3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(13):3404-3414. doi:10.7150/jca.76516 This issue Cite
Research Paper
Early detection of colorectal cancer somatic mutations using cfDNA liquid biopsies in a murine carcinogenesis model
1. Laboratorio Nacional en Salud, Diagnóstico Molecular y Efecto Ambiental en Enfermedades Crónico-Degenerativas, Facultad de Estudios Superiores Iztacala, Tlalnepantla 54090, México.
2. Unidad de Biomedicina, Facultad de Estudios Superiores Iztacala, UNAM, Tlalnepantla 54090, México.
3. Unidad de Investigación Biomédica en Cáncer INCan-UNAM, Instituto Nacional de Cancerología, Ciudad de México 14080, México.
4. Avenida Instituto Politécnico Nacional # 2508, Colonia San Pedro Zacatenco, Delegación Gustavo A. Madero, C.P. Departamento de Infectómica y Patogénesis Molecular, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), Mexico City 07360, Mexico
5. Instituto de Física, Universidad Nacional Autónoma de México, Ciudad de México 04510, México.
6. Subdirección de Investigación Básica, Instituto Nacional de Cancerología, Ciudad de México 14080, México.
Abstract
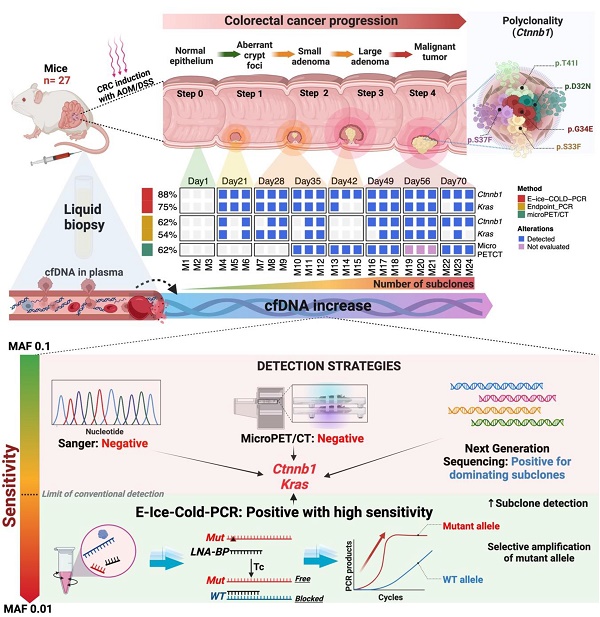
Colorectal cancer (CRC) is one of the top five cancers in incidence and mortality worldwide. The early detection of this neoplasm through analysis of circulating free DNA (cfDNA), which carries tumor genetic alterations, as a liquid biopsy, could have a major impact in enhancing early detection and reducing the mortality rate. The aim of this work was to demonstrate the feasibility of using cfDNA as a liquid biopsy for the early detection of CRC. For this purpose, we implemented an azoxymethane and dextran sodium sulfate-induced murine carcinogenesis model to detect oncogenic somatic mutations in Ctnnb1 and Kras during CRC development. To enhance the sensitivity in the detection, E-ice-COLD-PCR was utilized to selectively enrich for mutant alleles, followed by massively parallel sequencing. Driving somatic mutations were detected in Ctnnb1 and Kras in the liquid biopsies of early stages of tumor development, corresponding to the formation of aberrant crypt foci, the first histological alterations that can be identified throughout the formation of CRC. The concentration of cfDNA was increased along the carcinogenic process. Polyclonality in Ctnnb1 was found in tumor samples and cfDNA in this model. On the other hand, the use of cfDNA as a non-invasive test resulted in superior early detection compared to microPET/CT imaging. As a proof-of-principle, this study shows the great potential use of allelic-specific PCR for the detection and enrichment of pathogenic alleles present in cfDNA samples, as a test for early non-invasive detection of CRC. This work provides scientific evidence to set methodological bases that allow early detection of mutations in cfDNA obtained from plasma of CRC in humans.
Keywords: AOM/DSS model, cfDNA, liquid biopsy, early detection, biomarkers, colorectal cancer, polyclonality.

 Global reach, higher impact
Global reach, higher impact