3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(9):2844-2854. doi:10.7150/jca.73758 This issue Cite
Research Paper
LncRNA LINC00460 facilitates the proliferation and metastasis of renal cell carcinoma via PI3K/AKT signaling pathway
1. The Fourth Clinical College of Nanjing Medical University, Nanjing 211100, Jiangsu Province, China.
2. Cancer Institute, Xuzhou Medical University, Xuzhou 221002, Jiangsu Province, China.
3. Center of Clinical Oncology, the Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, Jiangsu Province, China.
4. Department of Radiotherapy, the Second Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, Jiangsu Province, China.
5. Hepatobiliary Center, The First Affiliated Hospital of Nanjing Medical University; Key Laboratory of Liver Transplantation, Chinese Academy of Medical Sciences; NHC Key Laboratory of Living Donor Liver Transplantation (Nanjing Medical University), Nanjing 210029, Jiangsu Province, China.
#These authors contributed equally to this work.
Received 2022-4-7; Accepted 2022-6-23; Published 2022-7-4
Abstract
Renal cell carcinoma (RCC) is one of the most prevalent cancers diseases in the worldwide. Long noncoding RNAs (LncRNAs) have been indicated as a mediator acted in tumorigenesis of RCC. LINC00460 has been reported to participate in many kinds of malignancies and promotes cancer progressions. However, the mechanism of LINC00460 on RCC is yet to be investigated. This study aimed to explore the potential function and regulation mechanism of LINC00460 in RCC. We analysed the LINC00460 expression and the prognosis in RCC patients using Gene Expression Profiling Interactive Analysis (GEPIA) and The Cancer Genome Atlas (TCGA) databases. LINC00460 level in normal renal cell line and RCC cell lines were examined by qRT-PCR. We study the effects of LINC00460 on proliferation, migration, invasion, apoptosis in RCC cells lines using a series of in vivo and in vitro experiments. RNA sequencing (RNA-seq) analysis was applied to searching potential LINC00460 related signal pathway in RCC. We identified the significant up-regulated expression of LINC00460 both in RCC tissues and cell. RCC patients with elevated LINC00460 expression have shorter survival. Up-expression of LINC00460 promoted cell proliferation, invasion and migration, meanwhile down-regulation of LINC00460 exerted inhibitory effect on these activities. We crucially identified that LNC00460 promotes development of RCC by influencing the PI3K/AKT pathway. Knockdown of LNC00460 decreased the phosphorylation of AKT and mTOR. The key finding of our study showed that LINC00460 functions as an oncogene in RCC pathogenesis by mediating the PI3K/AKT.
Keywords: RCC, LINC00460, proliferation, metastasis, PI3K/AKT pathway
Introduction
Renal cell carcinoma (RCC) is a typical malignant carcinoma originating from the renal tubules. The morbidity of RCC has been increased over the years, only next to prostate cancer and bladder cancer of adult cancer diseases [1]. When patients in the early phase of kidney cancer, the asymptomatic and covert symptoms always caused the awareness less of cancer screening, which made the patients often missed the diagnosis and treatment correspondingly. When the patients were diagnosed, they were already in the advanced clinical stage with local progression occurred [2]. Radical surgery is still a mainstay of treatment for patients in early-stage RCC so far [3]. The prognosis of RCC patients with advanced stage remains poor although progress in treatment has occurred through clinical development, and the targeted therapy effect is still limited due to drug resistance and adverse reactions [4, 5]. Therefore, unraveling the molecular mechanisms of RCC for screening novel biomarkers becomes an immediate concern.
Long noncoding RNAs (LncRNAs) have been considered as transcriptional, post-transcriptional or post-translational levels regulators of gene expression [6], LncRNAs with more than 200 nucleotides and no protein-coding capacity, participate in lots of physiological and pathological bio-behaviors [7, 8], complicating the gene regulation networks, such as gene transcription regulation, RNA processing, chromatin modification, especially in tumorigenic biological activities [9]. Accumulating evidences show that LncRNAs can caused occurrence of tumor and work as oncogenes [10-12]. Exactly as previous studies, LncRNA MALAT1, LncRNA XIST, LncRNA HOTAIR or LncRNA TUG1 facilitate tumor cell growth, migration or invasion in multiple types of tumors such as cervical cancer, triple-negative breast cancer, gastric cancer, multiple myeloma, pancreatic cancer, bladder cancer and thyroid cancer [13-17].
Long intergenic noncoding RNA 460 (LINC00460) is a novel carcinogenic LncRNA related in the development of different kinds of human malignancies, including lung cancer, thyroid cancer, nasopharyngeal carcinoma [10, 18, 19]. When we found that high expression of LINC00460 was associated with poor prognosis in RCC patients through TCGA database analysis, which caught our attention to study the biofunction of LINC00460 in RCC. Sadly, the more specific mechanism of the LINC00460 function in RCC is still unclear.
The phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway aberrant activation regulated cellular processes including proliferation, metabolism, apoptosis and metastasis in many cancer types [20-22]. Study reported that PI3K/AKT/mTOR signaling pathway promote human esophageal cancer tumorigenesis and metastasis, induce potential cell apoptosis of the esophageal squamous cell carcinoma [23]. This remarkable pathway is regulated by a number of upstream signaling and its cascades downstream signaling pathways collaborating with various compensatory effectors [24].
In this study, we analyzed data from the TCGA database using the online bioinformatics tool GEPIA and found the increased LINC00460 expression level in RCC tissues predicted poor survival of RCC patients. We evaluated the level of LINC00460 in RCC cells and its tumorigenesis on RCC cell vitality, migration, invasion and apoptosis in vitro and in vivo. RNA sequencing (RNA-seq) analysis proved that LINC00460 knockdown could mainly affect the genes involved in proliferation and apoptosis. In the viewpoint of the mechanism, our results indicated that LINC00460 mediates PI3K/AKT signaling to promote the progression of RCC cells. Thus, this study presented a perspective understanding of RCC pathogenesis. We deeply revealed the mechanism of LINC00460 function in RCC.
Materials and Methods
Bioinformatical analyses
The Gene Expression Profiling Interactive Analysis (GEPIA) and The Cancer Genome Atlas (TCGA) databases were used to analyzed LINC00460 expression in RCC and normal tissue. Clinicopathological features including overall survival (OS) and disease-free survival (DFS) was evaluated linked to RCC patients LINC00460 expression.
Cell Culture
Human embryo kidney epithelial cell line (HK-2) and renal cell carcinoma cell lines were purchased from cell bank of Chinese Academy of Sciences. ACHN, 786-O, OSRC-2, Ketr-3, and HK-2 were correspondingly cultured in RMPI-1640 or DMEM Medium, supplemented with 10% FBS, 100 U/mL each penicillin and streptomycin separately. Cell incubated condition is 37 °C humidified with 5% CO2.
Cell Transfection
For over-expressed LINC00460, sequences were constructed into pCDH-CMV-MCS-EF1-GreenPuro lentivirus vector (GenePharma Suzhou, China). shLINC00460#1 (5'-GCTAAGACCTAATAGCCAATA-3'), shLINC00460#2 (5'-ACCTTGGTCAAACGTTTAACC-3'), as well as the negative control shCtrl (5'-GTTCTCCGAACGTGTACGT-3') were constructed into pLKO.1 (GenePharma Suzhou, China) for knockdown of LINC00460. psPAX2, pMD2.G and pLKO-shRNA were co-transfected into HEK-293T cells for the lentiviruses until 48h later. Stable knockdown ACHN and 786-O cell lines with a treatment of 5 μg/mL puromycin for 1 week were obtained after lentiviruses infected.
RNA isolate, RT-PCR and qPCR
We prepared total RNA by using TRIzol Reagent (Invitrogen), synthesized cDNA with HiScript Q RT SuperMix for qPCR Kit (Vazyme Biotech). Quantitative realtime PCR was performed on ABI-7500 using UltraSYBR Mixture Kit (CWBIO Biotech). The primers using for qPCR analysis were listed as followed: LINC00460 Forward: ACGCAGTGGATGAGAACGAA, LINC00460 Reverse: GGGGTGACTTCAGAATGCGT, 18S rRNA Forward: GTAACCCGTTGAACCCCATT, 18S rRNA Reverse: CCATCCAATCGGTAGTAGCG.
Cell Counting Kit 8 (CCK-8)
CCK-8 (Beyotime, China) was used to measure cell proliferative capacity. 96-well plates planted with transfected cells (5×103 cells/group) were incubated at 37 °C with 5% CO2 for 24 h, 48 h, 72 h. 10 µl of CCK-8 solution was added to each cell well at each time point and incubate for 2h subsequently. The absorbance at 450 nm of each group was tested by the microplate reader.
Transwell Assays
Firstly, ACHN or 786-O cells were planted into the upper wells of chambers (BD Biosciences, USA) coated with or without Matrigel (BD Biosciences), then, cultured in a 200 μL FBS-free RMPI-1640 or DMEM medium. The lower wells of the chambers containing 390 μL culture medium and 10 μL FBS. 24 hours later, removed the medium of the upper wells, and fixed cells with 100% methanol and then stained with 0.1% crystal violet for 15 min. The stained cells were photographed using an Olympus microscope.
Wound Healing Assays
Gaps in seeded ACHN or 786-O cells were created by a sterilized pipette tip. After washing out the debris or the detached cells by PBS, the cells were cultured in RMPI-1640 or DMEM for another 24 hours. The calculation of wound healing width was using ImageJ Analysis Software.
Flow cytometry
Flow cytometry (BD, UA) was used to examine cell apoptosis. No stimulation was given to RCC cells to produce apoptosis. EDTA-free trypsin digested 786-O and ACHN cells. Then, the cells were suspended in 1× binding buffer at 3×106/mL. Subsequently, 100 μL of cells were moderately filled with each of 5 μL of APC and PI, followed by incubation with no light for 5 min at room temperature. Cell apoptosis was analyzed using the flow cytometer. The number of Annexin-APC positive cells represented cell apoptosis rate analyzed by FlowJo v10.6.2 software.
Western Blot
The total content of cellular protein harvested from RCC cells were extracted using RIPA lysis buffer (Keygen, Nanjing, China) and the protein amounts was determined using a BCA kit (Keygen, Nanjing, China). SDS-PAGE separated same volumes proteins of each lane, then blotted onto PVDF membranes. Primary antibodies were incubated overnight at 4 °C, and HRP-labeled secondary antibody (ABclonal, 1:5,000) 2 hours at RT after blocked with 5% BSA. Signal of immunoreactivities were photographed by ECL reagent (NCM Bio, China) on Tanon 5200 automatic chemiluminescence imaging system (Tanon, China). Antibody listed as follow: Anti-E-cadherin (BD Biosciences, 610181), Anti-Vimentin (Proteintech, 10366-1-AP), Anti-N-cadherin (BD Biosciences, 610920), Anti-PI3k (Proteintech, 67121-1-Ig), Anti-AKT (Proteintech, 101762-2-AP), Anti-p-AKT (Proteintech, 66444-1-Ig), Anti-p-mTOR (Cell Signaling Technology, 5536S), Anti-Bcl-2 (Cell Signaling Technology, 155071S), Anti-Cleved-Caspaes 9 (Cell Signaling Technology, 20750S), Anti-p53 (Proteintech, 10442-1-AP), and we took GAPDH(Santa Cruz, sc-32233) as control.
Animal Works
We used BALB/c mice (Vital River Laboratory Animal Technology. China) for research. We subcutaneously injected 786-O cells (5×106) of each group (shCtrl and shLINC00460) into the two side of mice. When tumors were visible, we recorded xenograft tumors volume (V) every 3 days by metering the long axis (L) and the short axis (W), and calculated the tumors' growth rate with the equation: V = (L × W2)/2. By day 27, the xenograft tumor tissues were harvested, weighed, photographed and subjected to subsequent analysis.
Meanwhile, shCtrl and shLINC00460 786-O-Luc cells (3 × 106) were injected intravenously via the mice tail vein. Bioluminescence images were filmed after 6 weeks (Night OWL II LB983; Berthold Technologies). Animal Care and Use Committee and Ethics Committee of Xuzhou Medical University approved all animal experiments.
Statistical Analysis
All collected data were analyzed using Statistical Product and Service Solutions (SPSS 23.0) (IBM, USA) and presented in form of mean ± standard deviation. Student's t-test were used to tested the differences between two groups. Comparison among three groups was analyzed using One-way ANOVA test. p<0.05 suggested statistically significant differences.
Results
LINC00460 expression was elevated and closely linked to poor prognosis in RCC
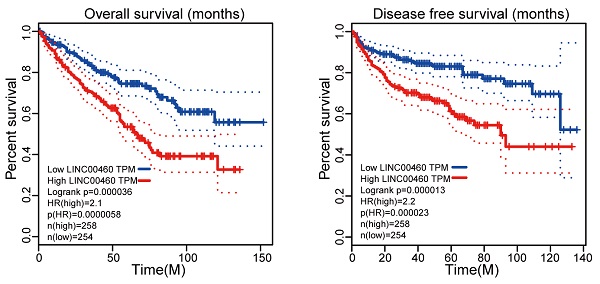
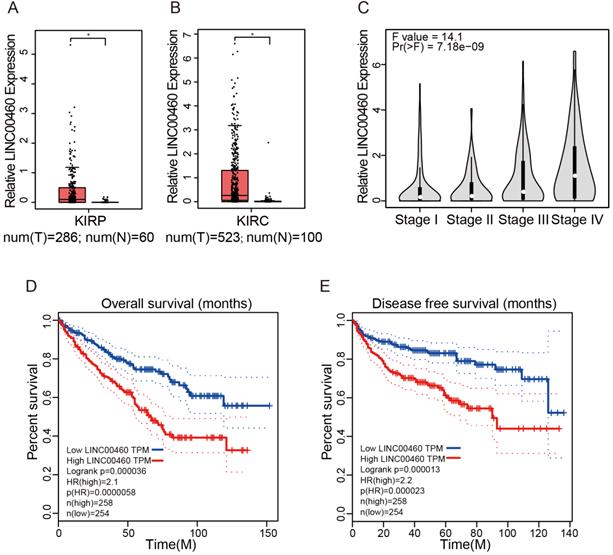
GEPIA database with TCGA datasets were used to obtain LINC00460 expression in RCC tumor tissues paired with normal tissues and the survival of RCC patients. GEPIA with TCGA database revealed that LINC00460 was higher level both in Kidney renal clear cell carcinoma (KIRP) and Kidney renal papillary cell carcinoma (KIRP) (Fig. 1A, 1B). In addition, clinicopathological staging are important prognostic factors for RCC patients, we investigated the LINC00460 expression patterns in different clinical pathological grade of RCC in GEPIA database, we identified a gradual increase in the expression of LINC00460 with the RCC pathological progression (Fig. 1C). Furthermore, RCC patients with high expression of LINC00460 exhibited a shorter OS and DFS than those with low expression of LINC00460 (Fig. 1D, 1E). The results indicated that aberrant expression of LINC00460 might be strongly associated with poor prognosis in RCC.
LINC00460 was elevated in RCC cell Lines and promoted cell proliferation
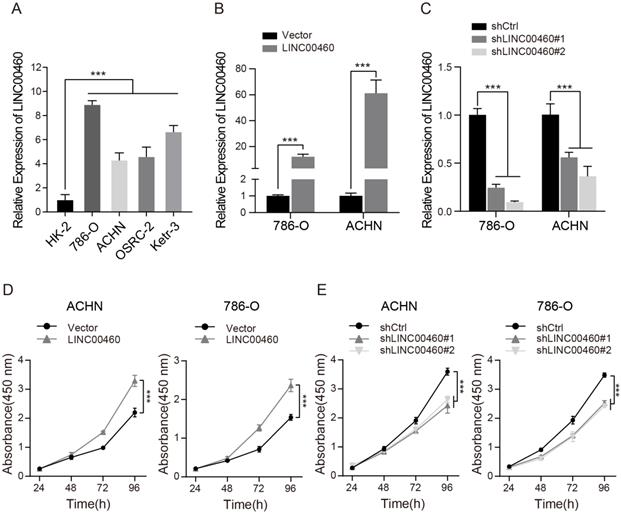
We detect the expression of LINC00460 in RCC cells to explore the effect of LINC00460 on RCC progression. qRT-PCR analysis demonstrated that LINC00460 was significantly upregulated in RCC cell lines compared with normal renal cell (Fig. 2A). Subsequently, qRT-PCR was used to measure the LINC00460 expression level after different transfection applied to the cells, and the results showed that LINC00460 was significantly upregulated in cells transfected with LINC00460 overexpression vector (Fig. 2B), while LINC00460 was remarkably downregulated in cells transfected with the pLKO.1-shRNA, compared with the control vector group (Fig. 2C). Next, we tested cell proliferation by CCK-8 assay. The data of CCK-8 revealed that upregulated LINC00460 expression increased cell proliferation as relative to the control groups (Fig. 2D), while downregulated LINC00460 expression suppressed cell proliferation (Fig. 2E).
LINC00460 facilitated RCC cell migration, invasion and induced EMT phenotype in vitro
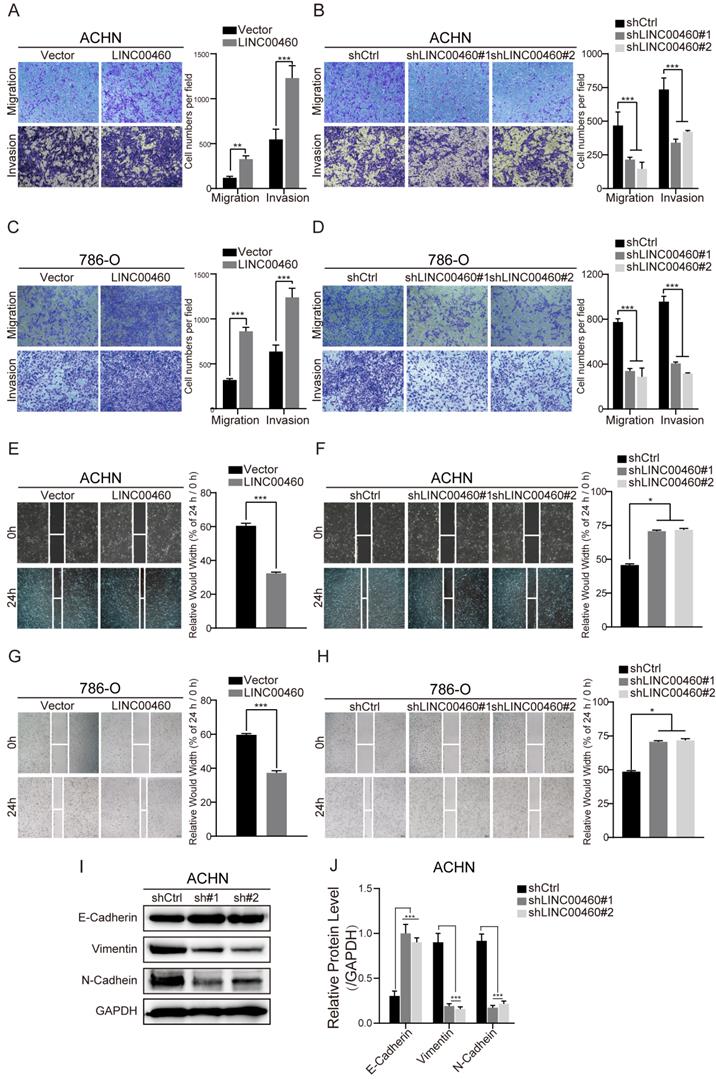
In transwell assays, we showed that LINC00460 overexpression enhanced ACHN and 786-O cells migration and invasion (Fig. 3A, 3C), LINC00460 knockdown inhibited migration and invasion of ACHN and 786-O cells (Fig. 3B, 3D). We conformed the same results that LINC00460 overexpression enhanced cell migration by wound healing assays (Fig. 3E, 3G), while LINC00460 knockdown suppressed cell migration (Fig. 3F, 3H) respectively.
Expression of LINC00460 in RCC tissues based on the data from the GEPIA and TCGA. (A, B) Expression level of LINC00460 in KIRP tissues (n=286) and normal tissues (n=60), KIRC tissues (n=523) and normal tissues (n=100), analyzed in TGCA database (fold change>2.0, * p<0.05). (C) LINC00460 was gradually elevated with advanced staging in RCC. (D, E) OS and DFS rate of RCC patients with lowly or highly LINC00460 analyzed using the Kaplan-Meier analyses and log-rank test.
LINC00460 expression level in RCC cell lines and effects on cell proliferation. (A) Expression patterns of LINC00460 in RCC cell lines and normal renal cell lines detected by qRT-PCR (*** p<0.001). (B, C) Expression patterns of LINC00460 in RCC cells treated with over-expressed or silenced LINC00460 detected by qRT-PCR (*** p<0.001). (D, E) CCK-8 assay of ACHN and 786-O cells transfected with over-expressed or silenced LINC00460 (*** p<0.001).
We detect the expression of EMT-related proteins inevitably, as the outcome obtained from Western blot analysis (Fig. 3I, 3J). Knockdown of LINC00460 increased the levels of EMT marker E-cadherin and decreased N-cadherin and Vimentin protein levels in ACHN cells. Consistent with the results, we can make the conclusion that LINC00460 could accelerate the invasion, migration and induced EMT progress of RCC cells.
LINC00460 affected the apoptosis of RCC cells
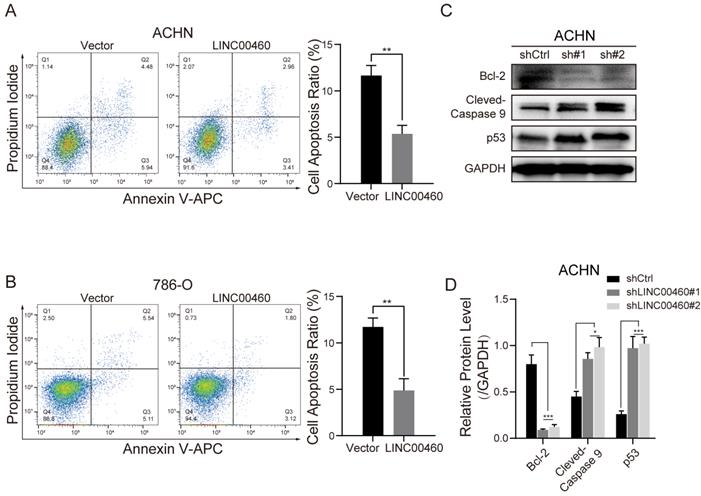
Flow cytometry and Western blot assay were associated to verify whether promotion of cell malignant progress by LINC00460 was associated with cell apoptosis. Compared with shCtrl group, apoptotic cells were decreased in the LINC00460 over-expressed ACHN or 786-O cells (Fig. 4A, 4B). Anti-apoptotic protein Bcl-2 was diminished after LINC00460 knockdown, conversely, pro-apoptotic protein Cleaved Caspase-9 and p53 expression were increased in ACHN cells (Fig. 4C, 4D). All of these results proved that LINC00460 affects the malignant progression of RCC cells by affecting apoptosis.
LINC00460 promoted RCC cell growth and metastasis in vivo
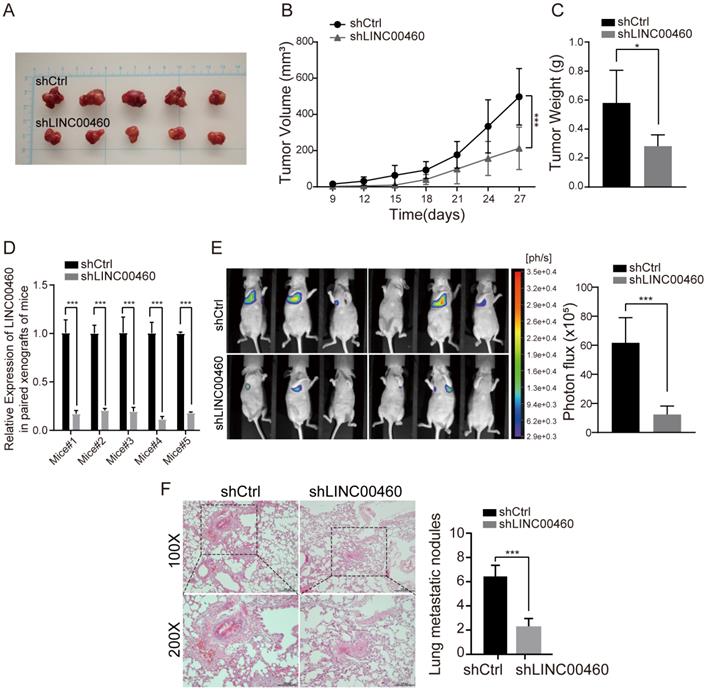
For the assessment of LINC00460 influence on tumor growth in vivo, nude mice were injected subcutaneously with transfected 786-O cells. Knockdown of LINC00460 noticeable suppressed proliferation of tumor cells (Fig. 5A), as presented by the markedly reduced tumor volume, size and weights in the knockdown group (Fig. 5B, 5C). Furthermore, the qRT-PCR results confirmed that the significantly lower level of LINC00460 in shLINC00460-treated groups of xenograft tumor tissues compared with shCtrl groups (Fig. 5D).
To further evaluate LINC00460 knockdown could produce inhibition effect on metastasis in vivo, we administered tail vein injections with 786-O-Luc cells stably transduced shLINC00460 or shCtrl vector into two groups of mice each respectively. Six weeks after tail vein injections, we used bioluminescence photograph system to assess the metastatic nodules formed on the lung surfaces of mice. Group with LINC00460 knockdown cells formed a smaller number of metastatic foci in lungs than the control group (Fig. 5E, 5F). Overall, the data demonstrated that LINC00460 downregulated could inhibit RCC cell growth and metastasis in vivo.
LINC00460 facilitated RCC cells migration, invasion in vitro. (A, B, C, D) The cell migration and invasion abilities of ACHN and 786-O cells transfected with overexpressed or silenced LINC00460 were determined by Transwell assays (*** p<0.001). (E, F, G, H) Wound healing assay was performed to examine the effect of LINC00460 overexpression or knockdown on ACHN and 786-O cells migration (* p<0.05, *** p<0.001). (I, J) EMT markers was detected by Western blot analysis when silenced LINC00460 was transfected in RCC cells. Data statistics was also shown (GAPDH as negative control, *p < 0.05, *** p<0.001).
Influence of LINC00460 on RCC cells apoptosis and EMT phenotype. (A, B) Cell apoptosis was determined using flow cytometry analysis after LINC00460 overexpression in RCC cell lines (** p<0.01). (C, D) Cell apoptosis markers were detected by Western blot analysis when silenced LINC00460 was transfected in RCC cells. Data statistics was also shown (GAPDH as negative control, *p < 0.05, *** p<0.001).
Dysregulation of LINC00460 was related to PI3K/AKT pathway
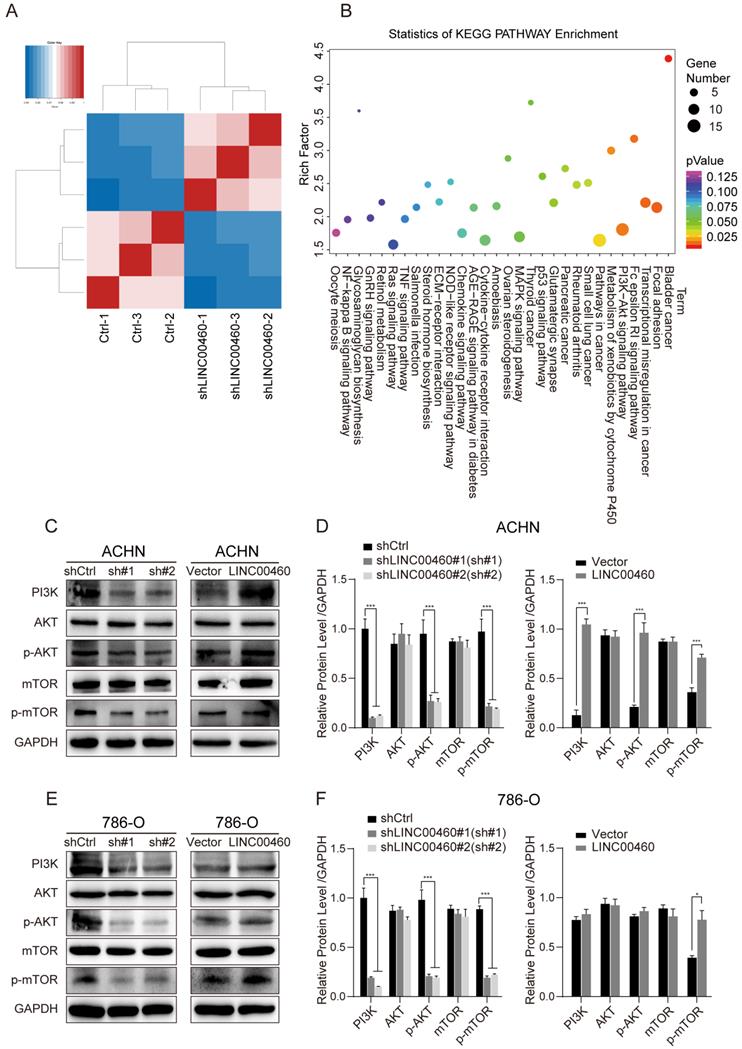
To explore more particular details about the LINC00460 involved pathway in RCC, RNA-seq was processed after LINC00460 knockdown in 786-O cells (Fig. 6A). To get further investigation of LINC00460 worked in RCC, Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis was conducted for looking the target genes of LINC00460 (>1.5 fold change). KEGG pathway clustering indicated that the significantly over-represented biological pathways mapping in cell motility, cell growth and death, cell repair and replication. Markedly, the dysregulated key genes that were related to the PI3K/AKT pathways (Fig. 6B).
PI3K/AKT pathway was involved in different cancer cell behaviors, especially cell proliferation and apoptosis [25]. Thus, we investigated whether LINC00460 control cancer cell progress by the PI3K/AKT pathway. The Western blot assay revealed a reduction of PI3K, p-AKT and p-mTOR expression in ANCH and 786-O cells transfected with shLINC00460 as relative to the control groups. Meanwhile, the expression of PI3K, p-AKT and p-mTOR were increased in cells transfected with overexpressed LINC00460 compared with the control groups (Fig. 6C-F). These results suggested that dysregulation of LINC00460 could regulate the PI3K/AKT pathway.
Discussion
Renal cell carcinoma (RCC) is a heterogenous cancer consisting of various different subtypes [26], and considered as a stepwise process involving the accumulation of multiple genetic and epigenetic alterations [27]. The most critical biological characteristics of RCC are uncontrolled cellular proliferation, abnormal apoptosis and metastasis, which are the leading cause of mortality [28]. Unfortunately, many patients have been in advanced RCC stage when diagnosed due to no effective diagnosis biomarker and a poor understanding of the mechanism.
To date, LncRNAs have been known as active biological marker in cancer diagnosis rather than transcriptional noise [29]. They were proved to play pivotal roles in carcinogenesis via regulating various important cellular biological behavior, including proliferation, apoptosis, angiogenesis, invasion and metastasis [30]. In cervical cancer, upregulation of LncRNA ZEB1-AS1 enhances cell epithelial to mesenchymal transition by elevating ZEB1 level [31]. LncRNA UCA1 is increased in thyroid cancer and represses cell proliferation and cell invasion by interacting with miR-204/IGFBP5 [32]. LncRNA CA3-AS1 restraint of colorectal cancer cell vital, invasion by miR-93/PTEN axis [33]. The downregulation of LINC00152 suppresses the development of gastric cancer through controlling miR-193b-3p [34]. To our knowledge, LINC00460 has been reported to participate in many kinds of malignancies and promotes cancer progressions.
In the present research, LINC00460 have been proved overexpressed in RCC according to the TCGA analysis of clinical specimens, higher LINC00460 expression level was correlated with poor OS and DFS and advanced clinicopathological staging in RCC patients, which is key finding of prognostic significance of LINC00460 for RCC patients. Our work is the initial presentation of LINC00460 biologic function, correlated with progress of RCC cells. A series of assays In vitro and in vivo revealed that LINC00460 over-expression enhanced cell proliferation, migration, and invasion, while its down-regulation suppressed tumor growth and reduced cell migration and invasion. Flow cytometry assays showed that overexpressed LINC00460 inhibited apoptosis in RCC cells. To routinely and precisely unravel the pathways that were related to the function of LINC00460 in tumorigenesis of RCC, RNA-seq and date analysis was conducted, and PI3K/AKT pathway was found to be regulated by LINC00460. Then we confirmed this finding via Western blot assays. These findings indicate that LINC00460 plays an oncogenic role in RCC development and is potential prognostic marker for RCC patients.
LINC00460 promoted RCC cells growth and metastasis in vivo. (A) 786-O cells were injected into nude mice after the transfection of shLINC00460 and shCtrl vector. (B) The growth curves of tumors from subcutaneously injected nude mice treated with shLINC00460 or shCtrl (*** p<0.001). (C) The weights of tumors from subcutaneously injected nude mice treated with shLINC00460 or shCtrl (* p<0.05). (D) The levels of LINC00460 expression in paired tumor tissues formed from subcutaneously injected nude mice treated with shLINC00460 or shCtrl, determined by qRT-PCR (*** p<0.001). (E) Representative bioluminescence images and statistical analysis of lung metastases in mice via tail vein injection of indicated cells (**p < 0.01, ***p < 0.001). (F) Representative H&E images and statistical analysis of resected lung xenografts with metastatic loci (*** p < 0.001).
Effect of LINC00460 knockdown on activation of PI3K/AKT pathway in RCC cell lines. (A) Hierarchically clustered heatmap of the upregulated and downregulated genes in 786-O cells after shLINC00460 and shCtrl transfections. (B) Pathway classification of differentially expressed genes (DEGs). Bubble plots represented the number of DEGs, x axis represented rich factor, y axis represented the functional classification of KEGG. (C, D, E, F) The Western blot assay was used to detect PI3K, AKT, p-AKT, mTOR and p-mTOR expression in cells transfection with shLINC00460 or overexpressed LINC00460 as relative to the control groups. (GAPDH as negative control, *p < 0.05, *** p<0.001).
Emerging studies have identified that the PI3K/AKT signaling pathway influences various pathophysiological progress that associated with cancer phenotypes, such as cell apoptosis and cell proliferation [35-40]. PI3K activation phosphorylates AKT and active AKT can lead to a number of downstream effects including the activation of mTOR, also in the form of phosphorylates mTOR, which in turn directly impacts cell growth and survival [41-43]. The crucial influence of the activation of AKT is the increasing cell viability [44], and involved in cellular tumorigenicity behaviors such as proliferation, metastasis, invasion, migration, and angiogenesis [45, 46]. Therefore, in accordance with our RNA-seq data analysis, we found that the knockdown of LINC00460 downregulated the expression of p-mTOR, p-AKT and PI3K, confirmed our hypothesis that LINC00460 might act as one upstream of PI3K/AKT pathway to control RCC progression. Furthermore, our findings displayed in Western blot assay indicating that LINC00460 could promote tumor migration and invasion via EMT.
Conclusions
In conclusion, our findings illustrated that LINC00460 expression was strongly upregulated in RCC tissues and cells, consequently, its higher expression might be associated with poor clinical prognosis in RCC patients, which enables it as an attractive prognostic factor for RCC. LINC00460 influenced tumor growth, metastasis and apoptosis in vitro and in vivo. In the aspect of signaling pathway, LINV00460 activating the PI3K/AKT pathway. Our findings might display a novel insight into the mechanism of LINC00460 in RCC. The mechanism about LINC00406 how to regulate PI3K/AKT signaling pathway are still needed our further study to verify. Further clinical experiments are still needed to verify.
Abbreviations
RCC: Renal cell carcinoma; KIRC: Kidney renal clear cell carcinoma; KIRP : Kidney renal papillary cell carcinoma; KICH: Kidney Chromophobe; LincRNA: Long intergenic noncoding RNA; GEPIA: Gene Expression Profiling Interactive Analysis; TCGA: The Cancer Genome Atlas; qRT-PCR: Quantitative real-time polymerase chain reaction; RNA-seq: RNA sequencing; OS: Overall survival; DFS: Disease-free survival; LncRNAs: Long noncoding RNAs; LINC00460: Long intergenic noncoding RNA 460; PI3K: The phosphatidylinositol 3-kinase; mTOR: Mammalian target of rapamycin; p-AKT: Phosphorylated AKT; p-mTOR: Phosphorylated mTOR; EMT: Epithelial-mesenchymal transition; PI: Propidium Iodide.
Acknowledgements
This work was financially supported by several grants from the National Natural Science Foundation of China (No. 82072649, 81872304), the Education Department of Jiangsu Province (No. 19KJA130001), the Outstanding Youth Foundation of Jiangsu Province (BK20200046).
Ethics approval and consent to participate
The present study was approved by the Animal Care and Use Committee of Xuzhou Medical University (approval no. 2019-92; Xuzhou, China). All experiments were performed in accordance with the approved National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Ethics approval
This study was performed under a protocol approved by the Institutional Review Boards of the Affiliated Hospital of Xuzhou Medical University. Animal experiments were performed in accordance with National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. All animal experiments were approved by the Animal Care and Use Committee and Ethics Committee of Xuzhou Medical University.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
PF Hou provided study concept and design. J Bai provided study concept. ML Li and S Meng collected the patients' samples and performed the experiments. SF Chu, S Meng and FJ Zhou collected, analyzed and interpreted the data. FJ Zhou and S Meng wrote the manuscript.
Availability of online data
Publicly available datasets were analyzed in this study. This data can be found here: http://gepia.cancer-pku.cn/detail.php?gene=LINC00460.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Fisher R, Gore M, Larkin J. Current and future systemic treatments for renal cell carcinoma. Semin Cancer Biol. 2013;23:38-45
3. Kenney PA, Wood CG. Integration of surgery and systemic therapy for renal cell carcinoma. Urol Clin North Am. 2012;39:211-31 vii
4. Powles T, Staehler M, Ljungberg B, Bensalah K, Canfield SE, Dabestani S. et al. European Association of Urology Guidelines for Clear Cell Renal Cancers That Are Resistant to Vascular Endothelial Growth Factor Receptor-Targeted Therapy. Eur Urol. 2016;70:705-6
5. Kotecha RR, Motzer RJ, Voss MH. Towards individualized therapy for metastatic renal cell carcinoma. Nat Rev Clin Oncol. 2019;16:621-33
6. Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177-86
7. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7-21
8. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139:269-80
9. Worku T, Bhattarai D, Ayers D, Wang K, Wang C, Rehman ZU. et al. Long Non-Coding RNAs: the New Horizon of Gene Regulation in Ovarian Cancer. Cell Physiol Biochem. 2017;44:948-66
10. Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77-84
11. Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y. et al. lncRNAs HIF1A-AS2 facilitates the up-regulation of HIF-1alpha by sponging to miR-153-3p, whereby promoting angiogenesis in HUVECs in hypoxia. Biomed Pharmacother. 2017;96:165-72
12. Webb A, Papp AC, Curtis A, Newman LC, Pietrzak M, Seweryn M. et al. RNA sequencing of transcriptomes in human brain regions: protein-coding and non-coding RNAs, isoforms and alleles. BMC Genomics. 2015;16:990
13. Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:279
14. Hu Y, Deng C, Zhang H, Zhang J, Peng B, Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/beta-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8:94554-68
15. Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8:101984-93
16. Wang F, Liang S, Liu X, Han L, Wang J, Du Q. LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR-342-3p in gastric cancer. Onco Targets Ther. 2018;11:6383-94
17. Di W, Li Q, Shen W, Guo H, Zhao S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. Am J Cancer Res. 2017;7:1298-309
18. Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y. et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80-90
19. Liang Y, Wu Y, Chen X, Zhang S, Wang K, Guan X. et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017 37
20. Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261-74
21. Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550-62
22. Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140-56
23. Zhang HB, Lu P, Guo QY, Zhang ZH, Meng XY. Baicalein induces apoptosis in esophageal squamous cell carcinoma cells through modulation of the PI3K/Akt pathway. Oncol Lett. 2013;5:722-8
24. Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD. et al. G protein-coupled receptor-mediated activation of p110beta by Gbetagamma is required for cellular transformation and invasiveness. Sci Signal. 2012;5:ra89
25. Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627-44
26. Akhtar M, Al-Bozom IA, Al Hussain T. Papillary Renal Cell Carcinoma (PRCC): An Update. Adv Anat Pathol. 2019;26:124-32
27. Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S. et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615-21
28. Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M. et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913-24
29. Diederichs S, Bartsch L, Berkmann JC, Frose K, Heitmann J, Hoppe C. et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med. 2016;8:442-57
30. Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S. et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89
31. Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J. et al. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer. 2017;8:4106-16
32. Liu H, Li R, Guan L, Jiang T. Knockdown of lncRNA UCA1 inhibits proliferation and invasion of papillary thyroid carcinoma through regulating miR-204/IGFBP5 axis. Onco Targets Ther. 2018;11:7197-204
33. Wei H, Yang Z, Lin B. Overexpression of long non coding RNA CA3-AS1 suppresses proliferation, invasion and promotes apoptosis via miRNA-93/PTEN axis in colorectal cancer. Gene. 2019;687:9-15
34. Wang H, Chen W, Yang P, Zhou J, Wang K, Tao Q. Knockdown of linc00152 inhibits the progression of gastric cancer by regulating microRNA-193b-3p/ETS1 axis. Cancer Biol Ther. 2019;20:461-73
35. Lee Y, Lee JY, Kim MH. PI3K/Akt pathway regulates retinoic acid-induced Hox gene expression in F9 cells. Dev Growth Differ. 2014;56:518-25
36. Holand K, Boller D, Hagel C, Dolski S, Treszl A, Pardo OE. et al. Targeting class IA PI3K isoforms selectively impairs cell growth, survival, and migration in glioblastoma. PLoS One. 2014;9:e94132
37. Thang ND, Yajima I, Kumasaka MY, Iida M, Suzuki T, Kato M. Deltex-3-like (DTX3L) stimulates metastasis of melanoma through FAK/PI3K/AKT but not MEK/ERK pathway. Oncotarget. 2015;6:14290-9
38. Arrighi N, Bodei S, Zani D, Michel MC, Simeone C, Cosciani Cunico S. et al. Different muscarinic receptor subtypes modulate proliferation of primary human detrusor smooth muscle cells via Akt/PI3K and map kinases. Pharmacol Res. 2013;74:1-6
39. Fang J, Xia C, Cao Z, Zheng JZ, Reed E, Jiang BH. Apigenin inhibits VEGF and HIF-1 expression via PI3K/AKT/p70S6K1 and HDM2/p53 pathways. FASEB J. 2005;19:342-53
40. He D, Zhang S. UNBS5162 inhibits the proliferation of esophageal cancer squamous cells via the PI3K/AKT signaling pathway. Mol Med Rep. 2018;17:549-55
41. King D, Yeomanson D, Bryant HE. PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol. 2015;37:245-51
42. Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348-61
43. Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol. 2011;93:182-203
44. Cucina A, Dinicola S, Coluccia P, Proietti S, D'Anselmi F, Pasqualato A. et al. Nicotine stimulates proliferation and inhibits apoptosis in colon cancer cell lines through activation of survival pathways. J Surg Res. 2012;178:233-41
45. Candeias E, Sebastiao I, Cardoso S, Carvalho C, Santos MS, Oliveira CR. et al. Brain GLP-1/IGF-1 Signaling and Autophagy Mediate Exendin-4 Protection Against Apoptosis in Type 2 Diabetic Rats. Mol Neurobiol. 2018;55:4030-50
46. Jiang C, Ma S, Hu R, Wang X, Li M, Tian F. et al. Effect of CXCR4 on Apoptosis in Osteosarcoma Cells via the PI3K/Akt/NF-kappabeta Signaling Pathway. Cell Physiol Biochem. 2018;46:2250-60
Author contact
![]() Corresponding authors: Jun-Nian Zheng, Cancer Institute, Xuzhou Medical University, 84 West Huaihai Road, Xuzhou, 221002, Jiangsu Province, China. E-mail: jnzhengedu.cn; Jin Bai, Cancer Institute, Xuzhou Medical University, 84 West Huaihai Road, Xuzhou, 221002, Jiangsu Province, China. E-mail: bjedu.cn.
Corresponding authors: Jun-Nian Zheng, Cancer Institute, Xuzhou Medical University, 84 West Huaihai Road, Xuzhou, 221002, Jiangsu Province, China. E-mail: jnzhengedu.cn; Jin Bai, Cancer Institute, Xuzhou Medical University, 84 West Huaihai Road, Xuzhou, 221002, Jiangsu Province, China. E-mail: bjedu.cn.

 Global reach, higher impact
Global reach, higher impact