Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(3):360-366. doi:10.7150/jca.81083 This issue Cite
Research Paper
Impact of tissue inhibitor of metalloproteinases-3 genetic variants on clinicopathological characteristics of urothelial cell carcinoma
1. Division of Urology, Department of Surgery, Tungs' Taichung MetroHarbor Hospital, Taichung, Taiwan
2. Department of Nursing, Jen-Teh Junior College of Medicine, Nursing and Management, Miaoli, Taiwan
3. School of Medicine, Chung Shan Medical University, Taichung, Taiwan
4. Department of Psychiatry, Chung Shan Medical University Hospital, Taichung, Taiwan
5. School of Medical Laboratory and Biotechnology, Chung Shan Medical University, Taichung, Taiwan
6. Department of Ophthalmology, Nobel Eye Institute, Taipei, Taiwan
7. School of Medicine, China Medical University, Taichung, Taiwan
8. Chinese Medicine Research Center, China Medical University, Taichung, Taiwan
9. Department of Medical Laboratory Science and Biotechnology, College of Medical and Health Science, Asia University, Taichung, Taiwan
10. Department of Mathematical Sciences, Florida Atlantic University, Boca Raton, FL, USA
11. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan
12. Department of Applied Chemistry, National Chi Nan University, Nantou, Taiwan
13. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
14. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
Received 2022-11-21; Accepted 2023-1-6; Published 2023-1-22
Abstract
To investigate the distribution of single nucleotide polymorphism (SNP) of tissue inhibitor of metalloproteinases-3 (TIMP-3) in patients with/without urothelial cell carcinoma (UCC), three loci of TIMP-3 SNPs (rs9862 C/T, rs9619311 T/C, rs11547635 C/T) were genotyped via TaqMan allelic discrimination for 424 UCC patients and 848 non-UCC participants. Furthermore, the TIMP-3 mRNA expression and its correlation with clinical characters of urothelial bladder carcinoma was analyzed using The Cancer Genome Atlas database (TCGA). The distribution of all 3 studied SNPs of TIMP-3 was insignificantly different between the UCC and non-UCC groups. However, significantly lower tumor T status was found in TIMP-3 SNP rs9862 CT + TT variant than the wild type (OR: 0.515, 95% CI: 0.289-0.917, P = 0.023). Moreover, the muscle invasive tumor type was significantly correlated to the TIMP-3 SNP rs9619311 TC + CC variant in the non-smoker subgroup (OR: 2.149, 95% CI: 1.143-4.039, P = 0.016). With the TIMP-3 expression data provided in TCGA, significantly higher TIMP-3 mRNA expression was observed in UCC with high tumor stage (P < 0.0001), high tumor T status (P < 0.0001) and high lymph node status (P = 0.0005). In conclusions, TIMP-3 SNP rs9862 variant is associated with lower tumor T status of UCC while TIMP-3 SNP rs9619311 variant is correlated to muscle invasive UCC development in non-smoker.
Keywords: single nucleotide polymorphism, urothelial cell carcinoma, tissue inhibitor of metalloproteinases-3, tobacco, tumor stage
Introduction
The urothelial cell carcinoma (UCC) is a common neoplasm with an annual incidence rates above 3 per 100,000 person-years in the eastern Asian region [1]. In the advanced form, the treatment options of UCC including surgery, platinum-based chemotherapy, immunotherapy and targeted therapy [2, 3]. About the risk factor of UCC, the tobacco consumption is the most important risk factor which may account for nearly half of UCC cases [4]. Also, the occupational exposure to substances like aromatic amines is a predictor for UCC development [5].
About the genetic aspect, several genes and their products are related to the development of UCC. The matrix metalloproteinases are a well-established genetic risk factor for the UCC occurrence especially for the matrix metalloproteinases-2 and matrix metalloproteinases-9 [6]. In addition, the expression of CCNA1 was significantly higher in the urine of UCC patients compared to the control group [7]. On the other hand, the single nucleotide polymorphism (SNP) of certain genes would influence the incidence or characters of UCC. For instance, certain SNP of AURKA including rs2064863 and rs6024836 made a prominent influence on the clinical characteristics of UCC which mainly retarded the tumor progression [8]. Besides, the patients with HMGB1 rs1045411T allele were under a lower risk of UCC development [9]. Accordingly, other gene or SNP may also affect the clinical status of UCC.
The tissue inhibitor of metalloproteinases-3 (TIMP-3) can alter the activity of the matrix metalloproteinases family and a higher concentration of TIMP-3 was observed in the patients with malignancies compared to those without such lesions [10-13]. In previous researches, the TIMP-3 and its genetic polymorphism would affect the clinical features of oral cancer and lung adenocarcinoma [14, 15]. However, there was rare research discussing the relationship between the genetic variant of TIMP-3 and UCC. In previous studies, both oral cancer and UCC are associated with epithelial agent [16-20], plus the TIMP-3 can alter the activity of the matrix metalloproteinases which is related to the UCC development [6]. Consequently, TIMP-3 and its SNP variant may influence UCC progression which needs further investigation.
The purpose of current study is to evaluate the correlation between the SNP of TIMP-3 and clinicopathological characters of UCC in a Taiwanese population. In addition, the results of urothelial bladder carcinoma from The Cancer Genome Atlas database were included and discussed.
Materials and Methods
Subject selection
This study was executed in Taichung Veterans General Hospital. Those who diagnosed with UCC in the Taichung Veterans General Hospital were selected and a total of 424 patients were included between Jan 2010 and Dec 2015 in the study group. Besides, subjects with history of cancer of any sites were excluded from the control group. The demographic data included age, gender, tobacco consumption history of these patients was taken from the medical document. The Tumor, Node, Metastasis (TNM) status, tumor stages were classified according to the American Joint Committee on Cancer. Patients would be dropped out from the current study of the blood samples degraded before the genetic variants analyses. The content of our study was adhered to declaration of Helsinki in 1964 and associated amendments. The Institutional Review Boards of Taichung Veterans General Hospital also approved our study (Project code: no. CF11094; 27 July 2011). A written informed consent was obtained from each participant after explaining the details of our study. For the polymorphism of TIMP-3, venous blood sample was taken and then preserved in the ethylene-diaminetetraacetic acid-containing tubes. The blood samples were then centrifuged and put in our laboratory refrigerator at -80 degree Celsius for our analyses.
Genomic DNA extraction and analyze TIMP-3 SNP via Real-Time PCR
Three SNPs of TIMP-3 including rs9862 (C/T), rs9619311 (T/C), rs11547635 (C/T) were picked out since our previous experience showed the effect of these SNPs on the oral cancer [15]. The genotyping procedure used in our study was similar as our previous research [21-23]. The genome was firstly taken from leukocytes of blood sample via the QIAamp DNA kits (Qiagen, Valencia, Valencia, CA, USA), and all the procedures with QIAamp DNA kits was adhered to the manufacture's guideline. We preserved theses isolated DNA in refrigerators under -20 degree Celsius. In the next step, the three TIMP-3 genetic polymorphisms we selected were analyzed with the use of ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, California). After all the procedures, the genetic polymorphisms about the three TIMP-3 SNPs were analyzed via TaqMan assay technique and SDS version 3.0 software (Applied Biosystems) to augment the completeness of Real-Time PCR in our study.
Bioinformatics analysis of TIMP-3 expression
For the potential association between TIMP-3 expression and clinical status of UCC, we use the data of urothelial bladder carcinoma obtained from The Cancer Genome Atlas (TCGA) to analyze this issue [24-26]. In this part of analysis, urothelial bladder carcinoma was divided into different subgroup according to the tumor stage and TNM statuses, then the mRNA level of TIMP-3 was compared between the subgroups.
Statistical analysis
The SAS version 9.4 (SAS Institute Inc, Cary, NC, USA) was used for the statistical analyses in the current study. Descriptive analysis including mean, standard deviation (SD) and percentage was used to showed the demography and laboratory data between the non-UCC and UCC groups. The student's t test or chi-squared test was used to compare different parameters between control group and patients with UCC. Then the logistic regression models were used to produce the odds ratio (OR) and associated 95% confidence interval (CI) about the polymorphism distribution between the non-UCC and UCC population. Moreover, the adjusted odds ratio (AOR) with 95% CI between the two groups was calculated via multiple logistic regression models after adjusting age, gender and tobacco consumption. For the subgroup analyses in the UCC population, the distribution frequencies between the different genotypes of TIMP-3 SNP rs9862 as well as rs9619311 and the clinical condition of UCC were presented as OR with 95% CI. Further, we divided the UCC population into non-smoker and smoker, and the distribution frequency between TIMP-3 SNP rs9619311 and clinicopathological characters of UCC was analyzed and then produced the AOR with 95% CI. The statistically significant level was set as P < 0.05 in the current study and those with P value lesser than 0.001 was presented as P < 0.001.
The distributions of demographical characteristics in 848 controls and 424 patients with UCC.
| Variable | Non-UCC (N=848)n (%) | UCC (N=424)n (%) | P value |
|---|---|---|---|
| Age (yrs) | |||
| Mean ± SD | 57.09 ± 10.04 | 68.58 ± 11.84 | <0.001 |
| Gender | 0.320 | ||
| Male | 554 (65.3%) | 265 (62.5%) | |
| Female | 294 (34.7%) | 159 (37.5%) | |
| Tobacco consumption | 0.151 | ||
| No | 558 (65.8%) | 296 (69.8%) | |
| Yes | 290 (34.2%) | 128 (30.2%) | |
| Stage | |||
| Non muscle invasive tumor (pTa-pT1) | 231 (54.5%) | ||
| Muscle invasive tumor (pT2-pT4) | 193 (45.5%) | ||
| Tumor T status | |||
| Ta | 87 (20.5%) | ||
| T1-T4 | 337 (79.5%) | ||
| Lymph node status | |||
| N0 | 374 (88.2%) | ||
| N1+N2 | 50 (11.8%) | ||
| Metastasis | |||
| M0 | 411 (96.9%) | ||
| M1 | 13 (3.1%) | ||
| Histopathologic grading | |||
| Low grade | 51 (12.0%) | ||
| High grade | 373 (88.0%) |
N: number; SD: standard deviation
Results
Basic characters between the non-UCC and UCC groups
The demography of the non-UCC and UCC groups are shown in Table 1. The mean age in the non-UCC group was 57.09 ± 10.04 years which was significant younger than that in the UCC group (68.58 ± 11.84, P < 0.001), while the gender and tobacco consumption distribution did not differ between the two groups (both P > 0.05). The tumor feature of the UCC group including tumor stage, TNM status and histopathologic grading are also available in Table 1.
Distribution frequencies of TIMP-3 SNPs between non-UCC and UCC groups
The genotype distribution of TIMP-3 SNPs between the non-UCC and UCC population are presented in the Table 2. Both the TIMP-3 SNP rs9862 CT+TT and TIMP-3 SNP rs9619311 TC+CC were numerically higher in the UCC group than the non-UCC group, while the TIMP-3 SNP rs11547635 CT+TT was numerically lower in the UCC group than the non-UCC group. Nevertheless, none of these values demonstrated significant difference between the UCC and non-UCC group regarding both the OR and AOR which adjusting age, gender and tobacco consumption (all P > 0.05) (Table 2).
Genotype Distributions of TIMP-3 Gene Polymorphisms in 848 Controls and 424 Patients with UCC.
| Variable | Non-UCC (N=848) n (%) | UCC (N=424) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs9862 | ||||
| CC | 293 (34.6%) | 125 (29.5%) | 1.000 (reference) | 1.000 (reference) |
| CT | 393 (46.3%) | 219 (51.7%) | 1.306 (0.997-1.706) | 1.256 (0.926-1.704) |
| TT | 162 (19.1%) | 80 (18.8%) | 1.158 (0.824-1.626) | 1.100 (0.747-1.619) |
| CT+TT | 555 (65.4%) | 299 (70.5%) | 1.124 (0.991-1.275) | 1.100 (0.952-1.271) |
| rs9619311 | ||||
| TT | 702 (82.8%) | 346 (81.6%) | 1.000 | 1.000 (reference) |
| TC | 135 (15.9%) | 76 (17.9%) | 1.142 (0.838-1.556) | 1.222 (0.859-1.739) |
| CC | 11 (1.3%) | 2 (0.5%) | 0.369 (0.081-1.673) | 0.490 (0.098-2.439) |
| TC+CC | 146 (17.2%) | 78 (18.4%) | 1.041 (0.895-1.212) | 1.082 (0.910-1.286) |
| rs11547635 | ||||
| CC | 374 (44.1%) | 192 (45.3%) | 1.000 (reference) | 1.000 (reference) |
| CT | 374 (44.1%) | 189 (44.6%) | 0.984 (0.769-1.260) | 0.949 (0.716-1.258) |
| TT | 100 (11.8%) | 43 (10.1%) | 0.838 (0.563-1.246) | 0.700 (0.444-1.101) |
| CT+TT | 474 (55.9%) | 232 (54.7%) | 0.976 (0.868-1.098) | 0.945 (0.827-1.080) |
N: number
OR: odds ratio
AOR: adjusted odds ratio with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, gender and tobacco consumption.
CI: confidence interval
Distribution frequency of the clinical status and TIMP-3 rs9862 genotype frequencies in 424 UCC patients.
| Variable | TIMP-3 (rs9862) | |||
|---|---|---|---|---|
| CC (%) (n=125) | CT + TT (%) (n=299) | OR (95% CI) | P value | |
| Stage | ||||
| Non muscle invasive tumor (pTa-pT1) | 63 (50.4%) | 168 (56.2%) | 1.000 (reference) | 0.275 |
| Muscle invasive tumor (pT2-pT4) | 62 (49.6%) | 131 (43.8%) | 0.792 (0.521-1.204) | |
| Tumor T status | ||||
| Ta | 17 (13.6%) | 70 (23.4%) | 1.000 (reference) | 0.023* |
| T1-T4 | 108 (86.4%) | 229 (76.6%) | 0.515 (0.289-0.917) | |
| Lymph node status | ||||
| N0 | 108 (86.4%) | 266 (89.0%) | 1.000 (reference) | 0.456 |
| N1+N2 | 17 (13.6%) | 33 (11.0%) | 0.788 (0.421-1.475) | |
| Metastasis | ||||
| M0 | 123 (98.4%) | 288 (96.3%) | 1.000 (reference) | 0.258 |
| M1 | 2 (1.6%) | 11 (3.7%) | 2.349 (0.513-10.755) | |
| Histopathologic grading | ||||
| Low grade | 15 (12.0%) | 36 (12.0%) | 1.000 (reference) | 0.991 |
| High grade | 110 (88.0%) | 263 (88.0%) | 0.996 (0.524-1.893) | |
N: number
OR: odds ratio
* denotes significant difference between the two groups
Distribution frequency of the clinical status and TIMP-3 rs9619311 genotype frequencies in 424 UCC patients.
| Variable | TIMP-3 (rs9619311) | |||
|---|---|---|---|---|
| TT (%) (n=346) | TC + CC (%) (n=78) | OR (95% CI) | P value | |
| Stage | ||||
| Non muscle invasive tumor (pTa-pT1) | 195 (56.4%) | 36 (46.2%) | 1.000 (reference) | 0.102 |
| Muscle invasive tumor (pT2-pT4) | 151 (43.6%) | 42 (53.8%) | 1.507 (0.920-2.467) | |
| Tumor T status | ||||
| Ta | 74 (21.4%) | 13 (16.7%) | 1.000 (reference) | 0.351 |
| T1-T4 | 272 (78.6%) | 65 (83.3%) | 1.360 (0.711-2.602) | |
| Lymph node status | ||||
| N0 | 306 (88.4%) | 68 (87.2%) | 1.000 (reference) | 0.755 |
| N1+N2 | 40 (11.6%) | 10 (12.8%) | 1.125 (0.536-2.361) | |
| Metastasis | ||||
| M0 | 336 (97.1%) | 75 (96.2%) | 1.000 (reference) | 0.658 |
| M1 | 10 (2.9%) | 3 (3.8%) | 1.344 (0.361-5.002) | |
| Histopathologic grading | ||||
| Low grade | 42 (12.1%) | 9 (11.5%) | 1.000 (reference) | 0.883 |
| High grade | 304 (87.9%) | 69 (88.5%) | 1.059 (0.492-2.278) | |
N: number
OR: odds ratio
Subgroup Analyses of TIMP-3 SNPs Distribution in the UCC group
In the subgroup analyses, the relationship between the clinical status of UCC and the TIMP-3 SNP rs9862 genotype is shown in Table 3. The TIMP-3 SNP rs9862 CT + TT variant owned a significantly lower tumor T status than the SNP rs9862 CC wild type (OR: 0.515, 95% CI: 0.289-0.917, P = 0.023) while the SNP rs9862 variant and SNP rs9862 wild type showed no difference in other tumor conditions (all P > 0.05) (Table 3). About the TIMP-3 rs9619311 genotype frequencies and the clinical characters of UCC, a similar tumor status was found in each parameter between the TIMP-3 rs9619311 TC + CC variant and TIMP-3 rs9619311 TT wild type (all P > 0.05) (Table 4). After dividing the UCC population into the non-smoker and smoker, the presence of muscle invasive tumor type was significantly correlated to the TIMP-3 SNP rs9619311 TC + CC variant in the non-smoker subgroup (OR: 2.149, 95% CI: 1.143-4.039, P = 0.016) (Table 5). On the other hand, the TIMP-3 SNP rs9619311 genotypes did not correlate to the change of UCC clinopathological characters in the smoker group (all P > 0.05) (Table 5).
Distribution frequency of the clinical status and TIMP-3 rs9619311 genotype frequencies in 424 UCC patients with cigarette smoking status.
| Variable | TIMP-3 (rs9619311) | |||||
|---|---|---|---|---|---|---|
| Non-Smoker (N=296) | Smoker (N=128) | |||||
| TT (%) (n=248) | TC + CC (%) (n=48) | P value | TT (%) (n=98) | TC + CC (%) (n=30) | P value | |
| Stage | ||||||
| Non muscle invasive tumor (pTa-pT1) | 145 (58.5%) | 19 (39.6%) | 0.016a | 50 (51.0%) | 17 (56.7%) | 0.588 |
| Muscle invasive tumor (pT2-pT4) | 103 (41.5%) | 29 (60.4%) | 48 (49.0%) | 13 (43.3%) | ||
| Tumor T status | ||||||
| Ta | 51 (20.6%) | 8 (16.7%) | 0.536 | 23 (23.5%) | 5 (16.7%) | 0.430 |
| T1-T4 | 197 (79.4%) | 40 (83.3%) | 75 (76.5%) | 25 (83.3%) | ||
| Lymph node status | ||||||
| N0 | 222 (89.5%) | 42 (87.5%) | 0.681 | 84 (85.7%) | 26 (86.7%) | 0.896 |
| N1+N2 | 26 (10.5%) | 6 (12.5%) | 14 (14.3%) | 4 (13.3%) | ||
| Metastasis | ||||||
| M0 | 245 (98.8%) | 46 (95.8%) | 0.146 | 91 (92.9%) | 29 (96.7%) | 0.451 |
| M1 | 3 (1.2%) | 2 (4.2%) | 7 (7.1%) | 1 (3.3%) | ||
| Histopathologic grading | ||||||
| Low grade | 32 (12.9%) | 3 (6.3%) | 0.191 | 10 (10.2%) | 6 (20.0%) | 0.156 |
| High grade | 216 (87.1%) | 45 (93.8%) | 88 (89.8%) | 24 (80.0%) | ||
N: number
OR: odds ratio
aOR and 95CI: 2.149 (1.143-4.039)
TIMP-3 mRNA expression in the urothelial bladder carcinoma from TCGA dataset
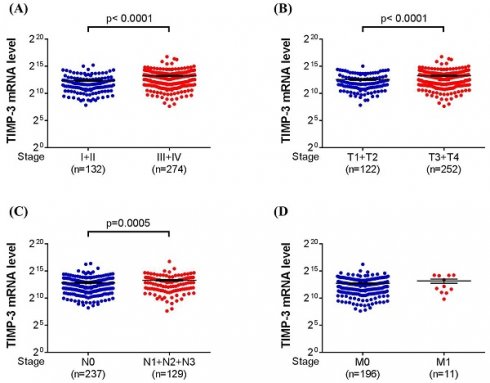
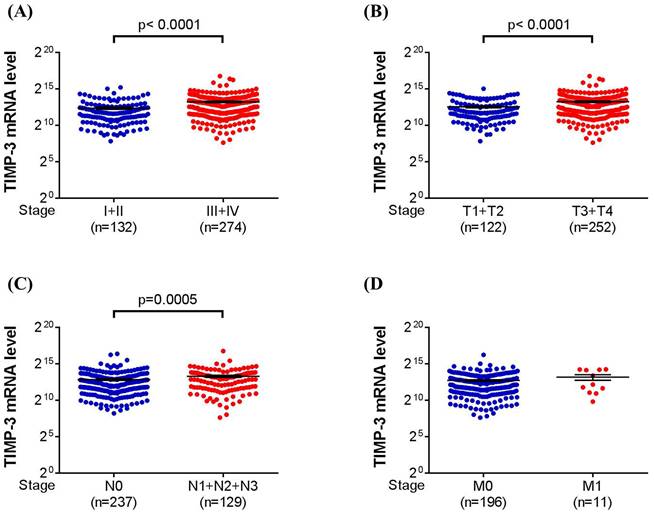
About the TIMP-3 expression in the database from TCGA, we categorized the urothelial bladder carcinoma into low tumor stage (stage I and II) and high tumor stage (stage III and IV), low tumor T status (T1 and T2) and high tumor T status (T3 and T4), no lymph node status (N0) and lymph node status (N1 to N3), and no metastasis (M0) and metastasis (M1). After the analyses, a significantly higher TIMP-3 mRNA level was found in high tumor stage (P < 0.0001), high tumor T status (P < 0.0001) and high lymph node status (P = 0.0005) (Figure 1A-1C). Still, the TIMP-3 mRNA level between no metastasis and metastasis form of urothelial bladder carcinoma was nearly identical (Figure 1D).
Discussion
In our study, the TIMP-3 SNP rs9862 variant is associated with lower tumor T status in patients with UCC. Moreover, the TIMP-3 SNP rs9619311 variant is correlated to higher tumor stage in the non-smoker population who diagnosed with UCC. Besides, the data from TCGA demonstrated that the TIMP-3 mRNA levels showed significant relationship to higher tumor stage, tumor T status and lymph node status in urothelial bladder carcinoma.
Many gene and related polymorphisms would influence the clinical course of UCC in preceding researches. The endothelial nitric oxide synthase rs1799983 GT + TT variants own higher risk of developing large tumor [27]. And the RAGE gene and its polymorphism have been demonstrated to cause high UCC incidence and worse disease-specific survival [28]. Other genetic predictors of UCC include the mutation on TP53/MDM2, RAS, FGFR3, hyper-mutated and triple negative transform [29]. On the other hand, there is also genetic protector for the UCC, in which low level of growth arrest-specific 5 expression in female with bladder urothelial carcinoma showed poorer overall survival rate [30]. About the TIMP-3, this gene is correlated to various diseases including several malignancies [31, 32]. In previous studies, the TIMP-3 would cause higher possibility of cardiovascular diseases development such as myocardial infarction and coronary arterial plaque [33, 34]. For the field of neoplasm, the TIMP-3 and its polymorphism showed significant association to the colorectal cancer and prostate cancer [35, 36]. Moreover, the character of TIMP-3 that can serve as tumor progression predictor let TIMP-3 own the potentiality to become a target for cancer therapy [37]. Because the TIMP-3 illustrated such characters on several tumors, and considering the effect of matrix metalloproteinases on UCC [6], we speculate that the genotype of TIMP-3 may affect the clinical condition of UCC, whether a predictor or protector. Our hypothesis was supported by the results of the current study at least to some degrees.
The TIMP-3 expression in the urothelial bladder carcinoma with different grade according to The Cancer Genome Atlas database. (A) The expression of TIMP-3 mRNA in different tumor stages. (B) The expression of TIMP-3 mRNA in different tumor T statuses. (C) The expression of TIMP-3 mRNA in different lymph node statuses. (D) The expression of TIMP-3 mRNA in different metastasis statuses
For the TIMP-3 SNP variants and the clinicopathological characteristics of UCC, none of the three TIMP-3 SNP variants analyzed in the current study showed significant difference of distribution frequencies between the non-UCC and UCC population. However, the TIMP-3 SNP rs9862 CT + TT genotype is associated with lower tumor T status in the UCC patients. About the percentage aspect, 76.6 percent of TIMP-3 SNP rs9862 CT + TT genotype showed advanced tumor T status while 86.4 percent of TIMP-3 SNP rs9862 CC genotype illustrated advanced tumor T status. In previous researches, the TIMP-3 has both tumorigenic and anti-tumorigenic properties [38, 39]. Accordingly, the genotype of TIMP-3 SNP rs9862 CT + TT may be a protector for the UCC in general population.
Concerning the subgroup analyses in the current study dependent on the existence of tobacco consumption, the UCC individuals who never smoke would experience higher tumor stage of the TIMP-3 SNP rs9619311 variant was existed. This is a relative novel finding in the field of UCC to our knowledge. The muscle invasive tumor is a high-risk form of UCC which needs more complicated therapy than the non-muscle invasive type which may be treated with a curative intent [40, 41]. The previous study showed that the five years survival rate of muscle invasive tumor was around 40 percent [42], which was significantly lower than that in the non-muscle invasive type [43]. Consequently, to find the patients who own higher risk of muscle invasive tumor development should be emphasized. Our study demonstrated that the non-smoker with TIMP-3 SNP rs9619311 TC + CC genotype may be under higher risk of muscle invasive tumor development, thus these patients may be suitable to receive aggressive therapy at the early stage while further experiments are needed to support this concept.
In the TCGA analysis, the TIMP-3 showed higher level of mRNA expression in the UCC with advanced tumor stage, tumor T status and lymph node status. TCGA database enrolled considerable patients with urothelial bladder carcinoma and related literature has been published before [44]. Because of our study design, we did not analyze the quantity of TIMP-3 mRNA expression, but the TCGA database can compensate this shortness in our study. The findings of TCGA data, combined with the results of our patients, illustrated that the TIMP-3 could lead to the higher incidence of advanced UCC development while the TIMP-3 SNP genotypes would alter this condition. This may further highlight the prominent influence of SNP on a tumor-aggregating gene. On the other hand, the existence of metastasis did not influence by the TIMP-3 mRNA expression in TCGA database, and we also found that none of TIMP-3 SNP is associated with the ratio of metastasis. The reasons that TIMP-3 has minimal effect on the UCC metastasis need further evaluation.
In conclusion, individuals with UCC are associated with lower level of tumor T status under the presence of TIMP-3 SNP rs9862 variant. Furthermore, the TIMP-3 SNP rs9619311 variant may lead to higher tumor stage of UCC in the non-smoker population, which is in accordance with the oncogenic effect of TIMP-3 for UCC according to the result of TCGA analysis. Consequently, the presence of TIMP-3 SNP rs9619311 variant might be screened for patients with UCC to find those with high possibility of muscle invasive tumor. Further population-based prospective study to survey whether the SNP variant of TIMP-3 would affect the therapeutic outcome and survival rate of UCC is mandatory.
Acknowledgements
This research was funded by Chung Shan Medical University and Tungs' Taichung Metro Harbor Hospital (CSMU-TTM-109-01). This work was also supported by grants from the Taichung Veterans General Hospital (Grant numbers: TCVGH-1105002B).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lin MY, Niu SW, Li WM, Lee HL, Chen LT, Wu WJ. et al. Incidence and survival variations of upper tract urothelial cancer in Taiwan (2001-2010). Int J Urol. 2022;29:121-127
2. Cathomas R, Lorch A, Bruins HM, Compérat EM, Cowan NC, Efstathiou JA. et al. The 2021 Updated European Association of Urology Guidelines on Metastatic Urothelial Carcinoma. Eur Urol. 2022;81:95-103
3. Freifeld Y, Krabbe LM, Clinton TN, Woldu SL, Margulis V. Therapeutic strategies for upper tract urothelial carcinoma. Expert Rev Anticancer Ther. 2018;18:765-774
4. Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. Jama. 2011;306:737-745
5. Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P. et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234-241
6. Chuang CK, Pang ST, Chuang TJ, Liao SK. Profiling of matrix metalloproteinases and tissue inhibitors of metalloproteinases proteins in bladder urothelial carcinoma. Oncol Lett. 2010;1:691-695
7. Maldonado L, Brait M, Michailidi C, Munari E, Driscoll T, Schultz L. et al. An epigenetic marker panel for recurrence risk prediction of low grade papillary urothelial cell carcinoma (LGPUCC) and its potential use for surveillance after transurethral resection using urine. Oncotarget. 2014;5:5218-5233
8. Huang CH, Chen CJ, Chen PN, Wang SS, Chou YE, Hung SC. et al. Impacts of AURKA Genetic Polymorphism on Urothelial Cell Carcinoma Development. J Cancer. 2019;10:1370-1374
9. Hung SC, Wang SS, Li JR, Chen CS, Yang CK, Chiu KY. et al. Effect of HMGB1 Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathological Characteristics. Int J Med Sci. 2018;15:1731-1736
10. Wieczorek E, Jablonowski Z, Tomasik B, Konecki T, Jablonska E, Gromadzinska J. et al. MMP, VEGF and TIMP as prognostic factors in recurring bladder cancer. Clin Biochem. 2015;48:1235-1240
11. Chung JF, Chen CL, Nassef Y, Shiu BH, Wang CH, Kuo FH. et al. Effect of tissue inhibitor of metalloproteinases-3 genetics polymorphism on clinicopathological characteristics of uterine cervical cancer patients in Taiwan. Int J Med Sci. 2022;19:1013-1022
12. Su CW, Chang YC, Chien MH, Hsieh YH, Chen MK, Lin CW. et al. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death Dis. 2019;10:793
13. Su CW, Su BF, Chiang WL, Yang SF, Chen MK, Lin CW. Plasma levels of the tissue inhibitor matrix metalloproteinase-3 as a potential biomarker in oral cancer progression. Int J Med Sci. 2017;14:37-44
14. Chang JH, Lai TC, Yang PJ, Shih PC, Yang YC, Lee KL. et al. Associations of TIMP-3 Genetic Polymorphisms with EGFR Statuses and Cancer Clinicopathologic Development in Lung Adenocarcinoma Patients. Int J Mol Sci. 2020;21:8023
15. Su CW, Huang YW, Chen MK, Su SC, Yang SF, Lin CW. Polymorphisms and Plasma Levels of Tissue Inhibitor of Metalloproteinase-3: Impact on Genetic Susceptibility and Clinical Outcome of Oral Cancer. Medicine (Baltimore). 2015;94:e2092
16. Alhalabi O, Rafei H, Shah A, Siefker-Radtke A, Campbell M, Gao J. Targeting advanced urothelial carcinoma-developing strategies. Curr Opin Oncol. 2019;31:207-215
17. Chu YH, Su CW, Hsieh YS, Chen PN, Lin CW, Yang SF. Carbonic Anhydrase III Promotes Cell Migration and Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Cells. 2020;9:704
18. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP. et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088-1099
19. Su SC, Lin CW, Yang WE, Fan WL, Yang SF. The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies. Expert Opin Ther Targets. 2016;20:551-566
20. Lin CW, Yang WE, Lee WJ, Hua KT, Hsieh FK, Hsiao M. et al. Lipocalin 2 prevents oral cancer metastasis through carbonic anhydrase IX inhibition and is associated with favourable prognosis. Carcinogenesis. 2016;37:712-722
21. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM. et al. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018;97:717-724
22. Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT. et al. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45:e222-226
23. Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS. et al. 3'UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. 2017;7:4466
24. Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol. 2016;1418:111-141
25. Lee HL, Chiou HL, Wang SS, Hung SC, Chou MC, Yang SF. et al. WISP1 genetic variants as predictors of tumor development with urothelial cell carcinoma. Urol Oncol. 2018;36:160 e115-160 e121
26. Tung MC, Wen YC, Wang SS, Lin YW, Liu YC, Yang SF. et al. Dopamine receptor D2 genetic variations is associated with the risk and clinicopathological variables of urothelial cell carcinoma in a Taiwanese population. Int J Med Sci. 2018;15:1187-1193
27. Tsay MD, Hsieh MJ, Wang SS, Wang WC, Chou YY, Shih CH. et al. Impact of endothelial nitric oxide synthase polymorphisms on urothelial cell carcinoma development. Urol Oncol. 2019;37:293.e291-293.e299
28. Hung SC, Wang SS, Li JR, Chen CS, Lin CY, Chang LW. et al. Impact of RAGE polymorphisms on urothelial cell carcinoma clinicopathologic characteristics and long-term survival. Urol Oncol. 2019;37:573.e579-573.e517
29. Fujii Y, Sato Y, Suzuki H, Kakiuchi N, Yoshizato T, Lenis AT. et al. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell. 2021;39:793-809.e798
30. Weng WC, Chen CJ, Chen PN, Wang SS, Hsieh MJ, Yang SF. Impact of Gene Polymorphisms in GAS5 on Urothelial Cell Carcinoma Development and Clinical Characteristics. Diagnostics (Basel). 2020;10:260
31. Anand-Apte B, Bao L, Smith R, Iwata K, Olsen BR, Zetter B. et al. A review of tissue inhibitor of metalloproteinases-3 (TIMP-3) and experimental analysis of its effect on primary tumor growth. Biochem Cell Biol. 1996;74:853-862
32. Fan D, Kassiri Z. Biology of Tissue Inhibitor of Metalloproteinase 3 (TIMP3), and Its Therapeutic Implications in Cardiovascular Pathology. Front Physiol. 2020;11:661
33. Horne BD, Camp NJ, Carlquist JF, Muhlestein JB, Kolek MJ, Nicholas ZP. et al. Multiple-polymorphism associations of 7 matrix metalloproteinase and tissue inhibitor metalloproteinase genes with myocardial infarction and angiographic coronary artery disease. Am Heart J. 2007;154:751-758
34. Zheng Z, He X, Zhu M, Jin X, Li C, Zhu F. et al. Tissue inhibitor of the metalloproteinases-3 gene polymorphisms and carotid plaque susceptibility in the Han Chinese population. Int J Neurosci. 2018;128:920-927
35. Wang N, Zhou S, Fang XC, Gao P, Qiao Q, Wu T. et al. MMP-2, -3 and TIMP-2, -3 polymorphisms in colorectal cancer in a Chinese Han population: A case-control study. Gene. 2020;730:144320
36. Srivastava P, Kapoor R, Mittal RD. Impact of MMP-3 and TIMP-3 gene polymorphisms on prostate cancer susceptibility in North Indian cohort. Gene. 2013;530:273-277
37. Su CW, Lin CW, Yang WE, Yang SF. TIMP-3 as a therapeutic target for cancer. Ther Adv Med Oncol. 2019;11:1758835919864247
38. Rai GP, Baird SK. Tissue inhibitor of matrix metalloproteinase-3 has both anti-metastatic and anti-tumourigenic properties. Clin Exp Metastasis. 2020;37:69-76
39. Furukawa A, Tsuji M, Nishitani M, Kanda K, Inoue Y, Kanayama H. et al. Role of the matrix metalloproteinase and tissue inhibitors of metalloproteinase families in noninvasive and invasive tumors transplanted in mice with severe combined immunodeficiency. Urology. 1998;51:849-853
40. Lopez-Beltran A, Cimadamore A, Montironi R, Cheng L. Molecular pathology of urothelial carcinoma. Hum Pathol. 2021;113:67-83
41. Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404-423
42. James ND, Hussain SA, Hall E, Jenkins P, Tremlett J, Rawlings C. et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;366:1477-1488
43. Iqbal U, Elsayed AS, Jing Z, Stöckle M, Wijburg C, Wiklund P. et al. Upstaging and Survival Outcomes for Non-Muscle Invasive Bladder Cancer After Radical Cystectomy: Results from the International Robotic Cystectomy Consortium. J Endourol. 2021;35:1541-1547
44. Cao J, Yang X, Li J, Wu H, Li P, Yao Z. et al. Screening and Identifying Immune-Related Cells and Genes in the Tumor Microenvironment of Bladder Urothelial Carcinoma: Based on TCGA Database and Bioinformatics. Front Oncol. 2019;9:1533
Author contact
![]() Corresponding authors: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan; E-mail: ysfedu.tw (Shun-Fa Yang) or Shian-Shiang Wang, MD., PhD. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan. E-mail: sswdoccom.tw (Shian-Shiang Wang)
Corresponding authors: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan; E-mail: ysfedu.tw (Shun-Fa Yang) or Shian-Shiang Wang, MD., PhD. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan. E-mail: sswdoccom.tw (Shian-Shiang Wang)

 Global reach, higher impact
Global reach, higher impact