Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(9):2775-2780. doi:10.7150/jca.73898 This issue Cite
Research Paper
Impact of carbonic anhydrase 9 gene polymorphism on the progression of colorectal cancer
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Emergency Medicine, Kuang Tien General Hospital, Taichung, Taiwan.
3. Department of Surgery, Chung Shan Medical University Hospital, Taichung, Taiwan.
4. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Mathematical Sciences, Florida Atlantic University, Florida, USA.
6. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
7. MingDao High School, Taichung, Taiwan.
8. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
9. Department of Dermatology, Drug Hypersensitivity Clinical and Research Center, Chang Gung Memorial Hospital, Linkou, Taiwan.
#Equal contributions as co-first authors.
Received 2022-4-11; Accepted 2022-6-2; Published 2022-6-21
Abstract
Colorectal cancer (CRC) is a commonly occurring tumor type worldwide, and its development is governed by a connection between genetic variations and acquired factors. Carbonic anhydrase 9 (CA9) is a cell-surface pH modulator that has been demonstrated to contribute to key steps of cancer progression. Here, we attempted to interrogate the effect of CA9 gene polymorphisms on the development of CRC in 470 cases and 470 gender- and age-matched non-cancer controls. We found that none of three CA9 single-nucleotide polymorphisms (SNPs) tested, including rs2071676, rs3829078, and rs1048638, was significantly associated with the occurrence of CRC. Yet, while evaluating the clinicopathological variables, cases carrying at least one reference allele (G allele) of rs2071676 tended to develop poorly differentiated tumors less frequently than those who are homozygous for the alternative allele (A allele) of rs2071676 (GA+GG vs AA; OR, 0.483; 95% CI, 0.242-0.963; p=0.036). Further stratification revealed that as compared to homozygous carriers of the alternative allele (AA), cases of colon cancer bearing at least one reference allele of rs2071676 (GA+GG) less frequently developed poorly differentiated tumors (OR, 0.449; 95% CI, 0.221-0.911; p=0.024) and lymphovascular invasion (OR, 0.570; 95% CI, 0.361-0.900; p=0.015). Such genetic effect was exclusively observed in colon cancer but not in rectal cancer. Our results indicate an anatomical site-specific impact of CA9 gene polymorphisms on modulating the progression of colorectal malignancies.
Keywords: Colorectal cancer, carbonic anhydrase 9, single-nucleotide polymorphism, tumor differentiation
Introduction
Nowadays, colorectal cancer (CRC) is among the most common neoplasms globally [1], and this burden is expected to reach 3.2 million new cases by 2040 [2]. In Taiwan, being the most frequent malignancy in men and the second in women, CRC remains a growing national public health challenge, accounting for a significant portion of cancer-related deaths [3]. In spite of the recent progresses in therapeutic approaches and cancer etiology, a consistent increase in the age-standardized death rate of colon cancer was observed over the years in Taiwan [4]. A plethora of risk parameters were known to contribute to such high prevalence and mortality. Diet and lifestyle choices with excess exposure of tumor-causing agents, such as smoking and alcohol drinking, have long been recognized as key environmental causes of CRC [5]. In addition, various inherited alterations that affect angiogenesis, adhesion, proteolysis, and cell growth have been shown to modulate CRC carcinogenesis [6]. Aside from host factors, studies of CRC etiology currently lay a greater emphasis on a shift of the gut commensal microbiome, which has been proposed to lie at the intersection of those potential risks mentioned above [7]. Given the heterogeneous nature of colorectal tumorigenesis, all these susceptibility factors seem to be interconnected and required to evaluate the disease prognosis.
Hypoxia is a unique feature of cancer niche during multistage carcinogenesis. In the absence of oxygen, cancer cells rely on aerobic glycolysis or the Warburg effect [8] for tumor expansion. Such metabolic reprogramming creates an acidic tumor microenvironment, where cells are needed to enhance the activity of pH-regulating machinery to avoid prolonged intracellular acidosis [9]. Carbonic anhydrase 9 (CA9), belonging to the α carbonic anhydrase family of zinc metalloenzymes that catalyze the reversible hydration of carbon dioxide to bicarbonate ions and protons, is a cell-surface glycoprotein that is upregulated by hypoxia and implicated in adaptation to acidosis [10]. Aside from acting as a pH modulator, CA9 can also function as an adhesion molecule, mediating the assembly and maturation of focal contacts during cell spreading and migration [11]. Converging observations have indicated that CA9 is functionally involved in diverse hallmarks of cancer, such as promotion of primary tumor growth and metastatic dissemination [12]. In the prognosis of CRC, high expression of CA9 was found to be associated with a poor outcome [13, 14], suggesting its translational value in CRC management. Recently, a growing number of studies have unveiled the associations of CA9 gene polymorphisms with the risk, progression, or therapy outcome of various malignancies [15-20]. Yet, the influence of CA9 gene variants on the susceptibility to colon cancer remains mostly unexplored. Here, we conducted a case-control study to interrogate how and to what extent CA9 single-nucleotide polymorphisms (SNPs) affect the progression of CRC.
Materials and Methods
Subjects
A total of 470 patients with CRC and 470 cancer-free controls were recruited in this investigation, with the approval by the institutional review board of Chung Shan Medical University Hospital in Taichung, Taiwan. All subjects, enrolled from 2016 to 2021, provided informed written consent at enrollment. Moreover, subjects with history of cancer of any sites and self-reported diseases such as cardiovascular, diabetes, and autoimmune diseases were excluded from the control group. Clinical staging of CRC was staged determined according to the TNM staging system of the American Joint Committee on Cancer (AJCC) [21] at the time of diagnosis. Cancer differentiation was evaluated by a pathologist and graded according to the AJCC classification. Demographic data on age and gender were recorded from each participant.
Genotyping
Genomic DNA derived from the whole blood was isolated by using QIAamp DNA blood mini kits (Qiagen, Valencia, CA, USA) [22, 23]. Based on the previous research, three genetic variants of CA9 SNPs (rs2071676, rs3829078, and rs1048638) were selected [15-17, 24, 25]. Discrimination of polymorphic alleles for three CA9 SNPs (rs2071676, rs3829078, and rs1048638) was performed through the TaqMan assay with an ABI StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and further assessed with SDS version 3.0 software (Applied Biosystems).
Statistical analysis
The Hardy-Weinberg equilibrium for biallelic markers was assessed using a chi-square goodness-of-fit test. The comparison of demographic factors between CRC patients and controls was carried out using Fisher's exact test. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) for the genetic association of CA9 SNPs with and the predisposition to CRC were evaluated by multiple logistic regression models after controlling for age and gender. Data were calculated by using SAS statistical software (Version 9.1, 2005; SAS Institute Inc., Cary, NC). A p value < 0.05 was considered significant.
Results
Demographic and clinical characteristics of study cohorts
To assess the influence of CA9 gene polymorphisms on the risk and progression of colorectal tumor, 470 patients with CRC were enrolled in this investigation. As age and gender are known to affect the predisposition to CRC [26], 470 cancer-free subjects who matched age and gender to the cases were recruited to exclude possible confounders. The comparison of demographic and clinical features between the case and control group was shown in Table 1. Within the case group, 107 and 363 suffered from tumors of the rectum and colon, respectively. 48.4% and 16.7% of cases developed lymphatic spread and distal metastasis, respectively.
The distributions of demographical and clinical characteristics in 470 controls and 470 patients with CRC
| Variable | Controls (N=470), n (%) | Patients (N=470) (%) | p value |
|---|---|---|---|
| Age (yrs) | |||
| <65 | 269 (57.2%) | 249 (53.0%) | 0.190 |
| ≥65 | 201 (42.8%) | 221 (47.0%) | |
| Gender | |||
| Male | 292 (62.1%) | 275 (58.5%) | 0.257 |
| Female | 178 (37.9%) | 195 (41.5%) | |
| Tumor location | |||
| Rectum | 107 (22.8%) | ||
| Left colon | 220 (46.8%) | ||
| Right colon | 143 (30.4%) | ||
| Stage | |||
| I+II | 223 (47.4%) | ||
| III+IV | 247 (52.6%) | ||
| Tumor T status | |||
| T1-T2 | 113 (24.0%) | ||
| T3-T4 | 357 (76.0%) | ||
| Lymph node status | |||
| N0 | 233 (49.6%) | ||
| N1+N2 | 237 (50.4%) | ||
| Metastasis | |||
| M0 | 394 (83.8%) | ||
| M1 | 76 (16.2%) | ||
| Lymphovascular invasion | |||
| No | 258 (54.9%) | ||
| Yes | 212 (45.1%) | ||
| Perineural invasion | |||
| No | 264 (56.2%) | ||
| Yes | 206 (43.8%) | ||
| Pathologic grading | |||
| Well | 6 (1.3%) | ||
| Moderately | 428 (91.0%) | ||
| Poorly | 36 (7.7%) | ||
CA9 gene polymorphisms were associated with the progression but not the occurrence of CRC
To explore how and to what extent CA9 gene variations influence the development of CRC, three CA9 SNPs (rs2071676, rs3829078, and rs1048638) were chosen according to their broad correlations with the risk of many malignancies [16-18] and genotyped in the present study. The genotype ratios for individual SNP in our cohorts were examined (Table 2). No deviation (p>0.05) from Hardy-Weinberg equilibrium in both study groups was identified for all three SNPs. We found that none of these CA9 variants was significantly associated with the occurrence of CRC in our cohorts. Additionally, we tested whether genetic polymorphisms of CA9 were connected to the clinicopathological features of CRC patients. We found that cases who possess at least one reference allele (G allele) of rs2071676 (GA+GG) tended to develop poorly differentiated tumors less frequently than those who are homozygous for the alternative allele (A allele) of rs2071676 (AA) (OR, 0.483; 95% CI, 0.242-0.963; p=0.036) (Table 3). Although not statistically significant, homozygotes for the alternative allele of rs2071676 (AA) are marginally associated with advanced cancers, with an inclination to develop large-size tumors and potentiate lymphovascular and perineural invasion. These results indicate a role of CA9 gene polymorphisms in CRC progression.
Genotype Distributions of CA9 Gene Polymorphisms in 470 Controls and 470 Patients with CRC
| Variable | Controls (N=470), n (%) | Patients (N=470), n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs2071676 | ||||
| AA | 130 (27.7%) | 137 (29.1%) | 1.000 (reference) | 1.000 (reference) |
| AG | 231 (49.1%) | 218 (46.4%) | 0.896 (0.661-1.212) | 0.774 (0.472-1.269) |
| GG | 109 (23.2%) | 115 (24.5%) | 1.001 (0.702-1.428) | 0.770 (0.436-1.359) |
| AG+GG | 340 (72.3%) | 333 (70.9%) | 0.929 (0.700-1.234) | 0.773 (0.487-1.226) |
| rs3829078 | ||||
| AA | 438 (93.2%) | 437 (93.0%) | 1.000 (reference) | 1.000 (reference) |
| AG | 32 (6.8%) | 33 (7.0%) | 1.034 (0.624-1.711) | 1.151 (0.504-2.629) |
| GG | 0 (0%) | 0 (0%) | --- | --- |
| AG+GG | 32 (6.8%) | 33 (7.0%) | 1.034 (0.624-1.711) | 1.151 (0.504-2.629) |
| rs1048638 | ||||
| CC | 411 (87.4%) | 408 (86.8%) | 1.000 (reference) | 1.000 (reference) |
| CA | 56 (11.9%) | 57 (12.1%) | 1.025 (0.692-1.520) | 0.636 (0.302-1.314) |
| AA | 3 (0.7%) | 5 (1.1%) | 1.679 (0.399-7.071) | 1.219 (0.142-10.475) |
| CA+AA | 59 (12.6%) | 62 (13.2%) | 1.059 (0.723-1.551) | 0.680 (0.337-1.372) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age and gender.
Distribution of the clinical status and CA9 rs2071676 genotype frequencies in 470 CRC patients
| Variable | AA (N=137) | AG + GG (N=333) | OR (95% CI) | p value |
|---|---|---|---|---|
| Stages | ||||
| I+II | 65 (47.4%) | 158 (47.4%) | 1.000 (reference) | p=1.000 |
| III+IV | 72 (52.6%) | 175 (52.6%) | 1.000 (0.671-1.326) | |
| Tumor T status | ||||
| T1+T2 | 28 (20.4%) | 85 (25.5%) | 1.000 (reference) | p=0.241 |
| T3+T4 | 109 (79.6%) | 248 (74.5%) | 0.749 (0.462-1.215) | |
| Lymph node status | ||||
| Negative | 69 (50.4%) | 164 (49.2%) | 1.000 (reference) | p=0.826 |
| Positive | 68 (49.6%) | 169 (50.8%) | 1.046 (0.702-1.557) | |
| Metastasis | ||||
| Negative | 116 (84.7%) | 278 (83.5%) | 1.000 (reference) | p=0.751 |
| Positive | 21 (15.3%) | 55 (16.5%) | 1.093 (0.632-1.889) | |
| Lymphovascular invasion | ||||
| No | 68 (49.6%) | 190 (57.1%) | 1.000 (reference) | p=0.142 |
| Yes | 69 (50.4%) | 143 (42.9%) | 0.742 (0.498-1.105) | |
| Perineural invasion | ||||
| No | 72 (52.6%) | 192 (57.7%) | 1.000 (reference) | p=0.311 |
| Yes | 65 (47.4%) | 141 (42.3%) | 0.813 (0.545-1.213) | |
| Cell differentiation | ||||
| Well/Moderately | 121 (88.3%) | 313 (94.0%) | 1.000 (reference) | p=0.036 |
| Poorly | 16 (11.7%) | 20 (6.0%) | 0.483 (0.242-0.963) | |
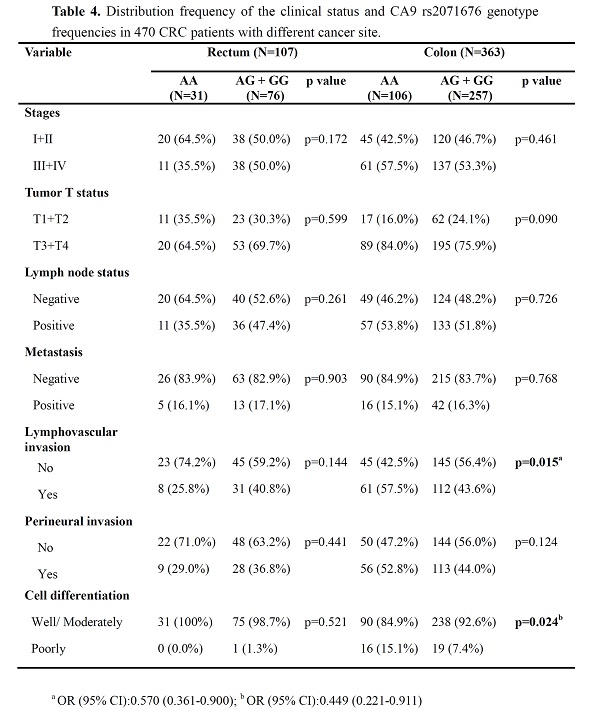
rs2071676 was associated exclusively with tumors of the colon, but not that of the rectum
Since a missense SNP, rs2071676, was noted to be associated with CRC differentiation, we next tested whether this genetic effect was specific to the tumor location. Our stratification analyses revealed that as compared to the homozygous carriers of the alternative allele of rs2071676 (AA), cases of colon cancer carrying at least one reference allele of rs2071676 (GA+GG) less frequently developed poorly differentiated tumors (OR, 0.449; 95% CI, 0.221-0.911; p=0.024) and lymphovascular invasion (OR, 0.570; 95% CI, 0.361-0.900; p=0.015) (Table 4). This genetic association was exclusively observed in colon cancer but not in rectal cancer. Even though there was no statistical significance, a marginally protective effect of rs2071676 G allele on the progression into advanced colon cancer (e.g. large-size tumors or perineural invasion) was seen. Overall, there data unveil a genetic interaction of CA9 with the tumor differentiation and lymphovascular invasion of colon adenocarcinoma.
Distribution frequency of the clinical status and CA9 rs2071676 genotype frequencies in 470 CRC patients with different cancer site
| Variable | Rectum (N=107) | Colon (N=363) | ||||
|---|---|---|---|---|---|---|
| AA (N=31) | AG + GG (N=76) | p value | AA (N=106) | AG + GG (N=257) | p value | |
| Stages | ||||||
| I+II | 20 (64.5%) | 38 (50.0%) | p=0.172 | 45 (42.5%) | 120 (46.7%) | p=0.461 |
| III+IV | 11 (35.5%) | 38 (50.0%) | 61 (57.5%) | 137 (53.3%) | ||
| Tumor T status | ||||||
| T1+T2 | 11 (35.5%) | 23 (30.3%) | p=0.599 | 17 (16.0%) | 62 (24.1%) | p=0.090 |
| T3+T4 | 20 (64.5%) | 53 (69.7%) | 89 (84.0%) | 195 (75.9%) | ||
| Lymph node status | ||||||
| Negative | 20 (64.5%) | 40 (52.6%) | p=0.261 | 49 (46.2%) | 124 (48.2%) | p=0.726 |
| Positive | 11 (35.5%) | 36 (47.4%) | 57 (53.8%) | 133 (51.8%) | ||
| Metastasis | ||||||
| Negative | 26 (83.9%) | 63 (82.9%) | p=0.903 | 90 (84.9%) | 215 (83.7%) | p=0.768 |
| Positive | 5 (16.1%) | 13 (17.1%) | 16 (15.1%) | 42 (16.3%) | ||
| Lymphovascular invasion | ||||||
| No | 23 (74.2%) | 45 (59.2%) | p=0.144 | 45 (42.5%) | 145 (56.4%) | p=0.015a |
| Yes | 8 (25.8%) | 31 (40.8%) | 61 (57.5%) | 112 (43.6%) | ||
| Perineural invasion | ||||||
| No | 22 (71.0%) | 48 (63.2%) | p=0.441 | 50 (47.2%) | 144 (56.0%) | p=0.124 |
| Yes | 9 (29.0%) | 28 (36.8%) | 56 (52.8%) | 113 (44.0%) | ||
| Cell differentiation | ||||||
| Well/Moderately | 31 (100%) | 75 (98.7%) | p=0.521 | 90 (84.9%) | 238 (92.6%) | p=0.024b |
| Poorly | 0 (0.0%) | 1 (1.3%) | 16 (15.1%) | 19 (7.4%) | ||
a OR (95% CI):0.570 (0.361-0.900); b OR (95% CI):0.449 (0.221-0.911).
Discussion
The progression of CRC is a multistep process governed by an intricate combination of environmental and genetic factors. Here, we demonstrated that CA9 gene polymorphisms, rs2071676, influenced colorectal cancer differentiation but did not affect the predisposition to CRC. Moreover, this genetic association was detected exclusively in colon cancer but not in rectal cancer, indicating an anatomical site-specific effect of CA9 gene polymorphisms on the progression of colorectal malignancies.
Through its catalytic and non-catalytic functions, CA9 is known to endow cancer cells with survival advantages in low-oxygen conditions and to confer a growing capability to disseminate [27]. In addition, associations of CA9 SNPs with different aspects of cancer development have been reported [15-20]. Among these, rs12553173, a synonymous variant, was shown to correlate with improved median survival in patients with metastatic kidney cancer [19]. Another non-coding variant located in the 3'-untranslated region of CA9 gene, rs1048638, has been demonstrated to affect the susceptibility to liver [16] and cervical cancer [17]. In the present study, we found that rs2071676 influenced colorectal cancer differentiation but did not affect the predisposition to CRC. This genetic variation (G>A) causes the substitution of valine by methionine at position 33 in the signal peptide of the protein product. Although the functional analysis of rs2071676 remain unavailable, this missense variation may disturb the removal of signal peptide after translocation of the nascent polypeptide into the endoplasmic reticulum (ER) lumen. Genetic alterations in either the signal peptidase recognition domain or hydrophobic region of signal peptides can impede cleavage of the signal peptide [28, 29]. As a consequence, some protein products could be accumulated or degraded due to deficient glycosylation, inaccurate protein folding, and attenuated transport from ER to Golgi [30]. In patients with retinitis pigmentosa, a pathogenic genetic variant that causes replacement of an arginine with a tryptophan in the signal sequence of the carbonic anhydrase 4 (CA4) gene resulted in a reduction of CA4 activity by virtue of a combination of decreased synthesis and accelerated turnover [31]. Thus, we speculate that polymorphic genotype of rs2071676 may lead to accumulation of misfolded forms of CA9 in the ER of epithelial cells within the colon. Such chronic ER stress, in turn, contributes to altered adhesion and catalytic activity of CA9, ultimately affecting the progression of CRC. Determining the functional impact of rs2071676 on CA9 enzyme activity, the rates of biosynthesis, conversion of unfolded to mature enzyme, and turnover of CA9 will require further investigation.
Furthermore, it is intriguing that rs2071676 was exclusively associated with clinical variables of colon cancer but not with that of rectal cancer, revealing an anatomical site-specific effect of CA9 gene polymorphisms on the progression of colorectal malignancies. Although both colon and rectal cancer develop in the large bowel and are often considered as a single tumor entity in all fields of basic and clinical research, substantial differences in molecular carcinogenesis exist [32]. Compared to rectal cancer, colon cancer more commonly exhibited higher activity of MAPK signaling pathways [33], elevated expression of HOX gene family [34], and constitutively active forms of BRAF [35, 36] and KRAS [37]. In addition to differential orchestration of many cancer hallmarks in tumors of the colon and rectum, these disturbed oncogenic signaling pathways also controlled the expression or stability of a major transcriptional inducer of CA9, hypoxia-inducible factor (HIF) [38-40], thereby leading to the fluctuations of local CA9 levels within CRC tumor microenvironment. These findings, in part, account for our observation that rs2071676 was associated with the progression of CRC in an anatomical site-specific manner.
This study unveiled a potential role of CA9 gene polymorphisms in many aspects of colon cancer progression. Yet, extra efforts are required to address several limitations in the study. One is that the influence of CA9 SNPs on CRC susceptibility might be underestimated due to the unavailability of data about the status of alcohol consumption and cigarette use for adjustment. Another caveat is that the mechanistic role of rs2071676 in tumor progression remains mostly unclear. Whether the substitution of valine by methionine due to the presence of polymorphic allele affects its expression, membrane translocation, or internalization needs to be further defined. Moreover, the results observed in this investigation might be not applicable to other racial groups except if replication studies are carried out.
In conclusion, our findings demonstrate an association of a CA9 SNP, rs2071676, with cancer differentiation and invasiveness in tumors of the colon, highlighting an anatomical site-specific effect of CA9 gene polymorphisms on the progression of colorectal malignancies.
Acknowledgements
We would like to thank the Human Biobank of Chung Shan Medical University Hospital for providing the biological specimen and related clinical data for our research. We thank Tissue Bank at Chang Gung Memorial Hospital, Keelung for sample preparation. This study was supported by a research grant from Chang Gung Memorial Hospital (BMRPE97).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
2. Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174
3. Kuo CN, Liao YM, Kuo LN, Tsai HJ, Chang WC, Yen Y. Cancers in Taiwan: Practical insight from epidemiology, treatments, biomarkers, and cost. J Formos Med Assoc. 2020;119:1731-41
4. Su SY, Huang JY, Jian ZH, Ho CC, Lung CC, Liaw YP. Mortality of colorectal cancer in Taiwan, 1971-2010: temporal changes and age-period-cohort analysis. Int J Colorectal Dis. 2012;27:1665-72
5. Hull R, Francies FZ, Oyomno M, Dlamini Z. Colorectal Cancer Genetics, Incidence and Risk Factors: In Search for Targeted Therapies. Cancer Manag Res. 2020;12:9869-82
6. Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185-92
7. Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016;70:395-411
8. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-33
9. Parks SK, Chiche J, Pouyssegur J. pH control mechanisms of tumor survival and growth. J Cell Physiol. 2011;226:299-308
10. Pastorek J, Pastorekova S, Callebaut I, Mornon JP, Zelnik V, Opavsky R. et al. Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene. 1994;9:2877-88
11. Csaderova L, Debreova M, Radvak P, Stano M, Vrestiakova M, Kopacek J. et al. The effect of carbonic anhydrase IX on focal contacts during cell spreading and migration. Front Physiol. 2013;4:271
12. Pastorekova S, Gillies RJ. The role of carbonic anhydrase IX in cancer development: links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev. 2019;38:65-77
13. Hong YS, Cho HJ, Kim SY, Jung KH, Park JW, Choi HS. et al. Carbonic anhydrase 9 is a predictive marker of survival benefit from lower dose of bevacizumab in patients with previously treated metastatic colorectal cancer. BMC Cancer. 2009;9:246
14. Korkeila E, Talvinen K, Jaakkola PM, Minn H, Syrjanen K, Sundstrom J. et al. Expression of carbonic anhydrase IX suggests poor outcome in rectal cancer. Br J Cancer. 2009;100:874-80
15. Chien MH, Yang JS, Chu YH, Lin CH, Wei LH, Yang SF. et al. Impacts of CA9 gene polymorphisms and environmental factors on oral-cancer susceptibility and clinicopathologic characteristics in Taiwan. PloS one. 2012;7:e51051
16. Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS. et al. 3'UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. 2017;7:4466
17. Shen HP, Hsiao YH, Yang SF, Liu YF, Ko JL, Wang PH. Single nucleotide polymorphisms and haplotypes of carbonic anhydrase 9 can predict invasive squamous cell carcinoma of uterine cervix. International journal of medical sciences. 2018;15:587-94
18. Yu YY, Chiou HL, Tsao SM, Huang CC, Lin CY, Lee CY. et al. Association of Carbonic Anhydrase 9 Polymorphism and the Epithelial Growth Factor Receptor Mutations in Lung Adenocarcinoma Patients. Diagnostics (Basel). 2020;10:266
19. de Martino M, Klatte T, Seligson DB, LaRochelle J, Shuch B, Caliliw R. et al. CA9 gene: single nucleotide polymorphism predicts metastatic renal cell carcinoma prognosis. J Urol. 2009;182:728-34
20. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Genetic polymorphism and carbonic anhydrase 9 expression can predict nodal metastatic prostate cancer risk in patients with prostate-specific antigen levels </=10 ng/ml at initial biopsy. Urol Oncol. 2019;37:814 e9- e16
21. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471-4
22. Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC, Hsieh YH. et al. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. Journal of surgical oncology. 2010;102:163-9
23. Chung TT, Pan MS, Kuo CL, Wong RH, Lin CW, Chen MK. et al. Impact of RECK gene polymorphisms and environmental factors on oral cancer susceptibility and clinicopathologic characteristics in Taiwan. Carcinogenesis. 2011;32:1063-8
24. Wang SS, Liu YF, Ou YC, Chen CS, Li JR, Yang SF. Impacts of CA9 gene polymorphisms on urothelial cell carcinoma susceptibility and clinicopathologic characteristics in Taiwan. PloS one. 2013;8:e82804
25. Yang SF, Liu YF, Cheng CW, Yang WE, Lin WL, Ko JL. et al. Impact of microRNA-34a and polymorphism of its target gene CA9 on susceptibility to uterine cervical cancer. Oncotarget. 2017;8:77860-71
26. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103
27. Pastorek J, Pastorekova S. Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: from biology to clinical use. Semin Cancer Biol. 2015;31:52-64
28. Ellgaard L, Helenius A. ER quality control: towards an understanding at the molecular level. Curr Opin Cell Biol. 2001;13:431-7
29. Chen X, VanValkenburgh C, Liang H, Fang H, Green N. Signal peptidase and oligosaccharyltransferase interact in a sequential and dependent manner within the endoplasmic reticulum. J Biol Chem. 2001;276:2411-6
30. Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891-4
31. Rebello G, Ramesar R, Vorster A, Roberts L, Ehrenreich L, Oppon E. et al. Apoptosis-inducing signal sequence mutation in carbonic anhydrase IV identified in patients with the RP17 form of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2004;101:6617-22
32. Paschke S, Jafarov S, Staib L, Kreuser ED, Maulbecker-Armstrong C, Roitman M. et al. Are Colon and Rectal Cancer Two Different Tumor Entities? A Proposal to Abandon the Term Colorectal Cancer. Int J Mol Sci. 2018;19:2577
33. Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403-8
34. Sanz-Pamplona R, Cordero D, Berenguer A, Lejbkowicz F, Rennert H, Salazar R. et al. Gene expression differences between colon and rectum tumors. Clin Cancer Res. 2011;17:7303-12
35. Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G. et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30:1288-95
36. Budinska E, Popovici V, Tejpar S, D'Ario G, Lapique N, Sikora KO. et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63-76
37. Slattery ML, Curtin K, Wolff RK, Boucher KM, Sweeney C, Edwards S. et al. A comparison of colon and rectal somatic DNA alterations. Dis Colon Rectum. 2009;52:1304-11
38. Kopacek J, Barathova M, Dequiedt F, Sepelakova J, Kettmann R, Pastorek J. et al. MAPK pathway contributes to density- and hypoxia-induced expression of the tumor-associated carbonic anhydrase IX. Biochim Biophys Acta. 2005;1729:41-9
39. Kikuchi H, Pino MS, Zeng M, Shirasawa S, Chung DC. Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1alpha and -2alpha in colon cancer. Cancer Res. 2009;69:8499-506
40. Qi L, Chen J, Yang Y, Hu W. Hypoxia Correlates With Poor Survival and M2 Macrophage Infiltration in Colorectal Cancer. Frontiers in oncology. 2020;10:566430
Author contact
![]() Corresponding authors: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan. Fax: 886-4-24723229; E-mail:ysfedu.tw (Shun-Fa Yang) or Shih-Chi Su, PhD. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan. E-mail: ssu1org.tw (Shih-Chi Su).
Corresponding authors: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan. Fax: 886-4-24723229; E-mail:ysfedu.tw (Shun-Fa Yang) or Shih-Chi Su, PhD. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan. E-mail: ssu1org.tw (Shih-Chi Su).

 Global reach, higher impact
Global reach, higher impact