Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(8):1293-1300. doi:10.7150/jca.83747 This issue Cite
Research Paper
WDR4 gene polymorphisms and Wilms tumor susceptibility in Chinese children: A five-center case-control study
1. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangdong Provincial Clinical Research Center for Child Health, Guangzhou 510623, Guangdong, China.
2. Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China.
3. Department of Pediatric Surgery, The Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, Shaanxi, China.
4. Department of Pediatric Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China.
5. Department of Hematology, The Key Laboratory of Pediatric Hematology and Oncology Diseases of Wenzhou, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou 325027, Zhejiang, China.
6. Department of Pathology, Children Hospital and Women Health Center of Shanxi, Taiyuan 030013, Shannxi, China.
#These authors contributed equally to this work.
Received 2023-2-22; Accepted 2023-4-27; Published 2023-5-8
Abstract
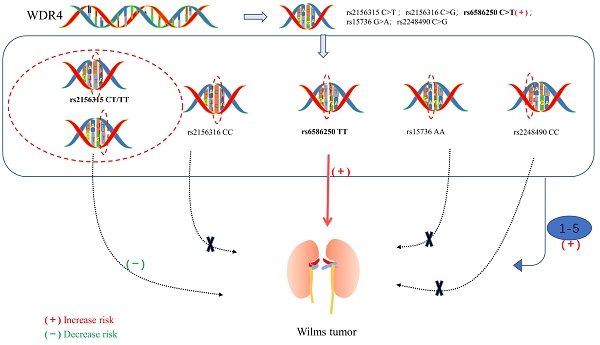
Wilms tumor is the most common embryonal renal malignancy in children. WDR4 is an indispensable noncatalytic subunit of the RNA N7-methylguanosine (m7G) methyltransferase complex and plays an essential role in tumorigenesis. However, the relationship between polymorphisms in the WDR4 gene and susceptibility to Wilms tumor remains to be fully investigated. We performed a large case-control study involving 414 patients and 1199 cancer-free controls to investigate whether single nucleotide polymorphisms (SNPs) in the WDR4 gene are associated with Wilms tumor susceptibility. WDR4 gene polymorphisms (rs2156315 C > T, rs2156316 C > G, rs6586250 C > T, rs15736 G > A, and rs2248490 C > G) were genotyped using the TaqMan assay. In addition, unconditioned logistic regression analysis was performed, odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the association between WDR4 gene SNPs and Wilms tumor susceptibility as well as the strength of the associations. We found that only the rs6586250 C>T polymorphism was significantly associated with an increased risk of Wilms tumor (adjusted OR=2.99, 95% CI = 1.28-6.97, P = 0.011 for the rs6586250 TT genotype; adjusted OR=3.08, 95% CI = 1.33-7.17, P = 0.009 for the rs6586250 CC/CT genotype). Furthermore, the stratification analysis revealed that patients with the rs6586250 TT genotype and carriers with 1-5 risk genotypes exhibited statistically significant associations with increased Wilms tumor risk in specific subgroups. However, the rs2156315 CT/TT genotype was identified as having a protective effect against Wilms tumor in the age >18 months subgroup compared with the rs2156315 CC genotype. In brief, our study demonstrated that the rs6586250 C > T polymorphism of the WDR4 gene was significantly associated with Wilms tumor. This finding may contribute to the understanding of the genetic mechanism of Wilms tumor.
Keywords: Wilms tumor, susceptibility, WDR4, polymorphism, m7G modification
Introduction
Wilms tumor is the most frequently occurring embryonal renal malignancy in children, representing approximately 7% of childhood tumors [1]. It often develops in children before the age of 15 years, and the incidence of Wilms tumor varies greatly by geography and ethnicity. The highest incidence rate of Wilms tumor is in the Black population in the United States, with over 10 cases per million children, while the East Asian population has the lowest incidence rate of 3 to 4 cases per million children [2, 3]. In China, the annual prevalence of Wilms tumor is approximately 3.3 cases per million children [4]. With advances in modern multimodality therapy, the overall survival rate for patients with Wilms tumor has reached more than 90%, except for patients with unfavorable histology, advanced disease, bilateral disease, and recurrent Wilms tumor [5]. The outcome is not positive for children with relapsed disease because of a lower long-term survival rate of less than 30% [6]. In addition, it has been reported that nearly 25% of Wilms tumor survivors suffer from chronic health problems owing to the side effects of cancer treatment, such as kidney failure, cardiac disease, and secondary malignancies [7, 8].
The interaction of environmental and genetic factors is a crucial cause of tumorigenesis. In previous publications, several perinatal and environmental factors were identified as risk factors for Wilms tumor, including exposure to pesticides during pregnancy, preterm birth, high birthweight and maternal hypertension [9]. Rapidly developing biotechnology has led to the identification of an increasing number of Wilms tumor susceptibility genes, including WT1, P53, TRIM28, BARD1, XPD, hOGG1, and FEN1 [10-15]. However, thus far, the well-established genetic alterations can only explain a small part of the tumorigenesis of Wilms tumor. It is imperative to validate additional genetic variants to better understand the pathogenesis of Wilms tumor and develop new drug-targeted therapies.
Misregulation in RNA modification has been found to be closely related to many human diseases, including genetic defects, neurological diseases, and cancers [16]. Currently, over 160 kinds of RNA modifications have been discovered, and tRNAs are the most heavily modified [17, 18]. The METTL1/WDR4 complex is an extremely critical tRNA N7-methylguanosine (m7G) methyltransferase that catalyzes m7G modifications not only in the cap of mRNAs but also in internal mRNAs and tRNAs, helping to stabilize the structure of tRNAs and enhance the efficiency of mRNA translation [19, 20]. Moreover, in the G-rich regions of miRNAs, m7G appeared to form an unstable structure of G-quadruplexes, facilitating the process of pre-miRNA and miRNA maturation [21]. Mammalian m7G modifications display additional complexity and critical physiological functions. METTL1 and WDR4 defects have been reported to cause impaired stem cell self-renewal ability and neural differentiation in mouse embryonic stem cells [22, 23]. Moreover, WDR4 was demonstrated to be a potential oncogene. Zeng et al. observed that WDR4 was aberrantly overexpressed in different tumors, and the expression level of WDR4 was closely related to poor prognosis in a human pan-cancer analysis of 33 different types of cancers [24]. In addition, mutations in the WDR4 gene cause not only microcephalic primordial dwarfism accompanied by different levels of severe growth retardation and microcephaly [25] but also Galloway-Mowat syndrome, characterized by neurodevelopmental defects and renal-glomerular disease [26]. METTL1 or WDR4 expression dysregulation has been detected in many tumors, including bladder cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma, and lung cancer [27-30]. To date, no research has investigated the association between functional single nucleotide polymorphisms (SNPs) in the WDR4 gene and Wilms tumor susceptibility.
Materials and Methods
Study population
This case-control study enrolled 414 patients with Wilms tumor and 1199 healthy controls as described previously (Table S1) [31-33]. All Wilms tumor cases were confirmed by histopathology. Age-, gender- and ethnicity-matched healthy controls were enrolled from the same hospitals as the Wilms tumor patients during the same period and had no family history of Wilms tumor. All procedures in this study adhered to the Declaration of Helsinki. We obtained approval from the Institutional Review Board of Guangzhou Women and Children's Medical Center (No. 202016601), as well as written informed consent from the subjects' parents or guardians.
Genotyping
According to the standard criteria described in previous studies, we selected five potentially functional single nucleotide polymorphisms in the WDR4 gene (rs2156315 C > T, rs2156316 C > G, rs6586250 C > T, rs15736 G>A, and rs2248490 C > G) from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and SNPinfo (http://snpinfo.niehs.nih.gov/snpfunc.htm) [32, 34]. Briefly, we selected the underlying SNPs in the 5' and 3' untranslated regions, 5' near regions, 3' near regions and exons of the WDR4 gene. Moreover, the selected SNPs had a minor allele frequency (MAF) ≥5% among Chinese Han subjects and low linkage disequilibrium (LD) (R2<0.8). Additionally, SNPs that affect the activity of transcription factor binding site (TFBS) activity or miRNA binding sites were included. By using SNPinfo Web Server (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html), we discovered that rs15736 G > A and rs2248490 C > G have the potential to affect the exonic splicing enhancer (ESE) or exonic splicing silencer (ESS), whereas rs2156315 C > T and rs2156316 C > G are likely to affect miRNA binding sites. Finally, rs6586250 C > T, rs15736 G > A and rs2248490 C > G may influence nonsynonymous coding SNPs (nsSNPs) (Table S2). DNA was extracted from the peripheral blood samples of participants by the Genomic DNA kit (Tian Gen Biotech Co. Ltd., Beijing, China), and genotyping of WDR4 gene polymorphisms was carried out by the TaqMan assay. Generally, to ensure the study's accuracy, 10% of all samples were randomly selected for regenotyping by a double-blind method, and the results of regenotyping were consistent with those of the original genotyping.
Statistical analysis
The consistency between the selected SNP genotype and Hardy-Weinberg Equilibrium (HWE) in controls was determined by a χ2 goodness-of-fit test. In addition, a two-sided χ2 test was applied to analyze the differences in the distribution of demographic characteristics and allele frequency between cases and cancer-free controls. We also calculated the adjusted odds ratio (OR), 95% confidence interval (CI), and a two-sided P value to assess the association between each genotype and the risk of Wilms tumor by multivariate logistic regression analysis. Additionally, expression quantitative trait locus (eQTL) analysis was conducted to evaluate the correlations between significant SNPs and the expression levels of their genes or nearby genes using released data from Genotype-Tissue Expression (GTEx) (https://gtexportal.org). The statistically significant threshold was defined as P<0.05. All statistical analyses were implemented with SAS 9.1 software (SAS Institute, Cary, NC, USA).
Results
Association between WDR4 gene polymorphisms and Wilms tumor susceptibility
In our study, 403 cases and 1198 healthy controls were successfully genotyped for the five SNPs out of a cohort of 414 cases and 1199 cancer-free controls. The correlation of WDR4 gene polymorphisms with Wilms tumor susceptibility is portrayed in Table 1. The frequency distribution of all the analyzed SNP genotypes obeyed the HWE in controls. Notably, only the rs6586250 C > T genotype was significantly associated with an increased risk of Wilms tumor (adjusted OR = 2.99, 95% CI = 1.28 - 6.97, P = 0.011 for the rs6586250 TT genotype; adjusted OR = 3.08, 95% CI = 1.33-7.17, P = 0.009 for the rs6586250 CC/CT genotype). According to the ORs, we subsequently defined rs2156315 TT, rs2156316 CC, rs6586250 TT, rs15736 AA, and rs2248490 CC as risk genotypes. However, the 1-5 combined risk genotypes had no statistically significant effects on Wilms tumor susceptibility compared to non-risk genotypes.
Stratification analysis
We further employed a stratification analysis divided by age, sex, and clinical stage to explore whether WDR4 gene polymorphisms were associated with Wilms tumor risk in a specific subgroup, and the corresponding results are shown in Table 2. We found a statistically significant association between the rs6586250 TT genotype and increased risk of Wilms tumor in the specific subgroups of children aged over 18 months (adjusted OR = 3.89, 95% CI = 1.33 - 11.37, P = 0.013), females (adjusted OR = 7.13, 95% CI = 1.37 - 37.06, P = 0.020), and patients with clinical stage I+II (adjusted OR=3.11, 95% CI = 1.19 - 8.15, P = 0.021) and clinical stage III+IV (adjusted OR = 3.28, 95% CI = 1.02 - 10.52, P = 0.046). Similarly, in children older than 18 months, carriers with 1-5 risk genotypes were more likely to develop Wilms tumor (adjusted OR = 1.45, 95% CI = 1.09-1.92, P = 0.011) than carriers without any risk genotype. However, the rs2156315 CT/TT genotype was identified as having a protective effect against Wilms tumor in the specific subgroup of children older than 18 months when compared with the rs2156315 CC genotype (adjusted OR = 0.73, 95% CI = 0.55 - 0.98, P = 0.036). This could be due to different SNP genotypes having different effects on the same tumor, or it could be due to chance because of the limited number of subjects. Expanded sample sizes and replicate experiments are strongly recommended.
Effect of rs6586250 C>T on the expression of the WDR4 gene and surrounding genes
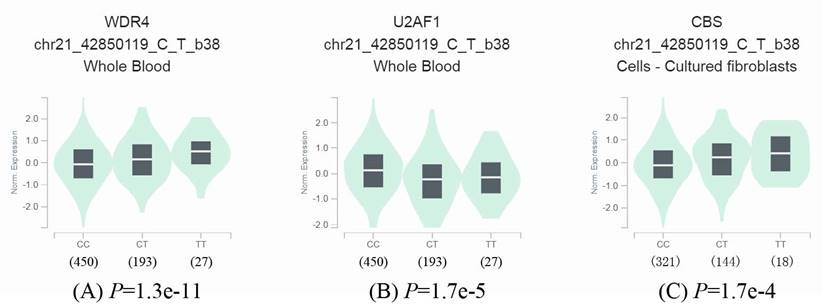
To further identify the effect of rs6586250 C > T on mRNA expression, we took full advantage of the released data from GTEx to perform an eQTL analysis of rs6586250 C > T. As shown in Figure 1, in whole blood, compared with the rs6586250 CC genotype, the rs6586250 CT/TT genotype was significantly associated with higher WDR4 mRNA expression (P=1.3e-11), whereas it was correlated with lower mRNA expression of the nearby gene U2AF1 (P = 1.7e-5). In addition, the rs6586250 CT/TT genotype showed enhanced mRNA expression of the nearby gene CBS in cultured fibroblast cells compared to the rs6586250 CC genotype (P = 1.7 e-4).
Effect of WDR4 gene rs6586250 C>T on the expression of the WDR4 gene and surrounding genes. (A) The expression of the WDR4 gene in whole blood; (B) The expression of the U2AF1 gene in whole blood; (C) The expression of the CBS gene in cultured fibroblasts cells.
Relationship of WDR4 gene polymorphisms with Wilms tumor susceptibility
| Genotype | Cases (N=403) | Controls (N=1198) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs2156315 C>T (HWE=0.151) | |||||||
| CC | 254 (63.03) | 700 (58.43) | 1.00 | 1.00 | |||
| CT | 129 (32.01) | 443 (36.98) | 0.80 (0.63-1.02) | 0.076 | 0.80 (0.63-1.02) | 0.074 | |
| TT | 20 (4.96) | 55 (4.59) | 1.00 (0.59-1.71) | 0.994 | 1.00 (0.58-1.69) | 0.985 | |
| Additive | 0.209 | 0.88 (0.72-1.07) | 0.210 | 0.88 (0.72-1.07) | 0.200 | ||
| Dominant | 149 (36.97) | 498 (41.57) | 0.104 | 0.83 (0.65-1.04) | 0.104 | 0.82 (0.65-1.04) | 0.100 |
| Recessive | 383 (95.04) | 1143 (95.41) | 0.760 | 1.09 (0.64-1.83) | 0.760 | 1.08 (0.64-1.82) | 0.779 |
| rs2156316 C>G (HWE=0.497) | |||||||
| CC | 187 (46.40) | 517 (43.16) | 1.00 | 1.00 | |||
| CG | 171 (42.43) | 548 (45.74) | 0.86 (0.68-1.10) | 0.227 | 0.87 (0.68-1.10) | 0.240 | |
| GG | 45 (11.17) | 133 (11.10) | 0.94 (0.64-1.36) | 0.729 | 0.94 (0.64-1.36) | 0.726 | |
| Additive | 0.407 | 0.93 (0.78-1.10) | 0.407 | 0.93 (0.79-1.11) | 0.414 | ||
| Dominant | 216 (53.60) | 681 (56.84) | 0.256 | 0.88 (0.70-1.10) | 0.256 | 0.88 (0.70-1.10) | 0.267 |
| Recessive | 358 (88.83) | 1065 (88.90) | 0.972 | 1.01 (0.70-1.44) | 0.972 | 1.00 (0.70-1.44) | 0.983 |
| rs6586250 C>T (HWE=0.296) | |||||||
| CC | 322 (79.90) | 945 (78.88) | 1.00 | 1.00 | |||
| CT | 70 (17.37) | 242 (20.20) | 0.85 (0.63-1.14) | 0.276 | 0.85 (0.64-1.15) | 0.295 | |
| TT | 11 (2.73) | 11 (0.92) | 2.94 (1.26-6.83) | 0.013 | 2.99 (1.28-6.97) | 0.011 | |
| Additive | 0.759 | 1.04 (0.81-1.34) | 0.757 | 1.05 (0.82-1.35) | 0.717 | ||
| Dominant | 81 (20.10) | 253 (21.12) | 0.663 | 0.94 (0.71-1.24) | 0.663 | 0.95 (0.72-1.25) | 0.700 |
| Recessive | 392 (97.27) | 1187 (99.08) | 0.007 | 3.03 (1.30-7.04) | 0.010 | 3.08 (1.33-7.17) | 0.009 |
| rs15736 G>A (HWE=0.246) | |||||||
| GG | 321 (79.65) | 941 (78.55) | 1.00 | 1.00 | |||
| GA | 74 (18.36) | 246 (20.53) | 0.88 (0.66-1.18) | 0.394 | 0.89 (0.67-1.19) | 0.422 | |
| AA | 8 (1.99) | 11 (0.92) | 2.13 (0.85-5.35) | 0.106 | 2.18 (0.87-5.47) | 0.098 | |
| Additive | 0.988 | 1.00 (0.77-1.29) | 0.988 | 1.01 (0.78-1.30) | 0.965 | ||
| Dominant | 82 (20.35) | 257 (21.45) | 0.639 | 0.94 (0.71-1.24) | 0.639 | 0.94 (0.71-1.25) | 0.678 |
| Recessive | 395 (98.01) | 1187 (99.08) | 0.087 | 2.19 (0.87-5.48) | 0.095 | 2.23 (0.89-5.59) | 0.087 |
| rs2248490 C>G (HWE=0.372) | |||||||
| CC | 187 (46.40) | 522 (43.57) | 1.00 | 1.00 | |||
| CG | 172 (42.68) | 548 (45.74) | 0.88 (0.69-1.11) | 0.279 | 0.88 (0.69-1.12) | 0.301 | |
| GG | 44 (10.92) | 128 (10.68) | 0.96 (0.66-1.41) | 0.832 | 0.96 (0.66-1.40) | 0.828 | |
| Additive | 0.496 | 0.94 (0.79-1.12) | 0.496 | 0.94 (0.80-1.12) | 0.509 | ||
| Dominant | 216 (53.60) | 676 (56.43) | 0.323 | 0.89 (0.71-1.12) | 0.323 | 0.90 (0.71-1.12) | 0.343 |
| Recessive | 359 (89.08) | 1070 (89.32) | 0.896 | 1.03 (0.71-1.47) | 0.895 | 1.02 (0.71-1.47) | 0.913 |
| Combined effect of risk genotypes | |||||||
| 0 | 182 (45.16) | 601 (50.17) | 1.00 | 1.00 | |||
| 1-5 | 221 (54.84) | 597 (49.83) | 0.082 | 1.22 (0.98-1.53) | 0.082 | 1.22 (0.97-1.53) | 0.089 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
a χ2 test for genotype distributions between Wilms tumor patients and controls.
b Adjusted for age and gender.
c Risk genotypes were rs2156315 TT, rs2156316 CC, rs6586250 TT, rs15736 AA and rs2248490 CC.
Stratification analysis for the association of WDR4 genotypes with Wilms tumor susceptibility
| Variables | rs2156315 (cases/controls) | AOR (95% CI) a | P a | rs6586250 (cases/controls) | AOR (95% CI) a | P a | Risk genotypes (cases/controls) | AOR (95% CI) a | P a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT/TT | CC/CT | TT | 0 | 1-5 | |||||||
| Age, month | ||||||||||||
| ≤18 | 80/273 | 57/192 | 1.03 (0.70-1.52) | 0.887 | 134/460 | 3/5 | 1.94 (0.46-8.26) | 0.370 | 67/215 | 70/250 | 0.89 (0.60-1.30) | 0.535 |
| >18 | 174/427 | 92/306 | 0.73 (0.55-0.98) | 0.036 | 258/727 | 8/6 | 3.89 (1.33-11.37) | 0.013 | 115/386 | 151/347 | 1.45 (1.09-1.92) | 0.011 |
| Gender | ||||||||||||
| Female | 112/297 | 75/224 | 0.89 (0.63-1.25) | 0.490 | 182/519 | 5/2 | 7.13 (1.37-37.06) | 0.020 | 87/258 | 100/263 | 1.13 (0.81-1.58) | 0.479 |
| Male | 142/403 | 74/274 | 0.77 (0.56-1.06) | 0.108 | 210/668 | 6/9 | 2.13 (0.75-6.05) | 0.157 | 95/343 | 121/334 | 1.29 (0.95-1.76) | 0.104 |
| Clinical stage | ||||||||||||
| I+II | 157/700 | 94/498 | 0.84 (0.64-1.12) | 0.234 | 244/1187 | 7/11 | 3.11 (1.19-8.15) | 0.021 | 117/601 | 134/597 | 1.13 (0.86-1.48) | 0.386 |
| III+IV | 86/700 | 49/498 | 0.79 (0.55-1.14) | 0.210 | 131/1187 | 4/11 | 3.28 (1.02-10.52) | 0.046 | 59/601 | 76/597 | 1.34 (0.93-1.92) | 0.115 |
AOR, adjusted odds ratio; CI, confidence interval.
a Adjusted for age and gender, omitting the corresponding factor.
Discussion
RNA modification has been proven to be dynamic and reversible and is involved in many important biological processes, such as cell differentiation, gene expression and protein synthesis [35, 36]. Thus, dysregulation of RNA modification may contribute to disease development by disrupting normal biological processes. WDR4, a member of the repeat protein family and the yeast ortholog of Trm82p, is located at human chromosome 21q22.3 [37]. WDR4 binds to METTL1 to form a heterodimer complex of m7G methyltransferase, which catalyzes the installation of m7G modification in different types of RNA molecules. m7G modification affects the metabolism and functions of RNA and ultimately controls gene expression and critical biological processes [38]. METTL1 plays the larger part in catalyzing the installation of m7G in target RNA, while WDR4 helps stabilize the active conformation and maintain the normal protein level of METTL1. The integrity of the METTL1/WDR4 complex is considered indispensable for the function of m7G modification [39]. The METTL1/WDR4 complex catalyzes the installation of m7G modification in a set of tRNAs and generates the so-called Arg-TCT tRNAs, which decode the corresponding homologous mRNA codons and thus selectively promote the translation of many cell cycle-associated and carcinogenic genes. This mechanism greatly contributes to oncogenic transformation [40].
Further studies revealed that METTL1/WDR4-mediated m7G modification played a double-edged role in the pathogenesis and progression of different tumors [41]. The main reason for this significant difference largely depended on whether it was mounted on the G46 site of tRNAs or the G-quadruplex structure of miRNAs [41]. According to previous research, the expression of WDR4 transcripts was obviously upregulated in hepatocellular carcinoma (HCC), along with increasing levels of m7G methylation. Overexpression of WDR4 promoted the development and metastasis of HCC by upregulating the cell cycle-associated factor CCNB1 and epithelial-mesenchymal transition (EMT) via the MYC/WDR4/CCNB1 signaling pathway. WDR4 plays an oncogenic role in HCC by positively regulating the expression of CCNB1 to induce the G2/M cell cycle transition and inhibit apoptosis, which promotes the proliferation ability of cancer cells [29]. In contrast, METTL1/WDR4-mediated m7G-modified microRNAs suppressed the progression of lung cancer. The complex directly methylated a series of tumor-suppressive miRNAs, particularly the let-7e family, and promoted the maturation of let-7e miRNAs; these miRNAs in return downregulated specific oncogenes such as high-mobility group AT-hook 2 (HMGA2), thereby suppressing the migration ability of lung cancer cells [21].
With tremendous advances in high-throughput sequencing technology, genome-wide association studies (GWASs) have successfully identified many predisposition loci that contribute to multiple complex human diseases, including cancers [42]. The underlying mechanistic elucidation of SNPs for cancer predisposition is that most nucleotide substitutions occur in noncoding regulatory regions, playing a crucial role in modulating gene expression by affecting RNA splicing and DNA methylation [42-44]. For instance, Wang et al. [45] discovered that the rs465663 polymorphism in the WDR4 gene, located in the intronic region, was related to asthenozoospermia (a clinical manifestation of male infertility) by regulating WDR4 gene expression and the DNA fragmentation level caused by oxidative stress. He et al. [34] conducted a case-control study of 313 hepatoblastoma patients and 1446 controls to explore whether patients with the WDR4 gene polymorphism were predisposed to hepatoblastoma in Chinese Han populations. They found that female carriers with 2-5 risk genotypes were more vulnerable to hepatoblastoma, demonstrating that the WDR4 gene polymorphism may be a risk factor for hepatoblastoma. However, no studies involving the relationship between genetic variants in the WDR4 gene and Wilms tumor risk have been reported thus far. Herein, our results suggested that WDR4 gene SNPs predispose patients to Wilms tumor. This finding may make a genetic contribution to the molecular mechanism of Wilms tumor.
In the current study, we demonstrated that the selected rs6586250 C > T polymorphism of the WDR4 gene was significantly associated with an increased risk of Wilms tumor. Further stratification analysis identified that the rs6586250 TT genotype was associated with a statistically significant increased risk of Wilms tumor in certain subgroups: children older than 18 months, females, and patients in clinical stage I+II and clinical stage III+IV. Similarly, carriers with 1-5 risk genotypes among children aged over 18 months were more likely to develop Wilms tumors compared to those without any risk genotypes. In conclusion, these data suggested that functionally selected SNPs in the WDR4 gene have a significant impact on increased risk for Wilms tumor. These findings have enriched our knowledge about the etiology of Wilms tumor and facilitated the identification of new biomarker molecules for early diagnosis and good prognosis. Moreover, we tried to further explore the possible mechanism of how significant SNPs of WDR4 conferred susceptibility to Wilms tumor. According to the eQTL analysis of WDR4 rs6586250 C>T, we determined that the rs6586250 T allele was significantly associated with higher WDR4 mRNA expression in whole blood. Previous studies also showed an overexpression of WDR4 in head and neck squamous cell carcinoma [28], hepatocellular carcinoma [29], intrahepatic cholangiocarcinoma [46], and so on, indicating that WDR4 serves as an oncogene for several cancers. To date, although there have been no studies on the direct effect of WDR4 on the risk of Wilms tumor, our results suggested that the WDR4 rs6586250 T genotype may increase the susceptibility to Wilms tumor by increasing WDR4 gene expression. Additionally, the eQTL analysis also identified that the WDR4 rs6586250 T allele was correlated with lower mRNA expression of the nearby gene U2AF1 in whole blood, whereas it showed increased mRNA expression of the nearby gene CBS in cultured fibroblast cells. These findings implied that the WDR4 rs6586250 T allele could regulate the expression of multiple surrounding genes. RNA splicing is essential for generating mature messenger RNA (mRNA) and regulating gene expression, thereby affecting cellular processes and cell fates, and is catalyzed by a ribonucleoprotein complex called the spliceosome [47]. U2AF1, the subunit of the U2 auxiliary factor (U2AF) heterodimer and an indispensable component of the spliceosome, has the ability to specifically recognize the AG dinucleotide of the 3' splice sites and promote the process of RNA splicing. Furthermore, mutations in U2AF1 were associated with hematological malignancies and myelodysplasia syndromes [47]. According to our analysis, a low expression level of U2AF1 might affect the activity or efficiency of RNA splicing, leading to the possibility of tumorigenesis. Moreover, we also found a significant association between rs6586250 and the enhanced expression of the neighboring gene CBS. Cystathionine β-synthase (CBS) is a mammalian enzyme that transfers sulfur, promoting the conversion of homocysteine to cysteine, and produces hydrogen sulfide (H2S), which plays a supportive role in maintaining tumor cell proliferation and survival in cancers with high CBS expression. CBS has been reported to be expressed at a high level in certain cancers, including kidney, colorectal, lung, and breast cancers, but at a low level in liver cancer and glioma. It is predicted that high CBS expression is associated with a poor survival rate for patients with high-CBS-expressing cancers [48]. These findings identified CBS as an oncogene in several types of tumors, which supports the results of the eQTL analysis. Therefore, it is reasonable to believe that the WDR4 rs6586250 C > T polymorphism is likely to alter Wilms tumor susceptibility by affecting the expression of nearby genes U2AF1 and CBS. Overall, we speculate that the WDR4 rs6586250 T allele may increase the risk of Wilms tumor by upregulating WDR4 gene expression or altering the expression of the surrounding genes U2AF1 and CBS. However, the exact mechanism of how WDR4 gene SNPs contribute to Wilms tumor susceptibility remains unclear. Further functional experiments are necessary to verify the underlying mechanism.
The strengths of our research include its scientific design, multicenter analysis, and innovativeness. However, we cannot ignore the accompanying disadvantages. First, the low prevalence of Wilms tumor limits the sample size and thus weakens the power of stratified analysis. Most importantly, intensive genetic studies require larger sample sizes and Bonferroni correction analysis to validate the relationship between selected SNPs and tumor risk. Second, the conclusion can only be applied to the Han Chinese, not other countries and ethnicities. Finally, the study focused only on the relationship between WDR4 gene SNPs and Wilms tumor risk. Therefore, further functional experiments are warranted to explore the unique mechanism by which SNPs in the WDR4 gene affect Wilms tumor susceptibility.
In brief, this large, five-center epidemiological study demonstrated a significant correlation between functional SNPs in the WDR4 gene and Wilms tumor susceptibility. The conclusion needs to be further validated in another well-designed scientific study of patients of different ethnicities without confounding factors.
Abbreviations
m7G: N7-methylguanosine; SNP: single nucleotide polymorphism; MAF: minor allele frequency; LD: linkage disequilibrium; TFBS: transcription factor binding site; ESE: exonic splicing enhancer; ESS: exonic splicing silencer; nsSNP: nonsynonymous coding SNP; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; eQTL: expression quantitative trait locus; GTEx: Genotype-Tissue Expression; GWAS: genome-wide association study; HCC: hepatocellular carcinoma; EMT: epithelial-mesenchymal transition; mRNA: messenger RNA; U2AF: U2 auxiliary factor; CBS: cystathionine β-synthase.
Supplementary Material
Supplementary tables.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (No: 82003523), Natural Science Foundation of Guangdong Province (No: 2021A1515010860), Science and Technology Project of Guangzhou (No: 202102010170), and Natural Science Foundation of Zhejiang Province (No: LGF21H260012).
Author Contributions
All authors contributed significantly to this work. JH, GL and WF designed the research study; LD, RXH, CD, JZhu, ZZ, JC, JZhang, HZ, SL, JR, JH, and WF performed the research study and collected the samples and clinical information; CD and JH analyzed the data; LD, RXH, JZhu and JH wrote the paper; and JH prepared all the tables. All authors included on the final manuscript have read and approved the final version to be published.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Servaes SE, Hoffer FA, Smith EA, Khanna G. Imaging of Wilms tumor: an update. Pediatr Radiol. 2019;49:1441-52
2. Stiller CA, Parkin DM. International variations in the incidence of childhood renal tumours. Br J Cancer. 1990;62:1026-30
3. Nakata K, Colombet M, Stiller CA, Pritchard-Jones K, Steliarova-Foucher E. Incidence of childhood renal tumours: An international population-based study. Int J Cancer. 2020;147:3313-27
4. Cunningham ME, Klug TD, Nuchtern JG, Chintagumpala MM, Venkatramani R, Lubega J. et al. Global Disparities in Wilms Tumor. J Surg Res. 2020;247:34-51
5. Liu L, Song Z, Gao XD, Chen X, Wu XB, Wang M. et al. Identification of the potential novel biomarkers as susceptibility gene for Wilms tumor. BMC Cancer. 2021;21:316
6. Malogolowkin M, Cotton CA, Green DM, Breslow NE, Perlman E, Miser J. et al. Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin. A report from the National Wilms Tumor Study Group. Pediatr Blood Cancer. 2008;50:236-41
7. Aldrink JH, Heaton TE, Dasgupta R, Lautz TB, Malek MM, Abdessalam SF. et al. Update on Wilms tumor. J Pediatr Surg. 2019;54:390-7
8. Wong KF, Reulen RC, Winter DL, Guha J, Fidler MM, Kelly J. et al. Risk of Adverse Health and Social Outcomes Up to 50 Years After Wilms Tumor: The British Childhood Cancer Survivor Study. J Clin Oncol. 2016;34:1772-9
9. Chu A, Heck JE, Ribeiro KB, Brennan P, Boffetta P, Buffler P. et al. Wilms' tumour: a systematic review of risk factors and meta-analysis. Paediatr Perinat Epidemiol. 2010;24:449-69
10. Lee SB, Haber DA. Wilms tumor and the WT1 gene. Exp Cell Res. 2001;264:74-99
11. Treger TD, Chowdhury T, Pritchard-Jones K, Behjati S. The genetic changes of Wilms tumour. Nat Rev Nephrol. 2019;15:240-51
12. Mahamdallie S, Yost S, Poyastro-Pearson E, Holt E, Zachariou A, Seal S. et al. Identification of new Wilms tumour predisposition genes: an exome sequencing study. Lancet Child Adolesc Health. 2019;3:322-31
13. Fu W, Zhu J, Xiong SW, Jia W, Zhao Z, Zhu SB. et al. BARD1 Gene Polymorphisms Confer Nephroblastoma Susceptibility. EBioMedicine. 2017;16:101-5
14. Zhu J, Fu W, Jia W, Xia H, Liu GC, He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol Ther Nucleic Acids. 2018;12:854-60
15. Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G. et al. Base Excision Repair Gene Polymorphisms and Wilms Tumor Susceptibility. EBioMedicine. 2018;33:88-93
16. Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. Rna. 2017;23:1754-69
17. Kadumuri RV, Janga SC. Epitranscriptomic Code and Its Alterations in Human Disease. Trends Mol Med. 2018;24:886-903
18. Katsara O, Schneider RJ. m(7)G tRNA modification reveals new secrets in the translational regulation of cancer development. Mol Cell. 2021;81:3243-5
19. Tomikawa C. 7-Methylguanosine Modifications in Transfer RNA (tRNA). Int J Mol Sci. 2018;19:4080
20. Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell. 2019;74:1304-16.e8
21. Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N. et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019;74:1278-90.e9
22. Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell. 2018;71:244-55.e5
23. Chen Y, Lin H, Miao L, He J. Role of N7-methylguanosine (m(7)G) in cancer. Trends Cell Biol. 2022;32:819-24
24. Zeng H, Xu S, Xia E, Hirachan S, Bhandari A, Shen Y. Aberrant expression of WDR4 affects the clinical significance of cancer immunity in pan-cancer. Aging (Albany NY). 2021;13:18360-75
25. Trimouille A, Lasseaux E, Barat P, Deiller C, Drunat S, Rooryck C. et al. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet. 2018;93:374-7
26. Braun DA, Shril S, Sinha A, Schneider R, Tan W, Ashraf S. et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am J Med Genet A. 2018;176:2460-5
27. Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X. et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. 2021;11:e675
28. Chen J, Li K, Chen J, Wang X, Ling R, Cheng M. et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond). 2022;42:223-44
29. Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang G. et al. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 2021;12:691
30. Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T. et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422-35
31. Hua RX, Liu J, Fu W, Zhu J, Zhang J, Cheng J. et al. ALKBH5 gene polymorphisms and Wilms tumor risk in Chinese children: A five-center case-control study. J Clin Lab Anal. 2020;34:e23251
32. Hua RX, Fu W, Lin A, Zhou H, Cheng J, Zhang J. et al. Role of FTO gene polymorphisms in Wilms tumor predisposition: A five-center case-control study. J Gene Med. 2021;23:e3348
33. Zhuo Z, Hua RX, Zhang H, Lin H, Fu W, Zhu J. et al. METTL14 gene polymorphisms decrease Wilms tumor susceptibility in Chinese children. BMC Cancer. 2021;21:1294
34. He S, Zhu J, Xiao Z, Liu J, Zhang J, Li Y. et al. WDR4 gene polymorphisms increase hepatoblastoma susceptibility in girls. J Cancer. 2022;13:3342-7
35. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707-19
36. Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell. 2014;56:5-12
37. Michaud J, Kudoh J, Berry A, Bonne-Tamir B, Lalioti MD, Rossier C. et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics. 2000;68:71-9
38. Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J Hematol Oncol. 2022;15:63
39. Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. Rna. 2005;11:821-30
40. Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W. et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323-38.e14
41. Cheng W, Gao A, Lin H, Zhang W. Novel roles of METTL1/WDR4 in tumor via m(7)G methylation. Mol Ther Oncolytics. 2022;26:27-34
42. Huang Q. Genetic study of complex diseases in the post-GWAS era. J Genet Genomics. 2015;42:87-98
43. Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190-5
44. Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3-22
45. Wang YJ, Mugiyanto E, Peng YT, Huang WC, Chou WH, Lee CC. et al. Genetic Association of the Functional WDR4 Gene in Male Fertility. J Pers Med. 2021;11:760
46. Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W. et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339-55 e8
47. Niño CA, Scotto di Perrotolo R, Polo S. Recurrent Spliceosome Mutations in Cancer: Mechanisms and Consequences of Aberrant Splice Site Selection. Cancers (Basel). 2022;14:281
48. Ascenção K, Szabo C. Emerging roles of cystathionine β-synthase in various forms of cancer. Redox Biol. 2022;53:102331
Author contact
![]() Corresponding authors: Wen Fu or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangdong Provincial Clinical Research Center for Child Health, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: lydia_fwcom (Wen Fu) or hejing198374com (Jing He).
Corresponding authors: Wen Fu or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangdong Provincial Clinical Research Center for Child Health, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: lydia_fwcom (Wen Fu) or hejing198374com (Jing He).

 Global reach, higher impact
Global reach, higher impact