3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(13):3463-3475. doi:10.7150/jca.72973 This issue Cite
Editorial Commentary
The most effective but largely ignored target for prostate cancer early detection and intervention
1. Department of Pathology, Hackensack Meridian School of Medicine, Nutley, NJ, USA
2. Tumor Immunology Laboratory, Jonsson Comprehensive Cancer Center, UCLA School of Dentistry and Medicine, Los Angeles, CA, USA
3. Department of Obstetrics and Gynecology, Changhua Christian Hospital, Changhua, Taiwan
4. Department of Pathology, Chinese PLA General Hospital 7 th Medical Center, Beijing, China
5. II ND Department of Gynecology, Lublin Medical University, Lublin, Poland
6. Pulmonary-Oncology Department, "Theageneio" Cancer Hospital, Thessaloniki, Greece
7. Laboratory of Biological Effects of Non-Ionizing Radiation, Institute of Cell Biophysics, Russian Academy of Sciences, Russian Federation
8. Section of Pathological Anatomy, Polytechnic University of the Marche Region, School of Medicine, United Hospitals, Ancona, Italy.
9. Department of Pathology, New York University School of Medicine, New York, NY, USA.
10. Department of Pathology, New York Harbor Healthcare System, New York, NY, USA.
11. Department of Morphological Sciences, Cordoba University Medical School, Cordoba, Spain
12. Molecular Medicine and Cell Therapy Foundation, Department of Clinical & Molecular Sciences, Polytechnic University of the Marche Region, Ancona, Italy
13. Medical Oncology, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy
14. Department of Biological Sciences, Boler-Parseghian Center for Rare and Neglected Diseases, Harper Cancer Research Institute, University of Notre Dame, Notre Dame, IN, USA.
15. Tumor Microenvironment and Metastasis Program, Indiana University Melvin and Bren Simon Comprehensive Cancer Center, Indianapolis, IN, USA.
16. Department of Pathology and Laboratory Medicine, Brown University Medical School | Lifespan Academic Medical Center, RI, USA.
Received 2022-3-17; Accepted 2022-10-1; Published 2022-10-17
Abstract
Over the past two decades, the global efforts for the early detection and intervention of prostate cancer seem to have made significant progresses in the basic researches, but the clinic outcomes have been disappointing: (1) prostate cancer is still the most common non-cutaneous cancer in Europe in men, (2) the age-standardized prostate cancer rate has increased in nearly all Asian and African countries, (3) the proportion of advanced cancers at the diagnosis has increased to 8.2% from 3.9% in the USA, (4) the worldwide use of PSA testing and digital rectal examination have failed to reduce the prostate cancer mortality, and (5) there is still no effective preventive method to significantly reduce the development, invasion, and metastasis of prostate cancer… Together, these facts strongly suggest that the global efforts during the past appear to be not in a correlated target with markedly inconsistent basic research and clinic outcomes. The most likely cause for the inconsistence appears due to the fact that basic scientific studies are traditionally conducted on the cell lines and animal models, where it is impossible to completely reflect or replicate the in vivo status. Thus, we would like to propose the human prostate basal cell layer (PBCL) as “the most effective target for the early detection and intervention of prostate cancer”. Our proposal is based on the morphologic, immunohistochemical and molecular evidence from our recent studies of normal and cancerous human prostate tissues with detailed clinic follow-up data. We believe that the human tissue-derived basic research data may provide a more realistic roadmap to guide the clinic practice and to avoid the potential misleading from in vitro and animal studies.
Keywords: Prostate basal cell layer, Tumor capsule, Cancer early detection and intervention.
Editorial Commentary
Over the past two decades, a wide variety of genetic and epigenetic factors, including the germline variants, somatic gene mutations, various SNPs and miRNAs, aberrant expression of tumor suppressor genes or oncogenes, tumor regulatory drivers or nominate novel transcription factors, phosphorylation, glycosylation, ubiquitination, acetylation, lipidation, bacterial or viral infections, dietary compounds, or changes in testosterone-estradiol ratio…, have been added to the list of risk factors for prostate carcinogenesis and progression [1-21]. Concurrently, a wide variety of detection and intervention methods, including artificial intelligence or nanotechnology -based devices, MRI-guided biopsy, bispecific antibodies, cancer vaccines, cytokine inhibitions, chimeric antigen receptor T-cells, immune checkpoint inhibition, high-intensity focal ultrasound, focal cryotherapy, photodynamic therapy, different sources of energy-based focal therapy, focal laser ablation, irreversible electroporation, DNA PSA-specific aptamers…, have been developed for clinic applications [22-32].
Unfortunately, the outcomes of global efforts for the early detection and intervention of prostate cancer are disappointing: (1) prostate cancer is still the most common cancer in Europe in man, (2) the prostate cancer incidence has increased in nearly all African and Asian countries, (3) the proportion of advanced cancers at diagnosis has increased from 3.9% to 8.2% in the US, (4) the worldwide use of PSA testing and digital rectal examination has failed to reduce prostate cancer mortality, and thus, is no longer recommended [33-44], and (5) there is still no effective preventive method to reduce the incidence, invasion, and metastasis of prostate cancer [22-32].
Together, these facts suggest that global efforts during the past appear to be not in a correlated target. The most likely cause appears due to the fact that basic scientific studies are traditionally conducted on the cell lines and animal models, which are impossible to completely reflect or replicate the in vivo status. Thus, we propose the prostate basal cell layer (PBCL) as “the most effective target for the early detection and intervention of prostate cancer” for the following reasons.
The PBCL is an essential component of the prostate gland
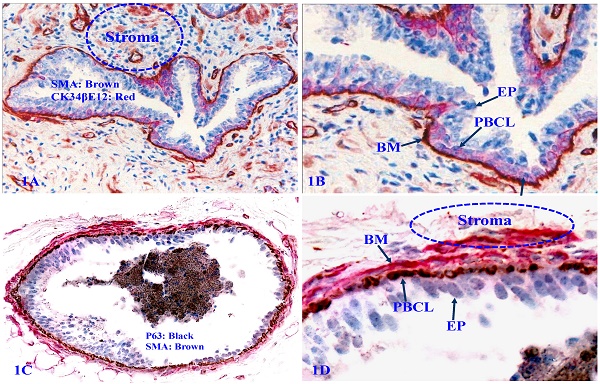
The human prostate gland consists of three components: the epithelium, capsule, and stroma. The epithelium is completely encircled by the capsule (a single layer of PBCL) and the basement membrane (BM, a thin sheet of extracellular fibers), and the stroma. Therefore, prostate cancer cells have to first pass through the PBCL and then BM to invade to the stroma [45-49] (Figure 1).
Structural relationships of prostate gland tissue components. Formalin-fixed and paraffin-embedded human prostate gland tissue sections were double immune-stained, 1A and 1B with antibodies to smooth muscle actin (SMA) that is reactive to the basement membrane (BM), smooth muscle, and endothelial cells, and CK34βE12, which is reactive to PBCL; 1C and 1D with SMA plus p63, which is reactive to PBCL. EP = Epithelium.
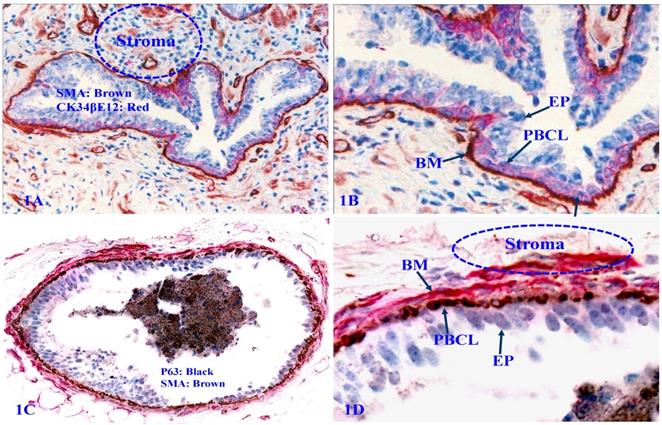
The PBCL is the source of several tumor suppressors
The PBCL produces several tumor suppressors, including Maspin and p63 [50-54] that exert significant regulatory functions on adjacent epithelial cells (Figure 2).
The focal PBCL degeneration is a triggering factor for tumor progression
The detailed dynamic cellular kinetics of the human prostate basal cell population remains elusive [55-61]. However, distinct senescence or degenerative changes of PBCL are frequently seen in some of normal and malignant prostate tissues. These changes include: focal disruptions (defined as the absence of basal cells resulting in a gap larger than the combined size of at least three basal cells in at least two or more consecutive sections), the loss of nuclear p63 expression, nuclear swelling, cell debris, and large clusters of cell debris. Luminal cells overlying focally disrupted PBCL often show markedly higher cellular density, proliferation rate, and morphology compared to those from their counterparts overlying non-disrupted PBCL (Figure 3).
Expression of tumor suppressors p63 and Maspin in PBCL. Formalin-fixed and paraffin-embedded human prostate gland sections were double immune-stained, 2A with CK34βE12 (red) and p63 (black); 2B with Maspin (red) and p63 (black). Please note that the normal PBCL is continuous and the basal cells express a high level of Maspin and p63. PBCL = Prostate basal cell layer. EP = Epithelium.
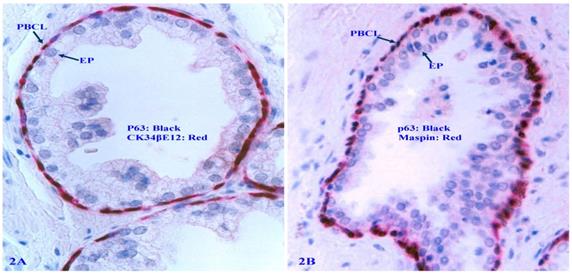
Focal PBCL senescence or degenerative changes. Formalin-fixed and paraffin-embedded human prostate gland tissue sections were double immune-stained (see the detailed labeling within individual figures). Circles identify focal PBCL disruptions; Squares identify large clusters of basal cell debris; Thick arrows in A and B identify basal cells with the loss of nuclear p63 expression; Thin arrows identify the residual PBCL; Red stars identify invasive lesions. Please note that the size of the focal disruptions in PBCL varies substantially, and that the epithelial cells overlying focally disrupted PBCL are morphologically more similar to the adjacent invasive cancer cells with a higher proliferation rate than their adjacent counterparts still enclosed by the residual PBCL.
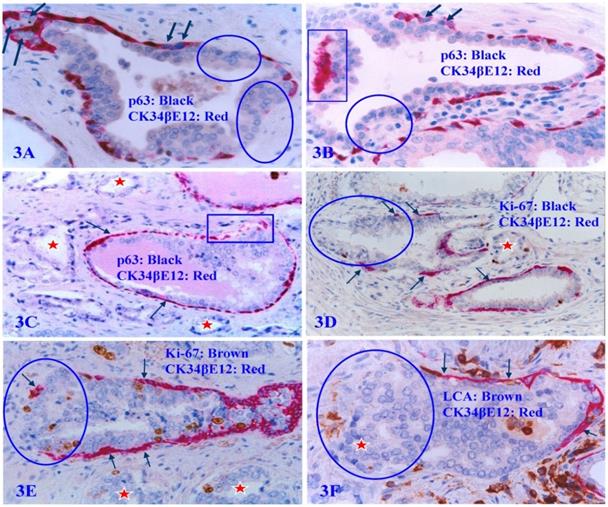
The focal PBCL disruption is absolutely needed for tumor invasion
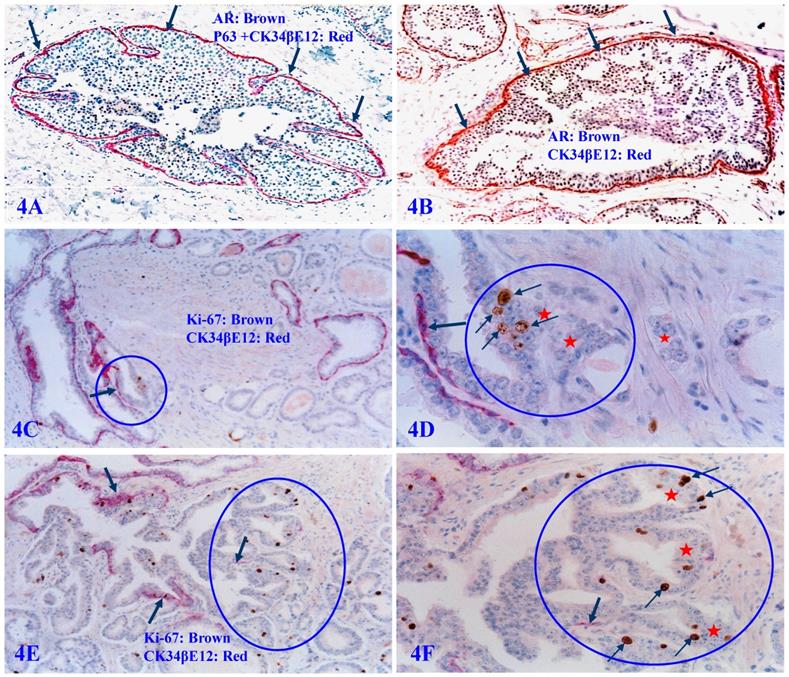
Our previous studies have consistently demonstrated that a focal disruption in a given PBCL is absolutely needed for the initiation of cancer invasion [62-68]. If the surrounding FBCL is intact, such tumors can grow to a very large size, but may remain at non-invasive state for many years (Figure 4A and 4B). In a sharp contrast, if the surrounding FBCL is focally disrupted, the cancer invasion could commence at a very early stage, despite a very small size (Figure 4C-4F). Double immunochemical staining with the cell proliferation specific marker Ki-67 and the PBCL specific marker CK34βE12 reveals that all invasion starts at the site of focal PBCL disruptions and a vast majority of proliferating cells are located at or near the site of focal PBCL disruptions. The “budding cells” from focally disrupted PBCL are often immediately adjacent to or in a direct continuity with the invasive cancer cells (Figure 4C-4F).
Focal PBCL disruptions and cancer invasion. Formalin-fixed and paraffin-embedded human prostate gland tissue sections were double immune-stained with several antibodies (see labeling within each individual picture). Circles identify focal disruptions in PBCL and the overlying epithelial cells. Thick arrows identify the normal or residual PBCL Thin arrows identify proliferating cells. Red starts identify invasive lesions. Please note that the budding cells overlying focally disrupted PBCL have a significantly higher proliferation rate than their counterparts enclosed by the residual PBCL, and that budding cells are immediately adjacent to or in a direct continuity with invasive lesions.
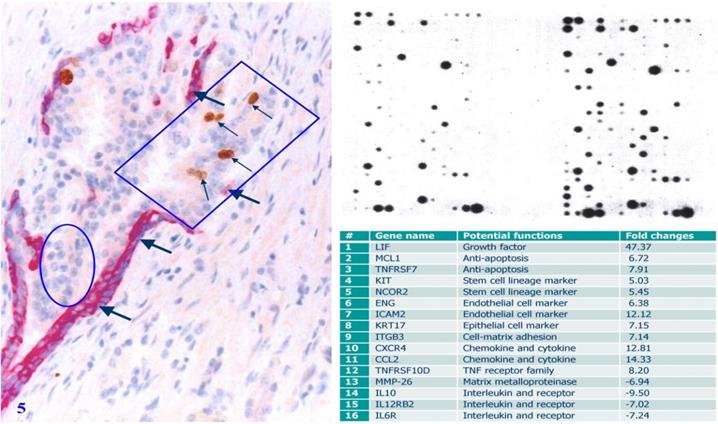
Selective gene expression profiling of micro-dissected human prostate gland tissue samples has consistently shown that cell clusters overlying focally disrupted PBCL have a significantly higher expression level of cell growth- and invasion-related molecules, including growth factors, stem cell lineage markers, anti-apoptosis-related genes, and endothelial cell makers than their morphologically similar counterparts enclosed by the residual PBCL [66] (Figure 5).
The PBCL itself is the target of a variety of pathologic alterations
It has been consistently documented that prostate basal cell carcinoma is very rare, accounting for only about 0.01% of the total prostate malignancies [69-73], which strongly suggests that the prostate basal cell population is very unlikely to be a major target of the prostate carcinogenesis. However, it has also been consistently demonstrated that the PBCL belongs to a self-renewal cell population [74-80]. For this and other reasons, the PBCL population itself also frequently suffers from a variety of degeneration- and regeneration-related pathologic alterations that includes, but is not limited to the following:
The loss or significant reduction of nuclear p63 expression in normal appearing PBCL
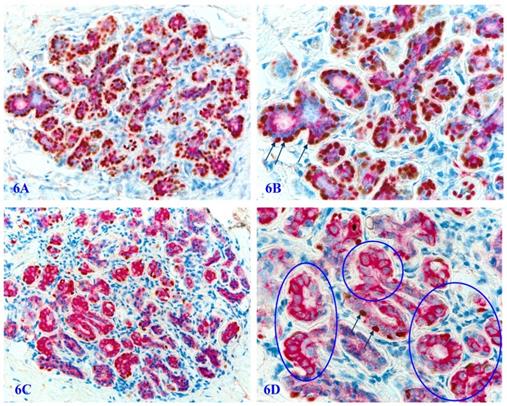
In autopsy, biopsy, and surgically resected human prostate gland tissues, a vast majority of the basal cells in normal or hyperplastic tissues are not only morphologically distinct, but also express a high level of p63 and Maspin, two well-documented tumor suppressors (Figure 6A-6B). However, about 6% to 8% of the cases harbor variable numbers of morphologically normal appearing basal cell clusters that are completely devoid of, or have a significantly reduced cells with p63 nuclear expression (Figure 6C-6D). Prostate cancer patients with such atypical basal cell clusters have a significantly more aggressive clinical courses and worse prognosis [81].
The loss of the expression of all PBCL specific markers
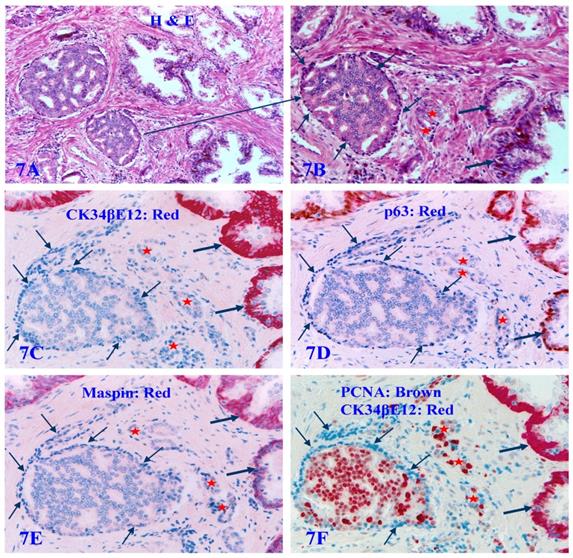
About 6% to 8% of the autopsy, biopsy, and surgically resected human prostate tissues also harbor a variable number of atypical pre-invasive cancer cell clusters, in which all the basal cell layers are largely non-disrupted and morphologically distinct, whereas they completely lack the expression of CK34βE12, p63, and Maspin. They are even completely devoid of the expression of the proliferating cell nuclear antigen (PCNA). These pre-invasive cancer cells enclosed by such PBCL are morphologically and immunohistochemically similar to adjacent invasive cancer cells, and have a significantly higher proliferation rate than their adjacent counterparts at the same stage [81] (Figure 7).
Gene expression profiling of cells overlying focally disrupted and enclosed by residual PBCL. Formalin-fixed and paraffin-embedded human prostate gland tissue sections were double immune-stained with antibodies for SMA and Ki-67. Thick arrows identify residual PBCL. Thin arrows identify proliferating epithelial cells. The square identifies epithelial cells overlying focally disrupted FBCL, and the circle identifies epithelial cells enclosed by the residual FBCL micro-dissected for gene expression profiling. Please note that morphologically comparable epithelial cells overlying focally disrupted PBCL and enclosed by the residual PBCL have different profiles of the gene expression.
Significantly reduced basal cell numbers of p63 expression in normal appearing PBCL. Formalin-fixed and paraffin-embedded human prostate gland tissue sections from two different biopsies with morphologically normal appearing basal cell layers associated with a morphologically normal appearing epithelial cell population were double immune-stained with p63 and Maspin. In the cases one, a vast majority of the normal appearing basal cells display the distinct expression of p63 within the cell nuclei. However, in a sharp contrast in the case two, only around 1-3% of the normal appearing basal cells show the normal localization of p63 expression within the cell nuclei. Circles identify normal appearing basal cells in the case 2, which display a high level of Maspin expression within the cell cytoplasm, whereas they are completely devoid of p63 expression within the cell nuclei despite the fact that the nuclei of these basal cells are clearly visible. 6B and 6D are the higher magnification of 6A and 6C, respectively.
The lack of all basal cell markers in the PBCL of pre-invasive prostate cancer clusters. Formalin-fixed and paraffin-embedded consecutive human prostate gland tissue sections from one case were subjected to H&E staining (7A-7B) and IHC staining (7C-7F) for different biomarkers (see detailed labeling within each individual picture). Red starts identify invasive lesions. Thin arrows identify atypical basal cells. Thick arrows identify normal basal cells. Please note that the PBCL overlying a walnut-like tumor nest is morphologically distinct in both H&E and IHC stained sections, but all basal cells are completely devoid of the expression of CK34βE12, p63, and Maspin. In addition, these morphologically distinct basal cells are also completely devoid of the expression of proliferating cell nuclear antigen (PCNA), in a sharp contrast to the enclosed epithelial cells, which are all strongly immunoreactive to PCNA.
Frequent apoptosis-related alterations
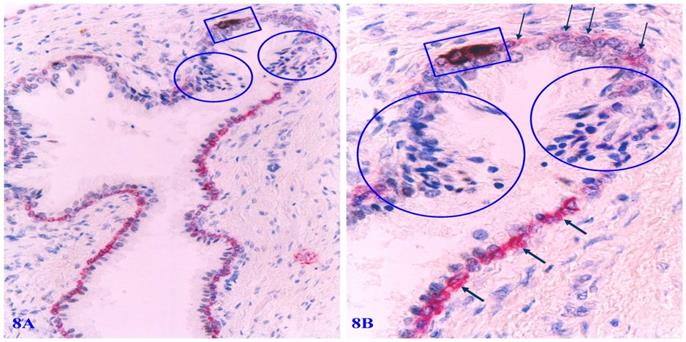
Distinct apoptosis-related alterations of basal cells are frequently seen in focally disrupted PBCL. Immunohistochemistry-based apoptotic assays clearly demonstrate that a vast majority of focally degenerated basal cells show high levels of apoptosis-related molecules. All apoptotic basal cells are physically located at or near focally disrupted PBCL. Epithelial cells adjacent to apoptotic basal cells have a much higher cell density than their adjacent counterparts (Figure 8).
The significant infiltration of immune cells
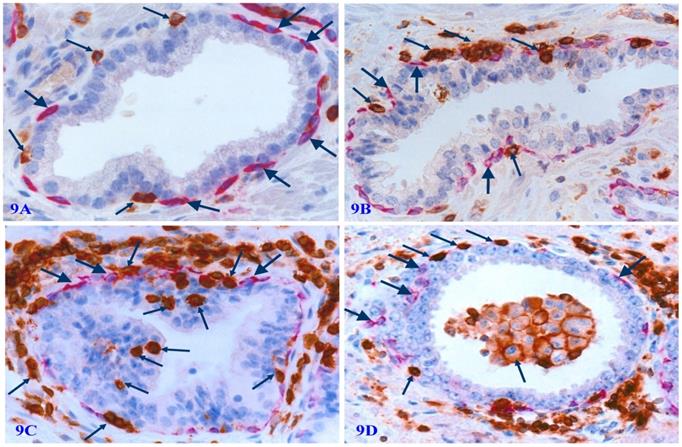
Degenerated basal cell products appear to act as cytokines to attract infiltration of immune-reactive cells to the physical sites of focally disrupted PBCL, and infiltrated lymphocytes are generally associated with cell debris or morphologically degenerated basal cells (Figure 9).
Apoptosis-related alterations in degenerated basal cells. Formalin-fixed and paraffin-embedded human prostate gland tissue sections were subjected to the apoptotic assays with a commercially available detection kit, and then, were immune-stained for CK34βE12 to elucidate PBCL. Arrows identify PBCL. Squares identify apoptotic basal cells. Circles identify epithelial cells overlying or near the focally disrupted PBCL.
Physical association of infiltrating immune cells with degenerated basal cells. Formalin-fixed and paraffin-embedded human prostate tissue sections from four different cases were subjected to double IHC staining with the basal specific marker CK34βE12 and infiltrating immune cell marker (leukocyte common antigen, LCA). Thick arrows identify residual basal cells. Thin arrows identify infiltrating immune cells. Please note that a vast majority of infiltrating immune cells are physically associated with or immediately adjacent to degenerative basal cells. The epithelial component or the acinar and ductal lumen in focally disrupted PBCL often harbor infiltrating immune cells. It is interesting to note that nearly all the normal, benign, and malignant epithelial cells show no distinct sign of degeneration-related alterations.
The high level of Tenascin C expression in focally disrupted PBCL
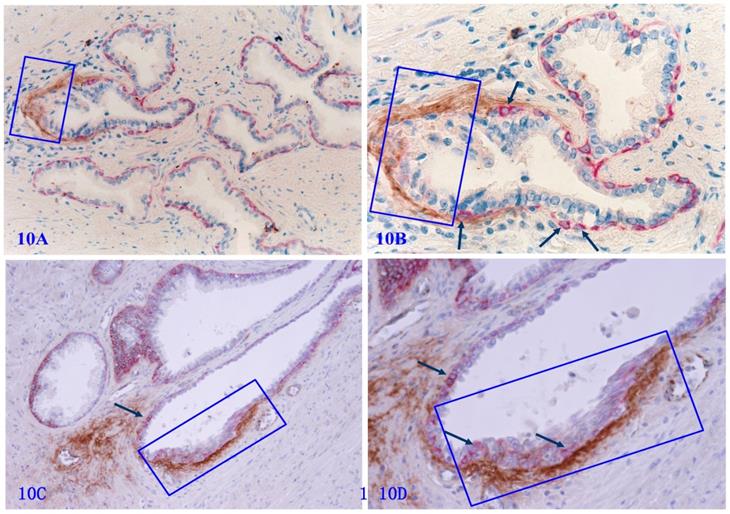
Tenascin C is an extracellular matrix glycoprotein, which paves the paths and facilitates the migration and metastasis of prostate cancer cells [82-86]. Our previous studies have shown that Tenascin C is highly expressed at the site of distinct degenerative basal cells, and epithelial cells in the vicinity of areas with elevated Tenascin C often lose the cohesion [66] (Figure 10).
Based on above findings, it is apparent that PBCL is not only an essential constitute of the prostate gland, but also an active producer of tumor suppressors, which exert significant decisive influence on adjacent epithelial and stromal cells. In addition, as the PBCL population belongs to a self-renewal population, it has to consistently undergo normal self-replenishment processes to replace aged and injured cells. Consequently, the PBCL population itself also suffers from a wide variety of degeneration- and regeneration-related normal and pathological alterations.
The detailed cellular and molecular mechanisms of focal disruptions or a total loss of the PBCL remains elusive. Furthermore, there is no solid evidence to determine whether the loss of basal cells is a direct trigger for the development of prostate adenocarcinoma, or the loss of basal cells is directly resulted from prostate cancer cells. However, it has been consistently concluded that focal disruptions or a total loss of the PBCL is statistically correlated with the invasion and metastasis of almost all types of prostate cancer (except prostate basal cell carcinoma) [62-73].
Based on above facts, the PBCL appears to be the most effective but largely ignored target for the early detection and intervention of prostate cancer. As the morphologic, pathological, and immuno-histochemical profiles of the basal cell population is far more easily recognizable and definable than its epithelial counterpart, the PBCL appears to be a more easily readable roadmap with the following specific scientific and clinic implications and applications:
- To use Maspin and PSA as independent risk factors for cancer screening. As Maspin is consistently expressed in basal cells [87-90], while the PSA is elevated in virtually all prostate malignancies [91-94], Maspin- and PSA-related signatures in the serum can be utilized for a population-based screening for the early detection of prostate cancer.
Tenascin expression at the site of focally disrupted PBCL. Formalin-fixed and paraffin-embedded human prostate gland tissue sections from two different cases were double immune-stained for CK34βE12 (red) and Tenascin C (brown). Arrows identify residual basal cells. Squares enclose Tenascin C overlying focally disrupted PBCL. Please note that strong Tenascin C positivity is seen only at or adjacent to the site of focally disrupted PBCL. Non-disrupted PBCLs are largely devoid of Tenascin C expression.
- To use p63 as a risk factor for a population-based screening to detect the predisposition of cancer susceptibility or tumor suppressing genes. Since p63 belongs to the p53 tumor suppressor family, and is normally expressed in the nucleus of the basal cells [95,96], an aberrant expression level or subcellular localization of p63 accompanying by an elevated PSA level may signify the predisposition of cancer susceptibility or mutated suppressing genes. Previous studies have clearly demonstrated that loss or cytoplasmic expression of p63 is associated with elevated cancer stem cells, enhanced cell migration and metastasis, and increased mortality in prostate cancer [97-99].
- To use p63, Maspin, and different RNA signatures as biomarkers for the non-invasive detection of early prostate cancer. Previous studies have revealed that p63, Maspin, RNA signatures, and PSA are detectable in the urine samples of prostate cancer patients [100-106]. Thus, a statistical comparison of the expression levels of p63, Maspin, and PSA in the urine samples may be used as a non-invasive clinic test for the detection of early prostate cancer.
- To use exfoliated basal cells combined with other urinary markers for early detection of prostate cancer progression and invasion. Previous studies have consistently shown that a variable number of exfoliated cancer cells are detectable in a majority of prostate cancer patients [107-110]. As the basal cells are localized at the base of the epithelial cell layer, the detection of exfoliated basal cells in the urine is likely to signify the disruptions of the PBCL and the stromal invasion of the prostate cancer.
- To use the PBCL physical integrity (disrupted vs non-disrupted) combined with PSA test results as a clinic marker for the differentiation diagnosis. As the disruption of the PBCL is a prerequisite for prostate cancer invasion and metastasis, while an elevated PSA level is detectable in both non-invasive and invasive prostate cancer, the physical integrity of the PBCL in patients with an elevated PSA could effectively differentiate between non-invasive and invasive prostate cancer.
- To use the expression of Tenascin in PBCL as a routine clinic test of the prostate biopsy or urine sample. Previous studies have consistently demonstrated that aberrant Tenascin expression is exclusively seen at or near focally disrupted PBCL and is also significantly correlated with prostate cancer invasion and metastasis [66, 82-86]. Thus, the assessment of the PBCL-associated Tenascin expression may lead to the identification of the specific cases at increased risk for prostate cancer progression.
- To use Maspin or CK34βE12 as biomarkers to discriminate prostate from non-prostate cancers. Previous studies have shown that (a) high levels of PSA is seen in patients with breast, lung, ovary, liver, kidney, adrenal, skin, salivary, and colorectal cancer [111-113], (b) smoking, asymptomatic inflammation, metformin use, chronic prostatitis can elevate the PSA expression level [114-117], and (c) age, ethnicity, triglyceride level, and BMI can also significantly impact the expression of PSA [118, 119]. In contrast, above factors have little impact on PBCL. Thus, normal expression status of Maspin or CK34βE12 in individuals with high levels of PSA may signify non-prostate lesions.
- To use focal PBCL disruptions as a localizer to identify cancer-stem cell clusters/specific precursors of invasive cancer. Our previous studies of multiple cancers have consistently shown that a focal disruptions of tumor capsules selectively facilitate clonal proliferation of overlying cancer stem cells to form distinct cell clusters. These newly formed clusters have significantly higher levels of cancer stem cell markers and invasion and metastasis-related genes than their morphologically comparable counters still enclosed by the non-disrupted tumor capsules [62-66, 120-123] (Figure 4 & 5). It is very likely that these cell clusters may represent the direct precursors of invasive lesions. Thus, micro-dissecting these clusters for further evaluation can potentially lead to the dentification of triggering molecules for the basal cell degenerations, tumor progression, and invasion.
- To use PBCL-associated immune-cell infiltration to monitor the tumor progression and treatment responses. Our previous studies have consistently revealed that the immune-cell infiltration is significantly associated with prostate tumor capsule disruptions which lead to the subsequent invasion and metastasis [62-68]. A great number of recent studies have not only confirmed our previous reports and conclusions, but have also consistently shown that immune-cell infiltration is also significantly correlated with the treatment responses in multiple cancer types [62-68, 124-131].
- To use anti-inflammatory drug aspirin or statin to repair the PBCL degeneration-related tumor capsule disruptions. Previous studies have consistently shown that aspirin or statin could significantly alter the immune milieu of prostate and to prevent cancer progression [132-134]. Therefore, the administration of aspirin or statin to individuals with focally disrupted PBCL associated with significant infiltration of the immune cells (as shown in Figure 9) and chronic prostatitis could potentially reduce the extent of associated immune cells and facilitate the repairing of focally disrupted tumor capsules.
- To administer stem cell specific molecules, inducers, or stimulators to burst the normal replenishment and the physical integrity of PBCL. Previous studies have suggested that BMP5, Zeb1, CD24, CD44, NANOG, and Nestin are prostate stem cell-specific markers that are essential for the maintenance of the normal replenishment and physical integrity of the PBCL [135-137]. Thus, the administration of these molecules or stem cell specific inducers or stimulators to patients at a high risk of prostate cancer progression may offer the promise of more effective approaches for prostate cancer early intervention.
- To use basal cells lacking the phenotypic and proliferation markers as targets to identify novel cell proliferation pathways or cell cycle regulators. As a subset of morphologically distinct and non-disrupted PBCL completely lack the expression of tumor suppressors, phenotypic markers, and cell proliferation specific markers (Figure 7), it is likely that the growth and expansion of these cells are regulated by previously undescribed mechanisms or pathways [138,139]. Therefore, microdissection of these PBCL for gene expression profiling may lead to the identification of novel cell proliferation pathways and novel cell cycle regulators.
In summary, above findings and analyses strongly suggest that the PBCL is the most effective but largely ignored target for the early detection and intervention of human prostate cancer. It is apparent that the human tissue-derived basic research data may provide a more realistic roadmap that allows to observe the direct interactions among different cell types and to avoid the potential misleading from in vitro and animal studies.
Acknowledgements
All studies from Dr. Yan-gao Man's laboratory were financially supported in part by grants from Armed Forces Institute of Pathology and American Registry of Pathology (AFIP & ARP 05AA), The US Military Cancer Institute and Henry M. Jackson Foundation (USMCI2008-02), The US Congressionally Directed Medical Research Programs (DAMD17-01-1-0129, DAMD17-01-1-0130, and PC0513080), The Susan G. Komen Breast Cancer Foundation (BCTR0706983), The Ministry of Chinese Science and Technology Department (2006CB910505), and The Natural Science Foundation of China (NSFC30801176) under the category of “Cancer Early Detection” to Dr. Yan-gao Man.
All pictures presented in the current commentary were taken from the above studies on human prostate tissues with detailed clinic parameters and follow-up data from the Armed Forces Institute of Pathology and American Registry of Pathology, which was the world's largest and most comprehensive second-opinion consultation center with the world's largest human tissue repository. As the basic medical scientific studies are traditionally conducted on cancer cell lines or animal models, where it is impossible to completely mirror or mimic the human in vivo status, we sincerely hope that these pictures would serve as standards for defining the intrinsic value of the experimental findings from cell lines or cell line-derived animal models.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ni Raghallaigh H, Eeles R. Genetic predisposition to prostate cancer: an update. Fam Cancer. 2022;21(1):101-114
2. Rebbeck TR. Prostate Cancer Genetics: Variation by Race, Ethnicity, and Geography. Semin Radiat Oncol. 2017;27(1):3-10
3. Kafka M, Surcel C, Heidegger I. Recent Insights on Genetic Testing in Primary Prostate Cancer. Mol Diagn Ther. 2021;25(4):425-438
4. Pandareesh MD, Kameshwar VH, Byrappa K. Prostate Carcinogenesis: Insights in Relation to Epigenetics and Inflammation. Endocr Metab Immune Disord Drug Targets. 2021;21(2):253-267
5. Howard N, Clementino M, Kim D, Wang L, Verma A, Shi X. et al. New developments in mechanisms of prostate cancer progression. Semin Cancer Biol. 2019;57:111-116
6. Mikhaylenko DS, Efremov GD, Strelnikov VV, Zaletaev DV, Alekseev BY. Somatic Mutation Analyses in Studies of the Clonal Evolution and Diagnostic Targets of Prostate Cancer. Curr Genomics. 2017;18(3):236-243
7. Doghish AS, Ismail A, El-Mahdy HA, Elkady MA, Elrebehy MA, Sallam AM. A review of the biological role of miRNAs in prostate cancer suppression and progression. Int J Biol Macromol. 2022;197:141-156
8. Abidi SH, Bilwani F, Ghias K, Abbas F. Viral etiology of prostate cancer: Genetic alterations and immune response. A literature review. Int J Surg. 2018;52:136-140
9. Kukkonen K, Taavitsainen S, Huhtala L. et al. Chromatin and Epigenetic Dysregulation of Prostate Cancer Development, Progression, and Therapeutic Response. Cancers (Basel). 2021;13(13):3325
10. Dhingra P, Martinez-Fundichely A, Berger A, Huang FW, Forbes AN, Liu EM. et al. Identification of novel prostate cancer drivers using RegNetDriver: a framework for integration of genetic and epigenetic alterations with tissue-specific regulatory network. Genome Biol. 2017;18(1):141
11. Yegnasubramanian S, De Marzo AM, Nelson WG. Prostate Cancer Epigenetics: From Basic Mechanisms to Clinical Implications. Cold Spring Harb Perspect Med. 2019;9(4):a030445
12. Mancarella D, Plass C. Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation. Genome Med. 2021;13(1):23
13. Samaržija I. Post-Translational Modifications That Drive Prostate Cancer Progression. Biomolecules. 2021;11(2):247
14. Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z. et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580(7801):93-99
15. Felgueiras J, Silva JV, Nunes A, Fernandes I, Patrício A, Maia N, Pelech S, Fardilha M. Investigation of spectroscopic and proteomic alterations underlying prostate carcinogenesis. J Proteomics. 2020;226:103888
16. Puhr M, De Marzo A, Isaacs W, Lucia MS, Sfanos K, Yegnasubramanian S, Culig Z. Inflammation, Microbiota, and Prostate Cancer. Eur Urol Focus. 2016;2(4):374-382
17. Prins GS. Developmental estrogenization: Prostate gland reprogramming leads to increased disease risk with aging. Differentiation. 2021;118:72-81
18. Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle-Encinas E, Lobo J. et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun. 2018;9(1):4900
19. Bellamri M, Turesky RJ. Dietary Carcinogens and DNA Adducts in Prostate Cancer. Adv Exp Med Biol. 2019;1210:29-55
20. Wang YZ, Wong YC. Oncogenes and tumor suppressor genes in prostate cancer: a review. Urol Oncol. 1997;3(2):41-6
21. Isaacs W, Kainu T. Oncogenes and tumor suppressor genes in prostate cancer. Epidemiol Rev. 2001;23(1):36-41
22. Shah U, Biswas MR, Alzubaidi MS, Ali H, Alam T, Househ M, Shah Z. Recent developments in artificial intelligence-based techniques for prostate cancer detection: A scoping review. Stud Health Technol Inform. 2022;289:268-271
23. Keeney E, Thom H, Turner E, Martin RM, Morley J, Sanghera S. Systematic review of cost-effectiveness models in prostate cancer: Exploring new developments in testing and diagnosis. Value Health. 2022;25(1):133-146
24. Kuppermann D, Calais J, Marks LS. Imaging Prostate Cancer: Clinical Utility of Prostate-Specific Membrane Antigen. J Urol. 2022;207(4):769-778
25. Eksi SE, Chitsazan A, Sayar Z, Thomas GV, Fields AJ, Kopp RP. et al. Epigenetic loss of heterogeneity from low to high grade localized prostate tumours. Nat Commun. 2021;12(1):7292
26. Ratajczak W, Lubkowski M, Lubkowska A. Heat Shock Proteins in Benign Prostatic Hyperplasia and Prostate Cancer. Int J Mol Sci. 2022;23(2):897
27. Hopstaken JS, Bomers JGR, Sedelaar MJP, Valerio M, Fütterer JJ, Rovers MM. An Updated Systematic Review on Focal Therapy in Localized Prostate Cancer: What Has Changed over the Past 5 Years? Eur Urol. 2022;81(1):5-33
28. Kayano PP, Klotz L. Current evidence for focal therapy and partial gland ablation for organ-confined prostate cancer: systematic review of literature published in the last 2 years. Curr Opin Urol. 2021;31(1):49-57
29. Runcie KD, Dallos MC. Prostate Cancer Immunotherapy-Finally in From the Cold? Curr Oncol Rep. 2021;23(8):88
30. Galgano SJ, Planz VB, Arora S, Rais-Bahrami S. MR-Guided High-Intensity Directional Ultrasound Ablation of Prostate Cancer. Curr Urol Rep. 2021;22(1):3
31. Upadhyay TK, Ali MI, Khan F, Goel H, Mathur M, Goyal K. et al. Nanoparticles mediated target-specific drug delivery in prostate cancer: In-depth review. Curr Med Chem. 2021 Epub ahead of print. PMID: 34939536
32. Campos-Fernández E, Oliveira Alqualo N, Moura Garcia LC, Coutinho Horácio Alves C, Ferreira Arantes Vieira TD. et al. The use of aptamers in prostate cancer: A systematic review of theranostic applications. Clin Biochem. 2021;93:9-25
33. Marhold M, Kramer G, Krainer M, Le Magnen C. The prostate cancer landscape in Europe: Current challenges, future opportunities. Cancer Lett. 2022;526:304-310
34. Culp MB, Soerjomataram I, Efstathiou JA, Bray F, Jemal A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur Urol. 2020;77(1):38-52
35. Taitt HE. Global Trends and Prostate Cancer: A Review of Incidence, Detection, and Mortality as Influenced by Race, Ethnicity, and Geographic Location. Am J Mens Health. 2018;12(6):1807-1823
36. Butler EN, Kelly SP, Coupland VH, Rosenberg PS, Cook MB. Fatal prostate cancer incidence trends in the United States and England by race, stage, and treatment. Br J Cancer. 2020;123(3):487-494
37. Marima R, Hull R, Mbeje M. et al. Role of Precision Oncology in Type II Endometrial and Prostate Cancers in the African Population: Global Cancer Genomics Disparities. Int J Mol Sci. 2022;23(2):628
38. Zhou CK, Check DP, Lortet-Tieulent J, Laversanne M, Jemal A, Ferlay J, Bray F, Cook MB, Devesa SS. Prostate cancer incidence in 43 populations worldwide: An analysis of time trends overall and by age group. Int J Cancer. 2016;138(6):1388-400
39. Dyba T, Randi G, Bray F, Martos C, Giusti F, Nicholson N, Gavin A, Flego M, Neamtiu L, Dimitrova N, Negrão Carvalho R, Ferlay J, Bettio M. The European cancer burden in 2020: Incidence and mortality estimates for 40 countries and 25 major cancers. Eur J Cancer. 2021;157:308-347
40. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7-33
41. Andriole GL, Crawford ED, Grubb RL 3rd, Buys SS, Chia D, Church TR. et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310-9
42. Halpern JA, Shoag JE, Mittal S, Oromendia C, Ballman KV, Hershman DL. et al. Prognostic Significance of Digital Rectal Examination and Prostate Specific Antigen in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Arm. J Urol. 2017;197(2):363-368
43. Pinsky PF, Prorok PC, Yu K, Kramer BS, Black A, Gohagan JK, Crawford ED, Grubb RL, Andriole GL. Extended mortality results for prostate cancer screening in the PLCO trial with median follow-up of 15 years. Cancer. 2017;123(4):592-599
44. Pinsky PF, Miller E, Prorok P, Grubb R, Crawford ED, Andriole G. Extended follow-up for prostate cancer incidence and mortality among participants in the Prostate, Lung, Colorectal and Ovarian randomized cancer screening trial. BJU Int. 2019;123(5):854-860
45. Timofte AD, Căruntu ID. The prostatic cellular and molecular kaleidoscope. Starting points for carcinogenesis. Rom J Morphol Embryol. 2018;59(1):43-48
46. Liu A, Wei L, Gardner WA, Deng CX, Man YG. Correlated alterations in prostate basal cell layer and basement membrane. Int J Biol Sci. 2009;5(3):276-85
47. Lee CH, Akin-Olugbade O, Kirschenbaum A. Overview of prostate anatomy, histology, and pathology. Endocrinol Metab Clin North Am. 2011;40(3):565-75
48. Packer JR, Maitland NJ. The molecular and cellular origin of human prostate cancer. Biochim Biophys Acta. 2016;1863(6 Pt A):1238-60
49. Strand DW, Aaron L, Henry G, Franco OE, Hayward SW. Isolation and analysis of discreet human prostate cellular populations. Differentiation. 2016;91(4-5):139-51
50. Zhou M, Shah R, Shen R, Rubin MA. Basal cell cocktail (34betaE12 + p63) improves the detection of prostate basal cells. Am J Surg Pathol. 2003;27(3):365-71
51. Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131(20):4955-64
52. Grisanzio C, Signoretti S. p63 in prostate biology and pathology. J Cell Biochem. 2008;103(5):1354-68
53. Pierson CR, McGowen R, Grignon D, Sakr W, Dey J, Sheng S. Maspin is up-regulated in premalignant prostate epithelia. Prostate. 2002;53(4):255-62
54. Lovrić E, Gatalica Z, Eyzaguirre E, Kruslin B. Expression of maspin and glutathionine-S-transferase-pi in normal human prostate and prostatic carcinomas. Appl Immunohistochem Mol Morphol. 2010;18(5):429-32
55. Lee SH, Shen MM. Cell types of origin for prostate cancer. Curr Opin Cell Biol. 2015;37:35-41
56. Lam JS, Reiter RE. Stem cells in prostate and prostate cancer development. Urol Oncol. 2006;24(2):131-40
57. Schalken JA, van Leenders G. Cellular and molecular biology of the prostate: stem cell biology. Urology. 2003;62(5 Suppl 1):11-20
58. van Leenders GJ, Schalken JA. Epithelial cell differentiation in the human prostate epithelium: implications for the pathogenesis and therapy of prostate cancer. Crit Rev Oncol Hematol. 2003;46(Suppl):S3-10
59. De Marzo AM, Meeker AK, Epstein JI, Coffey DS. Prostate stem cell compartments: expression of the cell cycle inhibitor p27Kip1 in normal, hyperplastic, and neoplastic cells. Am J Pathol. 1998;153(3):911-9
60. Tunn S, Nass R, Ekkernkamp A, Schulze H, Krieg M. Evaluation of average life span of epithelial and stromal cells of human prostate by superoxide dismutase activity. Prostate. 1989;15(3):263-71
61. Prins GS, Jung MH, Vellanoweth RL, Chatterjee B, Roy AK. Age-dependent expression of the androgen receptor gene in the prostate and its implication in glandular differentiation and hyperplasia. Dev Genet. 1996;18(2):99-106
62. Man YG, Shen T, Zhao Y, Amy Sang QX. Focal prostate basal cell layer disruptions and leukocyte infiltration are correlated events: A potential mechanism for basal cell layer disruptions and tumor invasion. Cancer Detect Prev. 2005;29(2):161-9
63. Liu A, Wei L, Gardner WA, Deng CX, Man YG. Correlated alterations in prostate basal cell layer and basement membrane. Int J Biol Sci. 2009;5(3):276-85
64. Song G, Hsiao H, Wang JL, Mannion C, Stojadinovic A. et al. Differential impact of tumor-infiltrating immune cells on basal and luminal cells: implications for tumor invasion and metastasis. Anticancer Res. 2014;34(11):6363-80
65. Man YG, Gardner WA. Focal degeneration of basal cells and the resultant auto-immunoreactions: a novel mechanism for prostate tumor progression and invasion. Med Hypotheses. 2008;70(2):387-408
66. Man YG, Fu SW, Liu AJ, Stojadinovic A, Izadjoo MJ, Chen L, Gardner WA. Aberrant expression of chromogranin A, miR-146a, and miR-146b-5p in prostate structures with focally disrupted basal cell layers: an early sign of invasion and hormone-refractory cancer? Cancer Genomics Proteomics. 2011;8(5):235-44
67. Man YG, Gardner WA. Bad seeds produce bad crops: a single stage-process of prostate tumor invasion. Int J Biol Sci. 2008;4(4):246-58
68. Jiang B, Mason J, Jewett A. Liu ML, Chen W, Qian J, et al. Tumor-infiltrating immune cells.
69. trigger for tumor capsule disruption and tumor progression? Int J Med Sci.2013;10(5):475-97
70. Ninomiya S, Kawahara T, Iwashita H, Iwamoto G, Takamoto D, Mochizuki T. et al. Prostate basal cell carcinoma: A case report. Case Rep Oncol. 2018;11:138-142
71. Chang K, Dai B, Kong Y. et al. Basal cell carcinoma of the prostate: clinicopathologic analysis of three cases and a review of the literature. World J Surg Oncol. 2013;11(1):193
72. Begnami MD, Quezado M, Pinto P. et al. Adenoid cystic/basal cell carcinoma of the prostate: Review and update. Arch Pathol Lab Med. 2007;131:637-640
73. McKenney JK, Amin MB, Srigley JR, Jimenez RE, Ro JY, Grignon DJ, Young RH. Basal cell proliferations of the prostate other than usual basal cell hyperplasia: a clinicopathologic study of 23 cases, including four carcinomas, with a proposed classification. Am J Surg Pathol. 2004;28(10):1289-98
74. He L, Metter C, Margulis V, Kapur P. A review leveraging a rare and unusual case of basal cell carcinoma of the prostate. Case Rep Pathol. 2021;2021:5520581
75. Peng YC, Levine CM, Zahid S, Wilson EL, Joyner AL. Sonic hedgehog signals to multiple prostate stromal stem cells that replenish distinct stromal subtypes during regeneration. Proc Natl Acad Sci U S A. 2013;110(51):20611-6
76. Gaisa NT, Graham TA, McDonald SA, Poulsom R, Heidenreich A, Jakse G, Knuechel R, Wright NA. Clonal architecture of human prostatic epithelium in benign and malignant conditions. J Pathol. 2011;225(2):172-80
77. Barclay WW, Axanova LS, Chen W, Romero L, Maund SL, Soker S, Lees CJ, Cramer SD. Characterization of adult prostatic progenitor/stem cells exhibiting self-renewal and multilineage differentiation. Stem Cells. 2008;26(3):600-10
78. Moad M, Hannezo E, Buczacki SJ, Wilson L, El-Sherif A, Sims D. et al. Multipotent Basal Stem Cells, Maintained in Localized Proximal Niches, Support Directed Long-Ranging Epithelial Flows in Human Prostates. Cell Rep. 2017;20(7):1609-1622
79. Le V, He Y, Aldahl J, Hooker E, Yu EJ, Olson A. et al. Loss of androgen signaling in mesenchymal sonic hedgehog responsive cells diminishes prostate development, growth, and regeneration. PLoS Genet. 2020;16(1):e1008588
80. Olson AW, Le V, Wang J, Hiroto A, Kim WK, Lee DH. et al. Stromal androgen and hedgehog signaling regulates stem cell niches in pubertal prostate development. Development. 2021;148(19):dev199738
81. Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25(11):2760-9
82. Man YG, Zhao C, Chen X. A subset of prostate basal cells lacks the expression of corresponding phenotypic markers. Pathol Res Pract. 2006;202(9):651-62
83. Ni WD, Yang ZT, Cui CA, Cui Y, Fang LY, Xuan YH. Tenascin-C is a potential cancer-associated fibroblasts marker and predicts poor prognosis in prostate cancer. Biochem Biophys Res Commun. 2017;486(3):607-612
84. Mishra P, Kiebish MA, Cullen J, Srinivasan A, Patterson A, Sarangarajan R, Genomic alterations of Tenascin C in highly aggressive prostate cancer. a meta-analysis. Genes Cancer. 2019;10(5-6):150-159
85. Qian Y, Liu X, Feng Y, Li X, Xuan Y. Tenascin C regulates cancer cell glycolysis and tumor progression in prostate cancer. Int J Urol. 2022;29(6):578-585
86. Lee YC, Lin SC, Yu G, Zhu M, Song JH, Rivera K. et al. Prostate tumor-induced stromal reprogramming generates Tenascin C that promotes prostate cancer metastasis through YAP/TAZ inhibition. Oncogene. 2022;41(6):757-769
87. Kato M. Editorial Comment to Tenascin C regulates cancer cell glycolysis and tumor progression in prostate cancer. Int J Urol. 2022;29(6):585-586
88. Lovrić E, Gatalica Z, Eyzaguirre E, Kruslin B. Expression of maspin and glutathionine-S-transferase-pi in normal human prostate and prostatic carcinomas. Appl Immunohistochem Mol Morphol. 2010;18(5):429-32
89. Pierson CR, McGowen R, Grignon D, Sakr W, Dey J, Sheng S. Maspin is up-regulated in premalignant prostate epithelia. Prostate. 2002;53(4):255-62
90. Shankar E, Pandey M, Verma S, Abbas A, Candamo M, Kanwal R, Shukla S, MacLennan GT, Gupta S. Role of class I histone deacetylases in the regulation of maspin expression in prostate cancer. Mol Carcinog. 2020;59(8):955-966
91. Tang S, Lian X, Jiang J, Cheng H, Guo J, Huang C, Meng H, Li X. Tumor Suppressive Maspin-Sensitized Prostate Cancer to Drug Treatment Through Negative Regulating Androgen Receptor Expression. Front Cell Dev Biol. 2020;8:573820
92. Van Poppel H, Roobol MJ, Chapple CR, Catto JWF, N'Dow J, Sønksen J, Stenzl A, Wirth M. Prostate-specific Antigen Testing as Part of a Risk-Adapted Early Detection Strategy for Prostate Cancer: European Association of Urology Position and Recommendations for 2021. Eur Urol. 2021;80(6):703-711
93. Pathirana T, Sequeira R, Del Mar C, Dickinson JA, Armstrong BK, Bell KJL, Glasziou P. Trends in Prostate Specific Antigen (PSA) testing and prostate cancer incidence and mortality in Australia: A critical analysis. Cancer Epidemiol. 2022;77:102093
94. Karunasinghe N, Minas TZ, Bao BY, Lee A, Wang A, Zhu S, Masters J, Goudie M, Huang SP, Jenkins FJ, Ferguson LR. Assessment of factors associated with PSA level in prostate cancer cases and controls from three geographical regions. Sci Rep. 2022;12(1):55
95. Flores JM, Bernie HL, Miranda E, Nascimento B, Schofield E, Benfante N, Carlsson S, Mulhall JP. The Relationship Between PSA and Total Testosterone Levels in Men With Prostate Cancer. J Sex Med. 2022;19(3):471-478
96. Grisanzio C, Signoretti S. p63 in prostate biology and pathology. J Cell Biochem. 2008;103(5):1354-68
97. Tomkova K, Tomka M, Zajac V. Contribution of p53, p63, and p73 to the developmental diseases and cancer. Neoplasma. 2008;55(3):177-81
98. Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH. et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A. 2012;109(38):15312-7
99. Ferronika P, Triningsih FX, Ghozali A, Moeljono A, Rahmayanti S, Shadrina AN. et al. p63 cytoplasmic aberrance is associated with high prostate cancer stem cell expression. Asian Pac J Cancer Prev. 2012;13(5):1943-8
100. Dhillon PK, Barry M, Stampfer MJ, Perner S, Fiorentino M, Fornari A. et al. Aberrant cytoplasmic expression of p63 and prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2009;18(2):595-600
101. Yoshida T, Okuyama H, Nakayama M, Endo H, Tomita Y, Nonomura N, Nishimura K, Inoue M. Dynamic Change in p63 Protein Expression during Implantation of Urothelial Cancer Clusters. Neoplasia. 2015;17(7):574-85
102. Stefan-van Staden RI, Bogea IM, Ilie-Mihai RM, Gheorghe DC, Coroş M, Pruneanu SM. Stochastic microsensors based on modified graphene for pattern recognition of maspin in biological samples. Anal Bioanal Chem. 2022;414(12):3667-3673
103. Stefan-van Staden RI, Ilie-Mihai RM, Gheorghe DC, Bogea IM, Badulescu M. 2D disposable stochastic sensors for molecular recognition and quantification of maspin in biological samples. Mikrochim Acta. 2022;189(3):101
104. Pannek J, Brands FH. Zusätzliche Hilfen bei der Erkennung von Prostatakarzinomen? PSA-Prostatavolumenquotient, PSA-Verdopplungszeit, altersabhängige PSA-Referenzwerte und PSA im Urin [Additional aids in detection of prostate carcinomas? PSA-prostatic volume quotient, PSA-doubling time, age-dependent PSA reference values and PSA in urine]. Urologe A. 2000;39(4):324-9
105. Occhipinti S, Mengozzi G, Oderda M, Zitella A, Molinaro L, Novelli F, Giovarelli M, Gontero P. Low Levels of Urinary PSA Better Identify Prostate Cancer Patients. Cancers (Basel). 2021;13(14):3570
106. Eskra JN, Rabizadeh D, Zhang J, Isaacs WB, Luo J, Pavlovich CP. Specific Detection of Prostate Cancer Cells in Urine by RNA In Situ Hybridization. J Urol. 2021;206(1):37-43
107. Juracek J, Madrzyk M, Stanik M, Slaby O. Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update. Cancers (Basel). 2022;14(13):3157
108. Guelfi G, Cochetti G, Stefanetti V, Zampini D, Diverio S, Boni A, Mearini E. Next
109. Generation Sequencing of urine exfoliated cells. an approach of prostate cancer microRNAs research. Sci Rep. 2018;8(1):7111
110. Eskra JN, Rabizadeh D, Mangold L, Fabian E, Brennen WN, Yeater DB, Pienta KJ, Partin AW, Isaacs WB, Pavlovich CP, Luo J. A novel method for detection of exfoliated prostate cancer cells in urine by RNA in situ hybridization. Prostate Cancer Prostatic Dis. 2021;24(1):220-232
111. Pal RP, Kockelbergh RC, Pringle JH, Cresswell L, Hew R, Dormer JP, Cooper C, Mellon JK, Barwell JG, Hollox EJ. Immunocytochemical detection of ERG expression in exfoliated urinary cells identifies with high specificity patients with prostate cancer. BJU Int. 2016;117(4):686-96
112. Guan Y, Wang X, Guan K, Wang D, Bi X, Xiao Z, Xiao Z, Shan X, Hu L, Ma J, Li C, Zhang Y, Shou J, Wang B, Qian Z, Xing N. Copy number variation of urine exfoliated cells by low-coverage whole genome sequencing for diagnosis of prostate adenocarcinoma: a prospective cohort study. BMC Med Genomics. 2022;15(Suppl 2):104
113. Diamandis EP. Prostate specific antigen-new applications in breast and other cancers. Anticancer Res. 1996;16(6C):3983-4
114. Wang HL, Wang SS, Song WH, Pan Y, Yu HP, Si TG, Liu Y, Cui XN, Guo Z. Expression of prostate-specific membrane antigen in lung cancer cells and tumor neovasculature endothelial cells and its clinical significance. PLoS One. 2015 15;10 (5):e0125924
115. Duraker N, Çaynak ZC, Trabulus DC. Free/Total Serum Prostate-Specific Antigen Ratio in Women with Colorectal Cancer Has Prognostic Significance. J Gastrointest Cancer. 2017;48(1):8-12
116. Burton AJ, Martin RM, Donovan JL, Lane JA, Davis M, Hamdy FC, Neal DE, Tilling K. Associations of lifestyle factors and anthropometric measures with repeat PSA levels during active surveillance/monitoring. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1877-85
117. Atalay E, Demir A, Eroglu HA. The Influences of Metformin on Prostate in Terms of PSA Level and Prostate Volume. Urol J. 2020;18(2):181-185
118. Sindhwani P, Wilson CM. Prostatitis and serum prostate-specific antigen. Curr Urol Rep. 2005;6(4):307-12
119. Nowroozi MR, Ayati M, Amini E, Aghamiri SM, Momeni SA, Ohadian Moghadam S, Valizadeh F. Role of Chronic Inflammation as a Predictor of Upstaging/Upgrading in Prostate Cancer: Finding a New Group Eligible for Active Surveillance. Urol J. 2020;17(4):370-373
120. Lim J, Bhoo-Pathy N, Sothilingam S, Malek R, Sundram M, Hisham Bahadzor B, Ong TA, Ng KL, Sivalingam S, Razack AH. Ethnicity is an independent determinant of age-specific PSA level: findings from a multiethnic Asian setting. PLoS One. 2014;9(8):e104917
121. Liu M, Wang JY, Zhu L, Wan G. Body mass index and serum lipid profile influence serum prostate-specific antigen in Chinese men younger than 50 years of age. Asian J Androl. 2011;13(4):640-3
122. Man YG, Sang QX. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: a paradigm shift from the "protease-centered" hypothesis. Exp Cell Res. 2004;301(2):103-18
123. Man YG, Tai L, Barner R, Vang R, Saenger JS, Shekitka KM, Bratthauer GL, Wheeler DT, Liang CY, Vinh TN, Strauss BL. Cell clusters overlying focally disrupted mammary myoepithelial cell layers and adjacent cells within the same duct display different immunohistochemical and genetic features: implications for tumor progression and invasion. Breast Cancer Res. 2003;5(6):R231-41
124. Man YG. Focal degeneration of aged or injured myoepithelial cells and the resultant auto-immunoreactions are trigger factors for breast tumor invasion. Med Hypotheses. 2007;69(6):1340-57
125. Man YG, Izadjoo M, Song G, Stojadinovic A. In situ malignant transformation and progenitor-mediated cell budding: two different pathways for breast ductal and lobular tumor invasion. J Cancer. 2011;2:401-12
126. Yuan H, Hsiao YH, Zhang Y, Wang J, Yin C, Shen R, Su Y. Destructive impact of T-lymphocytes, NK and Mast cells on basal cell layers: implications for tumor invasion. BMC Cancer. 2013;13:258
127. Kadota K, Yeh YC, Villena-Vargas J, Cherkassky L, Drill EN, Sima CS. et al. Tumor Budding Correlates With the Protumor Immune Microenvironment and Is an Independent Prognostic Factor for Recurrence of Stage I Lung Adenocarcinoma. Chest. 2015;148(3):711-721
128. Maibach F, Sadozai H, Seyed Jafari SM, Hunger RE, Schenk M. Tumor-Infiltrating Lymphocytes and Their Prognostic Value in Cutaneous Melanoma. Front Immunol. 2020;11:2105
129. Long X, Hou H, Wang X, Liu S, Diao T, Lai S. et al. Immune signature driven by ADT-induced immune microenvironment remodeling in prostate cancer is correlated with recurrence-free survival and immune infiltration. Cell Death Dis. 2020;11(9):779
130. Kim SW, Kim YI, Mustafa B, Kim MJ, Jeong G, Ahn SM. et al. Distribution pattern of tumor infiltrating lymphocytes and tumor microenvironment composition as prognostic indicators in anorectal malignant melanoma. Mod Pathol. 2021Jan;34(1):141-160
131. Fan C, Lu W, Li K, Zhao C, Wang F, Ding G, Wang J. Identification of immune cell infiltration pattern and related critical genes in metastatic castration-resistant prostate cancer by bioinformatics analysis. Cancer Biomark. 2021;32(3):363-377
132. Wu Y, Huang W, Xie Y, Wang C, Luo N, Chen Y. et al. Siglec-9, a Putative Immune Checkpoint Marker for Cancer Progression Across Multiple Cancer Types. Front Mol Biosci. 2022;9:743515
133. Lyu F, Li Y, Yan Z, He Q, Cheng L, Zhang P. et al. Identification of ISG15 and ZFP36 as novel hypoxia- and immune-related gene signatures contributing to a new perspective for the treatment of prostate cancer by bioinformatics and experimental verification. J Transl Med. 2022;20(1):202
134. Vidal AC, Freedland SJ. Aspirin and prostate cancer prevention. Aging (Albany NY). 2015;7(5):292-3
135. Joshi SN, Murphy EA, Olaniyi P, Bryant RJ. The multiple effects of aspirin in prostate cancer patients. Cancer Treat Res Commun. 2021;26:100267
136. Hurwitz LM, Kulac I, Gumuskaya B, Valle JABD, Benedetti I, Pan F. et al. Use of Aspirin and Statins in Relation to Inflammation in Benign Prostate Tissue in the Placebo Arm of the Prostate Cancer Prevention Trial. Cancer Prev Res (Phila). 2020;13(10):853-862
137. Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, He Zhu H. Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate. Nat Commun. 2020;11(1):706
138. Tremblay M, Viala S, Shafer ME, Graham-Paquin AL, Liu C, Bouchard M. Regulation of stem/progenitor cell maintenance by BMP5 in prostate homeostasis and cancer initiation. Elife. 2020;9:e54542
139. Costa CD, Justo AA, Kobayashi PE, Story MM, Palmieri C, Laufer Amorim R, Fonseca-Alves CE. Characterization of OCT3/4, Nestin, NANOG, CD44 and CD24 as stem cell markers in canine prostate cancer. Int J Biochem Cell Biol. 2019;108:21-28
140. Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y. et al. Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate. Nat Commun. 2020;11(1):706
141. Centonze A, Lin S, Tika E, Sifrim A, Fioramonti M, Malfait M. et al. Heterotypic cell-cell communication regulates glandular stem cell multipotency. Nature. 2020;584(7822):608-613
Author contact
![]() Corresponding authors: Yan-gao Man., MD., PhD. E-mail: ymanorg or Yanmanncom or Liang Cheng., MD. E-mail: liang_chengcom or liang_chengedu
Corresponding authors: Yan-gao Man., MD., PhD. E-mail: ymanorg or Yanmanncom or Liang Cheng., MD. E-mail: liang_chengcom or liang_chengedu

 Global reach, higher impact
Global reach, higher impact