Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(8):1301-1308. doi:10.7150/jca.83854 This issue Cite
Research Paper
miR-18a expression correlates with ATM and p53 levels and poor prognosis in lymphomas
1. Department of Hematology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
2. Department of Oncology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
3. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
4. Departments of Ultrasound Imaging, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
5. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
† These authors have contributed equally to this work.
* These authors jointly supervised this work.
Received 2023-2-25; Accepted 2023-4-27; Published 2023-5-8
Abstract
microRNAs (miRNAs) are non-coding, endogenous, small-molecule RNAs. They are involved in cell proliferation, differentiation, apoptosis, and metabolism. Additionally, they play an essential role in the development and progression of various malignancies. Recent research has revealed that miR-18a plays an important role in cancer development. However, its role in lymphoma is not yet fully understood. In this study, we investigated the clinicopathological characteristics and potential functional roles of miR-18a in lymphomas. First, we predicted the potential downstream genes of miR-18a using miRTarBase software and subjected these downstream genes to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to determine the potential mechanisms of action of these genes. We found that these target genes were closely related to cellular senescence, the p53 signaling pathway, and other signaling pathways. From the predicted downstream target genes, ATM and p53 were selected as the target genes; their deletion in patients with lymphoma was detected using the fluorescence in situ hybridization technique. The results showed that some patients with lymphoma have a deletion of the ATM and p53 genes. In addition, the deletion rates of ATM and p53 were positively correlated with the expression of miR-18a. Next, the expression levels of miR-18a and the deletion rates of ATM and p53 were used for correlation and prognostic analyses with patient clinical information. The findings revealed a significant difference in disease-free survival (DFS) between patients with lymphoma with ATM deletion and those with a normal ATM gene expression (p < 0.001). Moreover, a significant difference in overall survival (OS) and DFS between patients with p53 deletion and those with normal p53 expression was observed (p < 0.001). The results indicate that the deletion of ATM and p53 downstream of miR-18a is closely associated with the development of lymphoma. Thus, these biomarkers may serve as key prognostic biomarkers for lymphomas.
Keywords: miR-18a, Epstein-Barr virus, Lymphoma, Genomic instability, Biomarker
Introduction
Lymphoma is a malignant tumor that originates from the lymphopoietic system. The occurrence and development of lymphomas are complex processes involving multiple steps, stages, and factors [1]. Both environmental (such as physical, chemical, and biological) and host factors play essential roles in lymphoma development [2,3]. Pathogenic infections such as those caused by the Epstein-Barr virus (EBV) also play an important role in the development of lymphomas [4]. Therefore, complex interactions between environmental factors and host genetic susceptibility influence the occurrence and development of lymphomas.
Genomic instability has been found in most lymphomas using various molecular biological assays [5, 6]. Among these genomic instabilities, chromosomal instability is the most common manifestation in malignant tumors; genomic instability causes tumor cells to gain a growth advantage [7, 8]. The genotype of lymphoma is highly complex. Amplification of the 13q31 locus, which encodes the miR-17-92 cluster, is often observed in B-cell lymphomas. Moreover, members of the miR-17-92 cluster are overexpressed and amplified in lymphoma [9]. Our previous study found a significant correlation between the expression of miR-18a, a member of the miR-17-92 cluster, and the reactivation of EBV due to DNA damage [10]. However, the role of miR-18a in the development of lymphoma has not been elucidated. The aim of this study was to investigate the relationship between miR-18a interaction with its downstream target genes and genomic instability in lymphomas. In addition, we explored the clinical significance of these target genes in lymphoma development.
Materials and methods
Tissue samples and clinical data
The study included 100 patients who were diagnosed with lymphoma and 23 patients with inflammatory lymph nodes as the control group. All patients included in this study were diagnosed between January 2008 and December 2015 at the Xiangya Hospital, Central South University, China. The diagnosis of all patients with lymphoma was confirmed by clinical presentation, imaging, biochemistry, immunology, bone marrow morphology, and lymphoma biopsy. The sample included 59 cases of non-Hodgkin's B-cell lymphoma, 34 cases of NK/T-cell lymphoma, and 7 cases of Hodgkin's lymphoma. Clinical cases were classified based on sex, age, lymph node infiltration, ATM deletion, and p53 deletion with high miR-18a expression. All lymphomas were diagnosed and staged in accordance with the National Guidelines for the Treatment of Malignant Lymphoma in China [11]. Treatment effects were observed after 3-6 courses of conventional treatment for lymphoma. Patients were informed about the purpose of the study and signed informed consent. The study was approved by the ethical review committee of Xiangya Hospital, Central South University. Patient characteristics for the 100 lymphoma cases are presented in Table 1.
In situ hybridization
After dewaxing and hydrating the pathological sections, the slides were transferred to 3% H2O2 and protease buffer to inactivate the endogenous enzymes. The slides were then treated with pepsin diluted in 3% citric acid. After pepsin digestion, the slides were fixed with a fixative at room temperature for 10 min. The slides were hybridized to the probe overnight at 59 °C. Subsequently, the slides were rinsed and placed in a blocking solution at 37 °C for 30 min. Biotinylated murine anti-digoxin was added dropwise to the slides, incubated for 120 min at room temperature, and then washed with PBST buffer. The SABC was added dropwise, incubated for 30 min, and washed repeatedly with PBST buffer. Finally, the sections were developed using DAB color development rinses and hematoxylin re-staining. Following dehydration and mounting, sections were observed and imaged under a microscope (OLYMPUSBX-51, Japan). Two pathologists independently determined the in situ hybridization scores of the tissue samples.
Fluorescence in situ hybridization (FISH)
To prepare the lymph node tissue sections for analysis, they were first placed in an oven for 30 min at a temperature of 80 °C. Next, the sections were immersed in a preheated dewaxing agent at 68 °C for 15 min. After dewaxing, slides were washed twice for 5 min in 100% ethanol. Then, they were immersed in a permeabilizer for 20 min at 90 °C, followed by a 3 min wash with preheated deionized water at 37 °C. Subsequently, the slides underwent digestion through immersion in a preheated protease solution at 37 °C for a duration between 10 and 40 min. Following digestion, the tissue sections underwent two 5 min rinses with a washing solution. Gradient dehydration of the slides was accomplished through sequential immersion in 70%, 85%, and 100% ethanol concentrations for 2 min each, followed by air drying at room temperature. After dehydration, 10 μl of denatured probe mixture was injected onto each slide, and they were immediately covered with cover slips. For co-denaturation, the slides were exposed in a hybridizer at 85 °C for 5 min. Lastly, the slides were placed in a clean wet box at 42 °C overnight. After hybridization, the slides were placed in a pre-warmed 0.4× SSC/0.3% NP-40 solution at 67 °C for 30 s, and then dried at room temperature. The dried slides were re-stained with 15 µL DAPI and covered with a cover slip. Finally, the slides were observed under a fluorescence microscope by selecting the appropriate filter set. Two probes were used in this study - ATM and p53 probes, located at 14q32 and 17p13.1, respectively (all purchased from Beijing Golden Bodega Company, China). In normal interphase cells, the ATM probe showed two red signals and the p53 probe showed two green signals. To obtain 10 normal control specimens, 200 cells were selected under an oil microscope to observe the fluorescence signal and establish the threshold value. This threshold value was based on the results of the 23 control patients with lymph node inflammation. Two hundred cells from each member of the control group and from each patient were observed to calculate the average number of fluorescent particles in each cell. A fluorescent particle count of less than the threshold value (4-5 particles/cell) was considered negative. Patients with specimen fluorescence greater than the threshold were considered positive, whereas those below the threshold were considered negative.
Prediction and functional analysis of target genes
MiRTarbase is an experimentally validated miRNA-target interaction database (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) containing 4076 miRNAs and 23054 target genes based on experimental evidence (including reporter analysis, western blotting, microarray, or pSILAC). The database was used to construct and visualize miRNA-mRNA regulatory networks. GO and KEGG pathway analyses were performed using the online analysis website CancerMIRNome (http://bioinfo.jialab-ucr.org/CancerMIRNome/) to functionally annotate potential target genes.
Statistical analysis
The SPSS 26.0 statistical package was used for statistical analysis. Survival data were obtained using Kaplan-Meier analysis. The log-rank test was used to determine the differences between the survival curves. Pearson χ2-test was used to test the correlation between miR-18 expression and ATM and p53 gene deletions. A p value of less than 0.05 was considered significant.
Results
The expression level of miR-18a in patients with lymphoma
In a previous study, we detected the expression level of miR-18a using in situ hybridization and found that its expression level was increased in EBV-positive patients with lymphoma. This finding suggested that miR-18a expression was positively correlated with EBV infection. Further analysis revealed that miR-18a was highly expressed in more than half of the patients with lymphoma, with a positivity rate of 51%. In contrast, the positivity rate was only 4.6% in patients with inflammatory lymph nodes (Table 1). We found that the positivity rate and index of miR-8a were significantly higher in lymphoma specimens than in control lymphadenitis specimens (p < 0.01).
Analysis of potential downstream genes of miR-18a in lymphoma
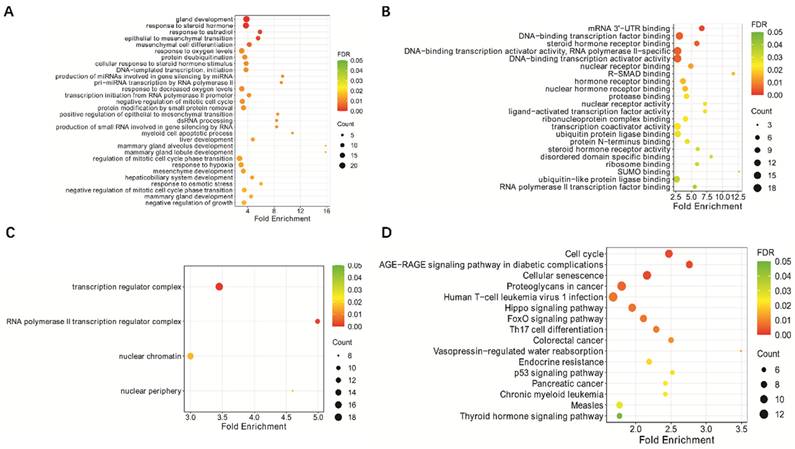
In a previous study, we found that miR-18a plays a role in EBV-associated lymphoma by inhibiting the DNA damage response and promoting EBV-associated genomic instability [10]. To further investigate the potential downstream genes of miR-18a, we used miRTarBase software to predict the potential downstream genes of miR-18a (Supplementary Table 1). The downstream genes were also subjected to GO and KEGG analyses to explore their potential mechanisms of action. We found that these genes were closely related to signaling sets such as cellular senescence and the p53 signaling pathway. Based on the above analyses, we selected ATM and p53 among the miR-18a downstream target genes for further study.
Expression levels of miR-18a in patients with lymphoma and lymphadenitis
| Lymphoma | Lymphadenitis | p Value | |
|---|---|---|---|
| miR-18a High | 51 | 1 | < 0.01 |
| miR-18a Low | 49 | 22 |
Detecting the deletion rate of ATM and p53 genes in lymphomas
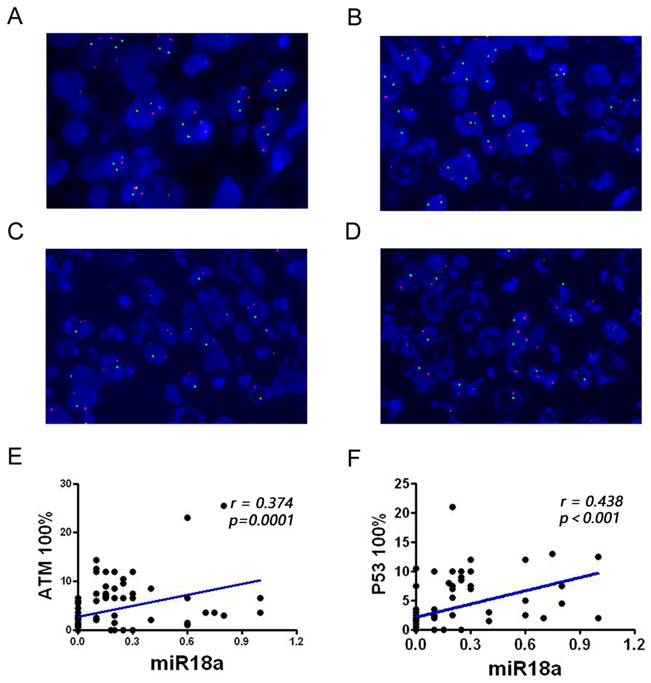
The deletion rates of ATM and p53 genes associated with genomic instability were examined in 100 patients with lymphoma and 23 control inflammatory specimens using FISH. The results showed that the ATM and p53 deletion rates were significantly higher in patients with lymphoma than in lymph node inflammatory specimens (P < 0.01). The two red and two green dots in Figure 2A indicate that both ATM and p53 are normally expressed. One red and two green lines in Figure 2B indicate that the ATM gene is absent and the p53 gene is normally expressed. Two reds and one green in Figure 2C indicate that the ATM gene is normal and the p53 gene is absent. One red and one green in Figure 2D indicate that the ATM gene is absent, and the p53 gene is absent.
Correlation analysis of miR-18a expression with p53 and ATM deletion rate
To further investigate the correlation between miRNA-18a and genomic instability in patients with lymphoma, we performed a correlation analysis of miR-18a expression with ATM and p53 deletion rate (Table 1, Figure 2E and 2F). miR-18a expression was positively correlated with ATM (r = 0.374, p = 0.001) and p53 deletion rates (r = 0.438, p < 0.001). These results also suggest that miR-18a expression levels are higher in patients with lymphoma and the molecular basis of lymphoma is associated with genomic instability.
Relationship between miR-18a expression and lymphoma clinicopathology and gene deletion characteristics
| Clinical and pathological features | N | miR-18a +high | miR-18a -low | p Value |
|---|---|---|---|---|
| Gender | ||||
| Male | 65 | 35 | 30 | |
| Female | 35 | 16 | 19 | 0.5304 |
| Age | ||||
| ≥50 | 55 | 28 | 27 | 1.000 |
| <50 | 45 | 23 | 22 | |
| Extranodal lymph node metastasis | ||||
| Yes | 71 | 43 | 28 | |
| No | 29 | 8 | 21 | 0.0039 |
| ATM | ||||
| + | 63 | 39 | 24 | |
| - | 37 | 12 | 25 | 0.0037 |
| p53 | ||||
| + | 51 | 32 | 19 | |
| - | 49 | 19 | 30 | 0.0018 |
Correlation between miR-18a expression and survival prognosis of patients with lymphoma
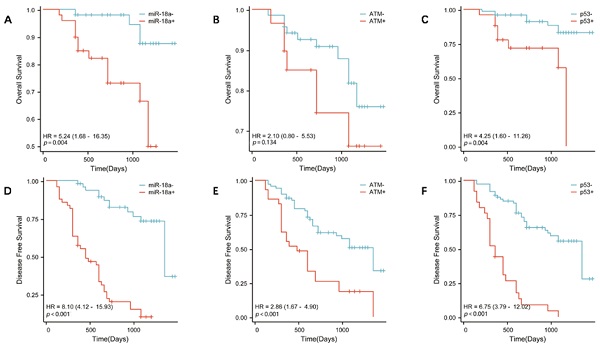
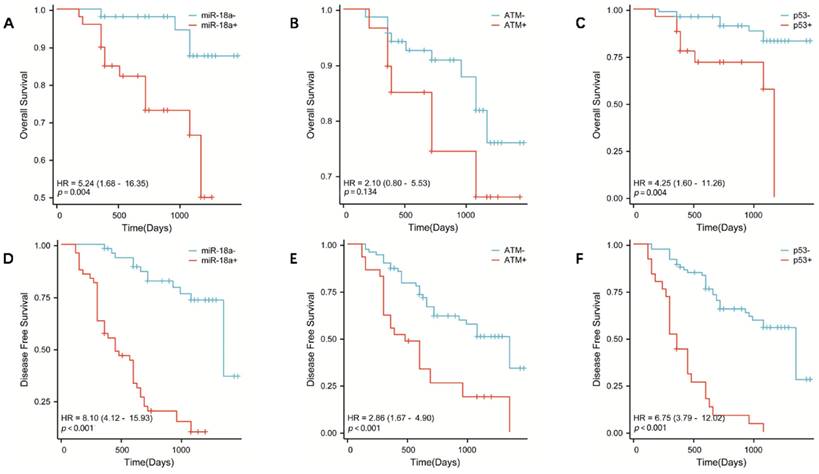
To further investigate the correlation between miR-18a expression and survival prognosis of patients with lymphoma, we collected clinical prognostic information from patients through regular and long-term telephone follow-up. The collected follow-up clinical data were statistically analyzed, and survival curves were plotted using SPSS 26.0. The results showed that the OS was lower in patients with miR-18a-positive expression of lymphoma than in patients with miR-18a-negative expression (p = 0.004) (Figure 3A). Moreover, DFS was lower in miR-18a-positive-expressing patients than in miR-18a-negative-expressing patients (p < 0.001) (Figure 3D). DFS in patients with ATM deletion was significantly different from DFS in those with normal ATM genes (p < 0.001) (Figure 3E). However, only patients with ATM deletion showed a trend towards worse OS (p = 0.128), compared with those with normal ATM genes (Figure 3B). Both OS and DFS in patients with p53 deletion were significantly different from those in patients with normal p53 (p = 0.004 and p < 0.001 for OS and DFS, respectively) (Figure 3 C and 3F).
Discussion
miRNAs are endogenous non-coding RNAs with an average length of approximately 22 nucleotides, and they have been found to regulate gene expression at the post-transcriptional level [12, 13]. They are widely present in nematodes, Drosophila, plants, and mammals [14-16]. Highly evolutionarily conserved miRNAs are fully or incompletely paired with the 3'-untranslated region (UTR) of target gene mRNAs, thereby inhibiting their translation. Subsequently, they exert physiological effects that regulate cell proliferation, differentiation, and individual development [17,18]. In addition, miRNAs play crucial roles in the occurrence and development of various diseases.
KEGG pathway enrichment analysis and GO functional enrichment analysis of hsa-miR-18a predicted target genes: (A) GO biological process; (B) GO molecular function; (C) GO cellular component; (D) KEGG analysis.
Detection of the ATM and p53 genes in pathological sections from patients with lymphoma, observed via FISH technique, and analysis of miR-18a and their correlation. (A) Normal ATM and p53 genes; (B) ATM gene deletion; (C) p53 gene deletion; (D) Simultaneous ATM and p53 gene deletions. (E-F) Correlation analysis of miR-18a expression with ATM or p53 deletion rates (magnification 1000 ×).
A growing body of evidence has highlighted the vital role of miRNAs in cancer progression [19-21]. The miR-17-92 cluster is one of the best-characterized novel non-coding RNA clusters [22]. It consists of seven members: miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92. This cluster of miRNAs is aberrantly amplified in lymphomas and other malignancies [23-26]. Upregulation of this gene cluster promotes lymphangiogenesis, especially in B-cell lymphomas. For example, overexpression of miR-17-92 has pro-oncogenic effects by promoting proliferation and decreasing apoptosis in mantle cell lymphoma tumor cells [27, 28]. Robaina et al. [29] found that miR-17 upregulation led to reduced OS in childhood Burkitt's lymphoma and was associated with a lack of expression of the pro-apoptotic gene BIM. In addition, Fassina et al. [30] reported that miR-17-92 was significantly overexpressed in germinal center-type diffuse large B-cell lymphoma and could be used as a reliable and standardized diagnostic tool for the subclassification of large B-cell lymphoid-like neoplasms. As the most representative miRNA in the miR-17-92 cluster, miR-18a plays a pro-tumorigenic role in a variety of malignancies. Our previous study demonstrated that miR-18a is highly expressed in EBV-positive lymphomas and mediates the growth of lymphoma cells by inducing EBV reactivation, which could promote malignancy of EBV-associated lymphomas. However, little is known regarding the role of miR-18a in lymphoma development. In this study, we further analyzed the data and found miR-18a overexpression and a significantly higher positivity rate and positivity index in lymphoma compared to that in lymph node inflammatory control specimens (p < 0.01).
OS in patients with lymphoma with miR-18a positive expression, ATM gene deletion, or p53 gene deletion. (A, B, C, respectively) DFS in patients with lymphoma with miR-18a-positive expression, ATM gene deletion, or p53 gene deletion. (D, E, F, respectively)
ATM and p53 are target genes that are closely associated with cellular senescence and the p53 signaling pathway. Activation of ATM, p53, and the DNA damage response (DDR) is an important mechanism to stop the proliferation of genomically altered cells. ATM is located on chromosome 11q22.3, with a full length of 184 kb and contains 66 exons. The coding region of the gene is flanked by variant 3′ and 5′-UTRs. These variants may be associated with the post-transcriptional regulation of ATM levels under physiological conditions [31]. ATM encodes a nuclear phosphoprotein, which is a member of the phosphatidylinositol 3-kinase (PI3K) family [32]. PI3K family members have been implicated in DNA repair after damage and cell cycle regulation. p53 is also an important anti-oncogene, commonly known as the "molecular police" in DNA damage and repair, which can play an important role in promoting apoptosis of cancer cells and preventing cellular carcinogenesis. p53 also plays an essential role in the repair of cellular DNA following damage [33]. Previous studies have reported that both ATM and p53 play important roles in lymphoma development. For example, Zhou et al. [34] found that activation of the ATM-Chk2-p53-p21 pathway blocked cell cycle progression and induced apoptosis in NK/T-cell lymphoma cell lines. Bhalla et al. [35] reported that ATM knockout induced mitochondrial deacetylase SIRT3 activity and disrupted the mitochondrial structure, thereby promoting the growth of diffuse large B-cell lymphoma. Based on these findings, ATM and p53 are two proteins that work closely together and may be involved in the construction of anti-cancer barriers. miRNAs play an important role in controlling gene expression by inhibiting protein translation or promoting messenger RNA degradation. miRTarBase software predicted potential miR-18a downstream target genes, which were then analyzed for GO and KEGG enrichment. The results indicated that miR-18a is closely associated with cellular senescence, the p53 signaling pathway, and other signaling pathways. We propose that miR-18a is likely to induce malignancy in lymphoma by affecting the expression of ATM and p53 genes.
Furthermore, the disruption of the ATM/p53 pathway can impact the synthesis of the corresponding proteins, which ultimately affects the survival and prognosis of individuals with lymphoma. Massive cell lymphoma (MCL) is a particularly aggressive subtype of lymphoma that has been linked to the deletion of ATM and p53; ATM has been identified as the most frequently mutated gene in MCL [36]. Interestingly, ATM inactivation did not significantly affect the survival of patients with MCL, whereas TP53 mutations had a substantial negative impact on OS in MCL [37]. Nevertheless, testing for ATM mutations remains important. Previous studies have demonstrated that cells with defective ATM function exhibit heightened radiosensitivity, which may be advantageous for treating highly chemo resistant lymphoma subtypes with radiotherapy [38]. In addition, chronic lymphocytic leukemia cells with ATM mutations have shown increased resistance to doxorubicin, likely due to their inability to activate the pro-apoptotic p53 pathway following drug administration [39]. It has been suggested that the deletion of the ATM/p53 pathway may be a common selection mechanism in malignant B lymphocytes [37]. In the present study, we utilized the FISH technique to investigate the frequency of ATM and p53 gene deletions in lymphoma. FISH is a molecular genetic technique that has been recently used in a wide range of clinical applications. It can directly reveal the relationship between DNA sequences in the nucleus or chromosomes of specific cells by in situ hybridization of labeled probes of specific molecules to chromosomes and the development of fluorescent color [40]. The basic principle of the FISH technique is qualitative localization and relative quantitative analysis of the nucleic acid targets in a specimen using a nucleic acid probe directly or indirectly labeled with fluorescein, based on the principle of base complementarity. The FISH technique overcomes limitations of traditional cytogenetic approaches by using sequence-specific probes to rapidly and accurately reveal structural abnormalities in chromosome number, identify chromosome origin, and analyze complex karyotypes. Owing to the disadvantages of poor chromosome morphology and difficulty in observation, numerous studies have shown high sensitivity and specificity of the FISH technique to examine multiple leukemia fusion genes and other site-specific gene deletions or mutations [41]. Currently, FISH technology is widely used for the detection of genes at the molecular level in China and abroad. In this study, FISH was used to detect the deletion rate of the genomic instability-related molecules ATM and p53. The data revealed that the rate of ATM and p53 gene deletion was significantly higher in patients with lymphoma than in inflamed lymph node tissues (P < 0.01). Combined with the analysis of clinical data, we found that the deletion of ATM and p53 correlated with the clinical grade of lymphoma and extra-nodal metastases, independently of age and sex. Moreover, the overall survival of patients with ATM and p53 deletions was significantly shorter. Correlation analysis of miR-18a expression with ATM and p53 deletion rates showed that miR-18a expression was positively correlated with ATM deletion rates (r = 0.374, p = 0.001) and p53 deletion rates (r = 0.438, p < 0.001). Based on the findings, it appears that miR-18a is strongly linked to the expression of ATM and p53 genes. Therefore, we propose that miR-18a may act as a regulator of ATM and p53, ultimately impacting the prognosis of individuals with lymphoma.
Conclusion
In conclusion, this study revealed that miR-18a is an upregulated biomarker of lymphoma and its expression level is positively correlated with the deletion of the ATM and p53 genes. miR-18a is likely to induce malignant behavior in lymphoma by targeting the downstream genes ATM and p53, which are associated with poor prognosis in lymphoma. It should be noted, however, that while we have established a clear association between miR-18a and lymphoma in clinical samples, we did not explore the relationship between individual subtypes and molecules. Thus, further in vitro and in vivo experiments on the pathogenesis of miR-18a are required to verify the role of its regulated molecular network in lymphoma.
Abbreviations
miRNA: microRNA; FISH: fluorescence in situ hybridization; GC-DLBCL: germinal central diffuse large B-cell lymphoma; DFS: disease-free survival time; OS: overall survival time; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PI3K: phosphatidylinositol 3-kinase; DDR: DNA damage response; UTR: untranslated region; MCL: massive cell lymphoma.
Supplementary Material
Supplementary table.
Acknowledgements
Funding
This study was supported by the Natural Science Foundation of Hunan Province (2019JJ40487, 2019JJ40497, and 2022JJ40784), the Changsha Municipal Natural Science Foundation (kq2202374) and the Central South University Innovation-Driven Research Programme (2023CXQD076).
Authors Contributions
Pengfei Cao and Xiaoyun He contributed to the conception and design of the study. Hao Zhou and Yuxiang He performed resource analysis, and wrote the first draft of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics statement
The research presented here has been performed in accordance with the Declaration of Helsinki and has been approved by the ethics committee of Xiangya Hospital, Central South University, China (reference number 201312484). The patients were informed about the sample collection and had signed informed consent forms.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Han Y, Wang D, Peng L, Huang T, He X, Wang J. et al. Single-cell sequencing: a promising approach for uncovering the mechanisms of tumor metastasis. J Hematol Oncol. 2022;15:59
2. Pearce N, McLean D. Agricultural exposures and non-Hodgkin's lymphoma. Scand J Work Environ Health. 2005;31(Suppl 1):18-25 discussion 5-7
3. Meloni F, Satta G, Padoan M, Montagna A, Pilia I, Argiolas A. et al. Occupational exposure to glyphosate and risk of lymphoma:results of an Italian multicenter case-control study. Environ Health. 2021;20:49
4. Mawson AR, Majumdar S. Malaria, Epstein-Barr virus infection and the pathogenesis of Burkitt's lymphoma. Int J Cancer. 2017;141:1849-55
5. Orsborne C, Byers R. Impact of gene expression profiling in lymphoma diagnosis and prognosis. Histopathology. 2011;58:106-27
6. Elenitoba-Johnson KSJ, Lim MS. New Insights into Lymphoma Pathogenesis. Annu Rev Pathol. 2018;13:193-217
7. Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML. et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995-9000
8. Wang D, Tang L, Wu Y, Fan C, Zhang S, Xiang B. et al. Abnormal X chromosome inactivation and tumor development. Cell Mol Life Sci. 2020;77:2949-58
9. Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE. et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520-5
10. Cao P, Zhang M, Wang L, Sai B, Tang J, Luo Z. et al. miR-18a reactivates the Epstein-Barr virus through defective DNA damage response and promotes genomic instability in EBV-associated lymphomas. BMC Cancer. 2018;18:1293
11. Health Commission Of The People's Republic Of China N. National guidelines for diagnosis and treatment of malignant lymphoma 2022 in China (English version). Chin J Cancer Res. 2022;34:425-46
12. He X, Kuang G, Wu Y, Ou C. Emerging roles of exosomal miRNAs in diabetes mellitus. Clin Transl Med. 2021;11:e468
13. Zhou H, He X, He Y, Ou C, Cao P. Exosomal circRNAs: Emerging Players in Tumor Metastasis. Front Cell Dev Biol. 2021;9:786224
14. Cui J, You C, Chen X. The evolution of microRNAs in plants. Curr Opin Plant Biol. 2017;35:61-7
15. Vieux KF, Prothro KP, Kelley LH, Palmer C, Maine EM, Veksler-Lublinsky I. et al. Screening by deep sequencing reveals mediators of microRNA tailing in C. elegans. Nucleic Acids Res. 2021;49:11167-80
16. Guo Z, Kuang Z, Deng Y, Li L, Yang X. Identification of Species-Specific MicroRNAs Provides Insights into Dynamic Evolution of MicroRNAs in Plants. Int J Mol Sci. 2022;23:14273
17. Wang Z. miRNA in the regulation of ion channel/transporter expression. Compr Physiol. 2013;3:599-653
18. Du S, Ling H, Guo Z, Cao Q, Song C. Roles of exosomal miRNA in vascular aging. Pharmacol Res. 2021;165:105278
19. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999-3004
20. He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S. et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. 2020;16:2628-47
21. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14:dmm047662
22. Fang LL, Wang XH, Sun BF, Zhang XD, Zhu XH, Yu ZJ. et al. Expression, regulation and mechanism of action of the miR-17-92 cluster in tumor cells (Review). Int J Mol Med. 2017;40:1624-30
23. Mihailovich M, Bremang M, Spadotto V, Musiani D, Vitale E, Varano G. et al. miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nat Commun. 2015;6:8725
24. Kral J, Korenkova V, Novosadova V, Langerova L, Schneiderova M, Liska V. et al. Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis. 2018;39:1359-67
25. Sun Y, Li S, Yu W, Zhao Z, Gao J, Chen C. et al. N6-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020;11:836
26. Wang J, Peng X, Li R, Liu K, Zhang C, Chen X. et al. Evaluation of Serum miR-17-92 Cluster as Noninvasive Biomarkers for Bladder Cancer Diagnosis. Front Oncol. 2021;11:795837
27. Navarro A, Beà S, Fernández V, Prieto M, Salaverria I, Jares P. et al. MicroRNA expression, chromosomal alterations, and immunoglobulin variable heavy chain hypermutations in Mantle cell lymphomas. Cancer Res. 2009;69:7071-8
28. Jiang C, Bi C, Jiang X, Tian T, Huang X, Wang C. et al. The miR-17~92 cluster activates mTORC1 in mantle cell lymphoma by targeting multiple regulators in the STK11/AMPK/TSC/mTOR pathway. Br J Haematol. 2019;185:616-20
29. Robaina MC, Faccion RS, Mazzoccoli L, Rezende LM, Queiroga E, Bacchi CE. et al. miR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosis. Ann Hematol. 2016;95:881-91
30. Fassina A, Marino F, Siri M, Zambello R, Ventura L, Fassan M. et al. The miR-17-92 microRNA cluster: a novel diagnostic tool in large B-cell malignancies. Lab Invest. 2012;92:1574-82
31. Sandoval N, Platzer M, Rosenthal A, Dörk T, Bendix R, Skawran B. et al. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum Mol Genet. 1999;8:69-79
32. Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711-38
33. Boutelle AM, Attardi LD. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021;31:298-310
34. Zhou J, Zhang C, Sui X, Cao S, Tang F, Sun S. et al. Histone deacetylase inhibitor chidamide induces growth inhibition and apoptosis in NK/T lymphoma cells through ATM-Chk2-p53-p21 signalling pathway. Invest New Drugs. 2018;36:571-80
35. Bhalla K, Jaber S, Reagan K, Hamburg A, Underwood KF, Jhajharia A. et al. SIRT3, a metabolic target linked to ataxia-telangiectasia mutated (ATM) gene deficiency in diffuse large B-cell lymphoma. Sci Rep. 2020;10:21159
36. Greiner TC, Dasgupta C, Ho VV, Weisenburger DD, Smith LM, Lynch JC. et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2006;103:2352-7
37. Mareckova A, Malcikova J, Tom N, Pal K, Radova L, Salek D. et al. ATM and TP53 mutations show mutual exclusivity but distinct clinical impact in mantle cell lymphoma patients. Leuk Lymphoma. 2019;60:1420-8
38. Haque W, Voong KR, Shihadeh F, Arzu I, Pinnix C, Mazloom A. et al. Radiation therapy is an effective modality in the treatment of mantle cell lymphoma, even in heavily pretreated patients. Clin Lymphoma Myeloma Leuk. 2014;14:474-9
39. Navrkalova V, Sebejova L, Zemanova J, Kminkova J, Kubesova B, Malcikova J. et al. ATM mutations uniformly lead to ATM dysfunction in chronic lymphocytic leukemia: application of functional test using doxorubicin. Haematologica. 2013;98:1124-31
40. Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116:2833-8
41. Zito Marino F, Brunelli M, Rossi G, Calabrese G, Caliò A, Nardiello P. et al. Multitarget fluorescence in situ hybridization diagnostic applications in solid and hematological tumors. Expert Rev Mol Diagn. 2021;21:161-73
Author contact
![]() Corresponding authors: Pengfei Cao. Department of hematology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: caopengfei66com; Xiaoyun He. Departments of Ultrasound Imaging, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: hexiaoyunedu.cn.
Corresponding authors: Pengfei Cao. Department of hematology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: caopengfei66com; Xiaoyun He. Departments of Ultrasound Imaging, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: hexiaoyunedu.cn.

 Global reach, higher impact
Global reach, higher impact