Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(12):3342-3347. doi:10.7150/jca.76255 This issue Cite
Research Paper
WDR4 gene polymorphisms increase hepatoblastoma susceptibility in girls
1. Department of Pediatric Surgery, Shengli Clinical Medical College of Fujian Medical University, Fuzhou 350001, Fujian, China.
2. Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China.
3. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
4. Department of Pediatric Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China.
5. Department of Pediatric Surgery, Hunan Children's Hospital, Changsha 410004, Hunan, China.
6. Department of Pediatric Surgery, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning, China.
7. Department of Pediatric Surgery, the Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, Shaanxi, China.
8. Department of Pathology, Children Hospital and Women Health Center of Shanxi, Taiyuan 030013, Shannxi, China.
9. Kunming Key Laboratory of Children Infection and Immunity, Yunnan Key Laboratory of Children's Major Disease Research, Yunnan Institute of Pediatrics Research, Yunnan Medical Center for Pediatric Diseases, Kunming Children's Hospital, Kunming 650228, Yunnan, China.
#These authors contributed equally to this work.
Received 2022-6-17; Accepted 2022-9-13; Published 2022-9-21
Abstract
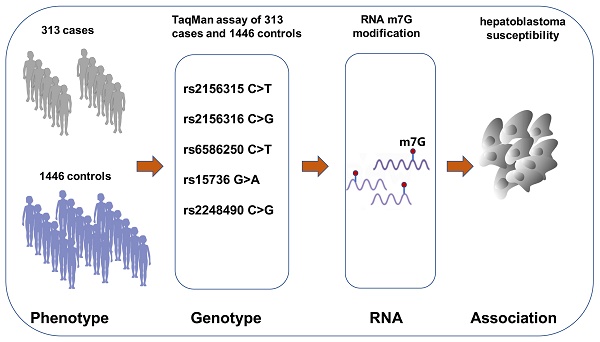
Hepatoblastoma, originating from hepatoblasts, is the most common hepatic malignancy. WD repeat domain 4 (WDR4) is a subunit of RNA N(7)-methylguanine (m7G) methyltransferase complex. Recently, WDR4 has shown oncogenic potential in various adult cancers, but its roles in pediatric cancers have not been reported. We performed a case-control study (313 cases vs. 1446 controls) to investigate whether genetic variants in the WDR4 gene influence hepatoblastoma susceptibility in the Chinese Han nationality. We first determine the genotypes of five WDR4 gene polymorphisms (rs2156315 C>T, rs2156316 C>G, rs6586250 C>T, rs15736 G>A, rs2248490 C>G) in participants, using the Taqman assay. And then, an unconditional logistic regression analysis was performed to assess the association between WDR4 gene polymorphisms and hepatoblastoma risk. Overall, we did not find any polymorphism significantly associated with the risk of developing hepatoblastoma. Instead, the stratified analysis revealed that the co-existence of 2-5 risk genotypes increased hepatoblastoma risk by 2.23 folds in girls (adjusted odds ratio=2.23, 95% confidence interval=1.17-4.23, P=0.014). In conclusion, our results demonstrate that single selected polymorphisms were too weak to exert a significant effect on the whole study population. However, in combination, two or more WDR4 gene polymorphisms significantly conferred increased hepatoblastoma risk in girls. Our findings may encourage more genetic association studies to discover significant polymorphisms in the WDR4 gene for hepatoblastoma.
Keywords: hepatoblastoma, susceptibility, WDR4, polymorphism, m7G
Introduction
Hepatoblastoma is an embryonal tumor arising from hepatoblast, a hepatocyte precursor cell, characterized by heterogeneous histological patterns with the presence of different stages of liver development cancer [1]. Even though the annual incidence is as low as 1.5 cases per million, hepatoblastoma is the most common hepatic malignancy in childhood. Most hepatoblastoma-affected children are below the age of 3 years, and the median age at the time of diagnosis is one year.
Epidemiology studies suggest prematurity, low birth weight, and parental smoking may be the risk for hepatoblastoma [2]. Apart from developmental processes and environmental factors, genetic predisposition is also a major factor influencing the risk of childhood cancer [3]. Several inherited syndromes have predisposed children to hepatoblastoma, including Familial Adenomatous Polyposis (FAP) with inactivating mutations of the APC gene, Beckwith-Wiedemann syndrome (BWS) with defective imprinting of the IGF2-H19 locus on chromosome 11p15, and Trisomy 18 (Edwards Syndrome) [2]. Besides the major predisposition syndromes, more subtle genetic factors are also involved in hepatoblastoma susceptibility. The Nagae group integratively and comprehensively analyzed the genetic and epigenetic landscape of hepatoblastoma in a Japanese cohort. They revealed telomerase reverse transcriptase (TERT) promoter mutations and the specific hypomethylated enhancers in the transcription factor ASCL2 targeted genes in hepatoblastoma [4]. Moreover, susceptibility loci (e.g., genetic polymorphisms) can be inherited, leading to an increased risk of diseases in descendants. Several teams and ours have shown that hereditary single nucleotide polymorphisms (SNPs) in a number of genes can alter hepatoblastoma susceptibility [5-11]. Due to the disease's rarity, molecular epidemiological studies on hepatoblastoma are extraordinarily few and lagged behind adult cancers. Therefore, it is imperative to identify more hepatoblastoma predisposing genes with case-control studies.
The human chromosome 21q22.3 gene WD repeat domain 4 (WDR4) encodes a subunit of a methyltransferase complex required for the formation of N(7)-methylguanine (m7G) [12, 13]. WDR4 complexes with and facilitates METTL1, the catalytic component, to install m7G in the cap structure of mRNA and specific sites internally within tRNA, rRNA, miRNA, and mRNA [14-19]. Accumulating evidence has suggested WDR4 as an oncogenic gene [14, 20-23]. Pan-cancer analysis of 33 types of cancer indicated WDR4 was aberrantly upregulated in various cancers [20]. Recently, two critical studies demonstrated that the METTL1/WDR4 complex increased oncogenic mRNA translation and drove malignant transformation by facilitating m7G modification of a subset of tRNAs [21, 22]. The implications of WDR4 in hepatoblastoma have not been reported, but it was reported to facilitate proliferation, metastasis, and sorafenib resistance in hepatocellular carcinoma [23]. Because of its tumorigenic role, we analyzed five WDR4 gene polymorphisms in 313 pediatric patients and 1446 controls to explore whether functional WDR4 gene variants can modify individuals' cancer susceptibility.
Materials and Materials
Study population
The pediatric cohort consisted of 313 children with hepatoblastoma collected from seven independent hospitals across China. All patients were children with newly diagnosed hepatoblastoma, which two or more pathologists confirmed. Hepatoblastoma patients were recruited based on the following inclusion and exclusion criteria: 1) Han Chinese descendants, 2) newly diagnosed and histopathologically validated hepatoblastoma, 3) lack of familial disorder or family history of cancer, and 4) ≤14 years old. Patients receiving the medical intervention or failing to provide signed informed consent would be excluded. The locations of these hospitals were as follows: Guangzhou, Kunming, Xi'an, Zhengzhou, Changsha, Taiyuan, and Shenyang. We also matched cases with 1446 healthy controls of Chinese Han nationality, who were children visiting the above hospitals for physical examination at the same periods as cases (Table S1). The clinical stages of the patients were determined following the PRETEXT classification [24]. Information on subject recruitments and participants' epidemiological and clinical characteristics were provided elsewhere [9, 11]. Signed informed consent by parents or guardians of each subject was a prerequisite for blood withdrawal. The study protocol was approved by the institutional review board of Guangzhou Women and Children's Medical Center (No: 202016601).
Selection and genotyping of SNPs
Based on standard criteria [25, 26], we selected five potential functional SNPs (rs2156315 C>T, rs2156316 C>G, rs6586250 C>T, rs15736 G>A, and rs2248490 C>G) in the WRD4 gene. Briefly, we obtained all WDR4 gene SNPs from the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and then selected common, potentially functional SNPs SNPinfo software (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html). These SNPs also are in low linkage disequilibrium (LD) (R2<0.8) with one another and have a minor allele frequency (MAF) equal to or larger than 5% in the Chinese Han population. The Tiangen Blood DNA Extraction kits (Tiangen Biotechnology, Beijing, China) were adopted to collect the Genomic DNA of all children, followed by genotyping using a TaqMan platform (Applied Biosystems, Foster City, CA). We routinely added both negative and positive control samples to each 384-well plate. Blinded laboratory workers finished all the experiments. Quality control was executed by regularly genotyping a certain fraction of the randomly selected sample. Concordance rates of 100% in duplicate tests were accepted.
Statistical analysis
To determine the significant differences in clinical variables between the case and control groups, t-tests and χ2 tests were performed for continuous or categorical variables, respectively. Hardy-Weinberg equilibrium (HWE) was checked in the control subjects using a goodness-of-fit χ2 test. The association of SNPs and hepatoblastoma susceptibility was estimated using unconditional logistic regression analysis, along with the generation odds ratios (ORs) and 95% confidence intervals (CIs). Children were also stratified regarding age, sex, and clinical stages, and the associations were reanalyzed in subgroups. The nominal significance level α was set at 0.05. All statistical analyses were carried out by SAS v9.1 (SAS Institute Inc., Cary, NC).
Results
Association of hepatoblastoma risk with WDR4 SNPs
We investigated five WDR4 gene SNPs in this study, including rs2156315 C>T, rs2156316 C>G, rs6586250 C>T, rs15736 G>A, and rs2248490 C>G. Out of the 313 hepatoblastoma cases and 1446 controls, the SNPs were successfully genotyped in 308 cases and 1444 controls. All genotyping results for hepatoblastoma patients and controls are displayed in Table 1. We confirmed the consistency with HWE genetic balance in control subjects for the five SNPs (HWE=0.063 for rs2156315 C>T, HWE=0.459 for rs2156316 C>G, HWE=0.897 for rs6586250 C>T, HWE=0.966 for rs15736 G>A, HWE=0.387 for rs2248490 C>G). However, neither single-locus nor combined analysis detected a significant association between the five SNPs and hepatoblastoma risk, suggesting that the effects of these SNPs on susceptibility are very weak.
Stratified analyses
We next performed stratified analysis for rs2156315 because it has the largest OR value of 1.42, although not significant (Table 2). However, this SNP did not show significant association in any subgroups in terms of age, gender, and stages. Regardless of significance, we defined genotypes with OR of >1 as risk genotype: rs2156315 TT, rs2156316 GG, rs6586250 TT, rs15736 AA, and rs2248490 CC/CG. The combined analysis of the five risk genotypes showed that 2-5 risk genotypes carriers were more prevalent in girls than in boys (30% vs. 19%). When compared with girls carrying 0-1 risk genotypes, girl carriers of 2-5 risk genotypes were associated with a 2.23-fold increased risk of hepatoblastoma (adjusted OR=2.23, 95% CI=1.17-4.23, P=0.014).
Discussion
Despite the extremely low incidence of hepatoblastoma, early diagnosis is a critical factor in achieving a high cure rate. Numerous studies have shown that small risk factors like genetic polymorphisms can modify cancer risk in conjunction with adverse environmental factors. It is also the case of hepatoblastoma [3, 27, 28]. However, the rarity of hepatoblastoma constitutes a severe obstacle to launching large case-control studies. As a result, case-control studies on hepatoblastoma are very few, and sample sizes are small. For example, Pakakasama et al. elucidated a significant association between MPO gene promoter SNP (G to A) and reduced risk of hepatoblastoma, but they only recruited 48 cases and 180 healthy controls of Caucasians [5]. Again, with only 84 children with hepatoblastoma, the same team studied found that a nonsynonymous SNP located (G to A) at codon 242 of CCND1 could significantly affect the age of the tumor onset [5]. Therefore, much more work is warranted to elucidate the significance of numerous genetic polymorphisms in hepatoblastoma risk with large case-control studies.
After have being engaged in genetic association studies in pediatric cancers for over a decade, our team has found a number of hepatoblastoma susceptibility genes with 313 cases and 1446 controls, including LINC00673 [29], NRAS [30], KRAS [30], TP53 [31], HMGA2 [32], miR-34b/c [31], as well as DNA repair genes [6, 7]. Moreover, we also identified genetic variants in RNA m6A-mediated genes (i.e., METTL3 [33], METTL14 [34], WTAP [9], YTHDF1 [11], YTHDC1 [10], and ALKBH5 [35]) associated with hepatoblastoma susceptibility. Over the past years, research gradually realized that, like RNA m6A modification, the abundant RNA m7G modifications also have fundamental importance to cellular homeostasis. However, molecular epidemiological studies of s WDR4 genetic variants and disease risk are still in infancy. There is a substantial gap in our knowledge of the relationship between WDR4 gene variants and disease susceptibility. Only one study suggests that WDR4 gene variants may be disease-causing. Wang et al. demonstrated that the WDR4 gene rs465663 polymorphism was associated with asthenozoospermia. Functional annotations using the GTEx portal indicated that the TT or TC genotype of SNP rs465663 was associated with decreased expression of WDR4 compared with the CC genotype. Furthermore, the functional study elucidated that mwdr4 heterozygous (+/-) mouse embryonic fibroblasts were more vulnerable to ROS-induced DNA fragmentation than wild-type counterparts [36]. In the current study, we analyzed five potential functional WDR4 gene SNPs with 313 cases and 1446 controls, and found that Chinese girls bearing two or more risk genotypes were more likely to develop hepatoblastoma than girls with few risk genotypes. Our results provided evidence that WDR4 may be a hepatoblastoma susceptibility gene.
Association between WDR4 gene polymorphisms and hepatoblastoma susceptibility
| Genotype | Cases (N=308) | Controls (N=1444) | Pa | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | Pb |
|---|---|---|---|---|---|---|---|
| rs2156315 C>T (HWE=0.063) | |||||||
| CC | 181 (58.77) | 858 (59.42) | 1.00 | 1.00 | |||
| CT | 109 (35.39) | 526 (36.43) | 0.98 (0.76-1.28) | 0.894 | 0.99 (0.76-1.28) | 0.908 | |
| TT | 18 (5.84) | 60 (4.16) | 1.42 (0.82-2.47) | 0.210 | 1.42 (0.82-2.46) | 0.213 | |
| Additive | 0.521 | 1.07 (0.87-1.32) | 0.520 | 1.07 (0.87-1.32) | 0.515 | ||
| Dominant | 127 (41.23) | 586 (40.58) | 0.833 | 1.03 (0.80-1.32) | 0.832 | 1.03 (0.80-1.32) | 0.822 |
| Recessive | 290 (94.16) | 1384 (95.84) | 0.192 | 1.43 (0.83-2.46) | 0.194 | 1.43 (0.83-2.46) | 0.198 |
| rs2156316 C>G (HWE=0.459) | |||||||
| CC | 136 (44.16) | 640 (44.32) | 1.00 | 1.00 | |||
| CG | 138 (44.81) | 652 (45.15) | 1.00 (0.77-1.29) | 0.976 | 1.00 (0.77-1.30) | 0.999 | |
| GG | 34 (11.04) | 152 (10.53) | 1.05 (0.70-1.60) | 0.809 | 1.05 (0.70-1.60) | 0.808 | |
| Additive | 0.870 | 1.02 (0.84-1.22) | 0.870 | 1.02 (0.84-1.23) | 0.857 | ||
| Dominant | 172 (55.84) | 804 (55.68) | 0.958 | 1.01 (0.79-1.29) | 0.958 | 1.01 (0.79-1.30) | 0.936 |
| Recessive | 274 (88.96) | 1292 (89.47) | 0.791 | 1.06 (0.71-1.56) | 0.791 | 1.05 (0.71-1.56) | 0.798 |
| rs6586250 C>T (HWE=0.897) | |||||||
| CC | 248 (80.52) | 1152 (79.78) | 1.00 | 1.00 | |||
| CT | 56 (18.18) | 275 (19.04) | 0.95 (0.69-1.30) | 0.732 | 0.95 (0.69-1.30) | 0.744 | |
| TT | 4 (1.30) | 17 (1.18) | 1.09 (0.37-3.28) | 0.874 | 1.08 (0.36-3.25) | 0.888 | |
| Additive | 0.822 | 0.97 (0.73-1.29) | 0.823 | 0.97 (0.73-1.29) | 0.826 | ||
| Dominant | 60 (19.48) | 292 (20.22) | 0.768 | 0.95 (0.70-1.30) | 0.768 | 0.96 (0.70-1.30) | 0.777 |
| Recessive | 304 (98.70) | 1427 (98.82) | 0.859 | 1.10 (0.37-3.31) | 0.859 | 1.09 (0.37-3.27) | 0.874 |
| rs15736 G>A (HWE=0.966) | |||||||
| GG | 244 (79.22) | 1146 (79.36) | 1.00 | 1.00 | |||
| GA | 60 (19.48) | 281 (19.46) | 1.00 (0.74-1.37) | 0.986 | 1.01 (0.74-1.38) | 0.964 | |
| AA | 4 (1.30) | 17 (1.18) | 1.11 (0.37-3.31) | 0.858 | 1.10 (0.37-3.29) | 0.867 | |
| Additive | 0.924 | 1.01 (0.77-1.34) | 0.924 | 1.02 (0.77-1.34) | 0.909 | ||
| Dominant | 64 (20.78) | 298 (20.64) | 0.955 | 1.01 (0.75-1.37) | 0.955 | 1.01 (0.75-1.37) | 0.936 |
| Recessive | 304 (98.70) | 1427 (98.82) | 0.859 | 1.10 (0.37-3.31) | 0.859 | 1.10 (0.37-3.28) | 0.869 |
| rs2248490 C>G (HWE=0.387) | |||||||
| CC | 137 (44.48) | 643 (44.53) | 1.00 | 1.00 | |||
| CG | 140 (45.45) | 652 (45.15) | 1.01 (0.78-1.31) | 0.953 | 1.01 (0.78-1.31) | 0.936 | |
| GG | 31 (10.06) | 149 (10.32) | 0.98 (0.64-1.50) | 0.914 | 0.98 (0.64-1.50) | 0.914 | |
| Additive | 0.960 | 1.00 (0.83-1.20) | 0.960 | 1.00 (0.83-1.20) | 0.970 | ||
| Dominant | 171 (55.52) | 801 (55.47) | 0.988 | 1.00 (0.78-1.28) | 0.988 | 1.00 (0.78-1.29) | 0.972 |
| Recessive | 277 (89.94) | 1295 (89.68) | 0.894 | 0.97 (0.65-1.46) | 0.895 | 0.97 (0.65-1.46) | 0.889 |
| Combined effect of risk genotypesc | |||||||
| 0-1 | 282 (91.56) | 1364 (94.46) | 1.00 | 1.00 | |||
| 2-5 | 26 (8.44) | 80 (5.54) | 0.053 | 1.57 (0.99-2.49) | 0.054 | 1.57 (0.99-2.49) | 0.056 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
a χ2 test for genotype distributions between hepatoblastoma patients and controls.
b Adjusted for age and gender.
c Risk genotypes were carriers with rs2156315 TT, rs2156316 GG, rs6586250 TT, rs15736 AA and rs2248490 CC/CG.
Stratification analysis of risk genotypes with hepatoblastoma susceptibility
| Variables | rs2156315 (cases/controls) | AOR (95% CI) a | Pa | Risk genotypes (cases/controls) | AOR (95% CI) a | Pa | ||
|---|---|---|---|---|---|---|---|---|
| CC/CT | TT | 0-1 | 2-5 | |||||
| Age, month | ||||||||
| <17 | 154/608 | 10/33 | 1.21 (0.59-2.52) | 0.603 | 149/600 | 15/41 | 1.50 (0.81-2.78) | 0.202 |
| ≥17 | 136/776 | 8/27 | 1.67 (0.74-3.76) | 0.214 | 133/764 | 11/39 | 1.61 (0.81-3.23) | 0.177 |
| Gender | ||||||||
| Females | 116/567 | 10/28 | 1.75 (0.83-3.69) | 0.145 | 111/561 | 15/34 | 2.23 (1.17-4.23) | 0.014 |
| Males | 174/817 | 8/32 | 1.16 (0.53-2.57) | 0.713 | 171/803 | 11/46 | 1.12 (0.57-2.20) | 0.749 |
| Clinical stages | ||||||||
| I+II | 149/1384 | 10/60 | 1.53 (0.76-3.05) | 0.231 | 146/1364 | 13/80 | 1.50 (0.82-2.77) | 0.191 |
| III+IV | 83/1384 | 6/60 | 1.69 (0.71-4.03) | 0.237 | 80/1364 | 9/80 | 1.93 (0.93-3.99) | 0.076 |
AOR, adjusted odds ratio; CI, confidence interval.
a Adjusted for age and gender, omitting the corresponding factor.
WDR4 is a non-catalytic component of the METTL1/WDR4 methyltransferase complex, essential for the stability and conformational alterations of METTL1, the catalytic subunit [12]. METTL1/WDR4 complex-medicated m7G modification is indispensable in controlling cell fate and growth, and WDR4 has been implicated in carcinogenesis. A missense mutation in WDR4, impairing tRNA m (7) G46 methylation, was identified as a causal variant for microcephalic primordial dwarfism [37]. Knockout of METT1 or WDR4 was detrimental to self-renewal and neural differentiation of mouse embryonic stem cells [16]. Pan-cancer analysis based on the TCGA database revealed aberrant expression of WDR4 in a broad spectrum of cancers and the association with inferior prognosis as well as tumor immunity [20]. WDR4 coupled with METTL1 facilitates the mounting of m7G modification at position 46 in tRNAs. Ma's group found that both WDR4 and METTL1 were significantly upregulated in human lung cancer. In vitro and in vivo evidence indicated that defective m7G tRNA modification resulting from the knockdown of WDR4/METTL1 suppressed cell proliferation, invasion, and tumorigenic capacity, while overexpression of these two methyltransferases promoted tumorigenesis [15]. Xia and colleagues substantiated the upregulated WDR4 in hepatocellular carcinoma (HCC), accompanied by increased m7G methylation levels. Overexpressed WDR4 boosted proliferation and the G2/M cell cycle progression but inhibited apoptosis of HCC cells by enhancing mRNA stability and translation of CCNB1, which encodes the cell cycle protein cyclin B1 [23]. Moreover, elevated METTL1/WDR4 and m7G tRNA modification were also observed in intrahepatic cholangiocarcinoma [22]. Mechanistically, m7G tRNA modification discriminatively enhanced mRNA translation of genes in cell-cycle and epidermal growth factor receptor (EGFR) [22]. Our results indicate WDR4 as a hepatoblastoma predisposition gene. In the future, the role of METTL1 should be investigated in the tumorigenesis of hepatoblastoma.
Some limitations of this study should be discussed. First, several factors may increase hepatoblastoma risk, including defective development (e.g., maturation and low birth weight), harmful environmental exposures, and susceptibility genes. Here, we failed to include the former two factors in the study. Second, we did not find significant SNPs in the single-locus analysis. Many more potential functional SNPs in the WDR4 gene should be interrogated. Third, the number of cases was relatively small, and our results should be interpreted carefully. Finally, biologically relevant evidence is lacking that WDR4 has a role in hepatoblastoma pathogenesis due to the novelty of this gene. In vitro and in vivo functional analyses are warranted to explore the implication of WDR4 in hepatoblastoma tumorigenesis in the future direction.
Overall, we demonstrated that the presence of two more selected WDR4 gene SNPs might increase the risk of hepatoblastoma in girls. The findings should be validated in large case-control studies.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by grants from the Natural Science Foundation of Fujian Province (No: 2021J01386), and Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (No: 2019B030301004).
Author Contributions
All authors contributed significantly to this work. JL, JZhang, YL, ZY, JC, SL, LL and JH performed the research study and collected the samples and data; JL, and JH analyzed the data; DX and JH designed the research study; SH, JZhu, ZX, JH and DX wrote the paper; JH prepared all the Tables. All authors have read and approved the final manuscript to be published.
Data Availability Statement
All the data are available upon request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-61
2. Spector LG, Birch J. The epidemiology of hepatoblastoma. Pediatr Blood Cancer. 2012;59:776-9
3. Pakakasama S, Tomlinson GE. Genetic predisposition and screening in pediatric cancer. Pediatr Clin North Am. 2002;49:1393-413
4. Nagae G, Yamamoto S, Fujita M, Fujita T, Nonaka A, Umeda T. et al. Genetic and epigenetic basis of hepatoblastoma diversity. Nat Commun. 2021;12:5423
5. Pakakasama S, Chen TT, Frawley W, Muller CY, Douglass EC, Lee R. et al. CCND1 polymorphism and age of onset of hepatoblastoma. Oncogene. 2004;23:4789-92
6. Zhuo Z, Lin A, Zhang J, Chen H, Li Y, Yang Z. et al. Genetic variations in base excision repair pathway genes and risk of hepatoblastoma: a seven-center case-control study. Am J Cancer Res. 2021;11:849-57
7. Zhuo Z, Miao L, Hua W, Chen H, Yang Z, Li Y. et al. Genetic variations in nucleotide excision repair pathway genes and hepatoblastoma susceptibility. Int J Cancer. 2021;149:1649-58
8. Pakakasama S, Chen TT, Frawley W, Muller C, Douglass EC, Tomlinson GE. Myeloperoxidase promotor polymorphism and risk of hepatoblastoma. Int J Cancer. 2003;106:205-7
9. Zhuo ZJ, Hua RX, Chen Z, Zhu J, Wang M, Yang Z. et al. WTAP Gene Variants Confer Hepatoblastoma Susceptibility: A Seven-Center Case-Control Study. Mol Ther Oncolytics. 2020;18:118-25
10. Chen H, Li Y, Li L, Zhu J, Yang Z, Zhang J. et al. YTHDC1 gene polymorphisms and hepatoblastoma susceptibility in Chinese children: A seven-center case-control study. J Gene Med. 2020;22:e3249
11. Luo Z, Li G, Wang M, Zhu J, Yang Z, Li Y. et al. YTHDF1 rs6090311 A>G polymorphism reduces Hepatoblastoma risk: Evidence from a seven-center case-control study. J Cancer. 2020;11:5129-34
12. Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253-66
13. Chen Y, Lin H, Miao L, He J. Role of N7-methylguanosine (m(7)G) in cancer. Trends Cell Biol. 2022;32:819-824
14. Chen J, Li K, Chen J, Wang X, Ling R, Cheng M. et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond). 2022;42:223-44
15. Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T. et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422-35
16. Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell. 2018;71:244-55.e5
17. Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell. 2019;74:1304-16.e8
18. Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N. et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019;74:1278-90.e9
19. Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R. et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022;13:1478
20. Zeng H, Xu S, Xia E, Hirachan S, Bhandari A, Shen Y. Aberrant expression of WDR4 affects the clinical significance of cancer immunity in pan-cancer. Aging (Albany NY). 2021;13:18360-75
21. Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W. et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323-38.e14
22. Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W. et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339-55.e8
23. Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang G. et al. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 2021;12:691
24. Roebuck DJ, Aronson D, Clapuyt P, Czauderna P, de Ville de Goyet J, Gauthier F. et al. 2005 PRETEXT: a revised staging system for primary malignant liver tumours of childhood developed by the SIOPEL group. Pediatr Radiol. 2007;37:123-32 quiz 249-50
25. Bian J, Zhuo Z, Zhu J, Yang Z, Jiao Z, Li Y. et al. Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: A nine-centre case-control study. J Cell Mol Med. 2020;24:9280-6
26. Zhang Z, Zhang R, Zhu J, Wang F, Yang T, Zou Y. et al. Common variations within HACE1 gene and neuroblastoma susceptibility in a Southern Chinese population. Onco Targets Ther. 2017;10:703-9
27. Nie H, Liao Z, Wang Y, Zhou J, He X, Ou C. Exosomal long non-coding RNAs: Emerging players in cancer metastasis and potential diagnostic biomarkers for personalized oncology. Genes Dis. 2021;8:769-80
28. Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou J. et al. The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics. 2020;10:11049-62
29. Yang T, Li J, Wen Y, Tan T, Yang J, Pan J. et al. LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front Genet. 2019;10:506
30. Yang T, Wen Y, Li J, Tan T, Yang J, Pan J. et al. NRAS and KRAS polymorphisms are not associated with hepatoblastoma susceptibility in Chinese children. Exp Hematol Oncol. 2019;8:11
31. Liu P, Zhuo ZJ, Zhu J, Yang Z, Xin Y, Li S. et al. Association of TP53 rs1042522 C>G and miR-34b/c rs4938723 T>C polymorphisms with hepatoblastoma susceptibility: A seven-center case-control study. J Gene Med. 2020;22:e3182
32. Li L, Zhuo Z, Yang Z, Zhu J, He X, Yang Z. et al. HMGA2 Polymorphisms and Hepatoblastoma Susceptibility: A Five-Center Case-Control Study. Pharmgenomics Pers Med. 2020;13:51-7
33. Chen H, Duan F, Wang M, Zhu J, Zhang J, Cheng J. et al. Polymorphisms in METTL3 gene and hepatoblastoma risk in Chinese children: A seven-center case-control study. Gene. 2021;800:145834
34. Chen H, Chen Z, Wang M, Zhang J, Li Y, Li L. et al. METTL14 gene polymorphisms influence hepatoblastoma predisposition in Chinese children: Evidences from a seven-center case-control study. Gene. 2022;809:146050
35. Ren H, Zhuo ZJ, Duan F, Li Y, Yang Z, Zhang J. et al. ALKBH5 Gene Polymorphisms and Hepatoblastoma Susceptibility in Chinese Children. J Oncol. 2021;2021:6658480
36. Wang YJ, Mugiyanto E, Peng YT, Huang WC, Chou WH, Lee CC. et al. Genetic Association of the Functional WDR4 Gene in Male Fertility. J Pers Med. 2021;11:760
37. Shaheen R, Abdel-Salam GM, Guy MP, Alomar R, Abdel-Hamid MS, Afifi HH. et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015;16:210
Author contact
![]() Corresponding authors: Di Xu, Department of Pediatric Surgery, Shengli Clinical Medical College of Fujian Medical University, 134 Dongjie Road, Fuzhou 350001, Fujian, China, Tel./Fax: (+86-0591) 88217420, Email: fjsleykcom; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (+86-020) 38076560, Email: hejing198374com or hejingorg.
Corresponding authors: Di Xu, Department of Pediatric Surgery, Shengli Clinical Medical College of Fujian Medical University, 134 Dongjie Road, Fuzhou 350001, Fujian, China, Tel./Fax: (+86-0591) 88217420, Email: fjsleykcom; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (+86-020) 38076560, Email: hejing198374com or hejingorg.

 Global reach, higher impact
Global reach, higher impact