast growth factors are present in glioblastoma multiforme. Cancer Res. 1991;51:5760-5
23. el-Hariry I, Pignatelli M, Lemoine N. Fibroblast growth factor 1 and fibroblast growth factor 2 immunoreactivity in gastrointestinal tumours. J Pathol. 1997;181:39-45
24. Takase N, Koma Y, Urakawa N, Nishio M, Arai N, Akiyama H. et al. NCAM- and FGF-2-mediated FGFR1 signaling in the tumor microenvironment of esophageal cancer regulates the survival and migration of tumor-associated macrophages and cancer cells. Cancer Lett. 2016;380:47-58
25. Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L. et al. A Network Map of FGF-1/FGFR Signaling System. J Signal Transduct. 2014;2014:962962
Author contact
![]() Corresponding authors: Yi Li & Zhihua Liu are co-corresponding authors that jointly supervised this work. Yi Li, State Key Lab of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China. Tel: +86 10 87788979. Email: liyiac.cn; Zhihua Liu, State Key Lab of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China. Tel: +86 10 87788490. Email: liuzhac.cn.
Corresponding authors: Yi Li & Zhihua Liu are co-corresponding authors that jointly supervised this work. Yi Li, State Key Lab of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China. Tel: +86 10 87788979. Email: liyiac.cn; Zhihua Liu, State Key Lab of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China. Tel: +86 10 87788490. Email: liuzhac.cn.

 Global reach, higher impact
Global reach, higher impact