3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(9):1479-1485. doi:10.7150/jca.83395 This issue Cite
Research Paper
Porphyromonas gingivalis in Colorectal Cancer and its Association to Patient Prognosis
1. Department of Medical Biosciences, Pathology, Umeå University, Umeå, Sweden.
2. Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden.
†Shared last authorship
Received 2023-2-9; Accepted 2023-5-5; Published 2023-5-21
Abstract
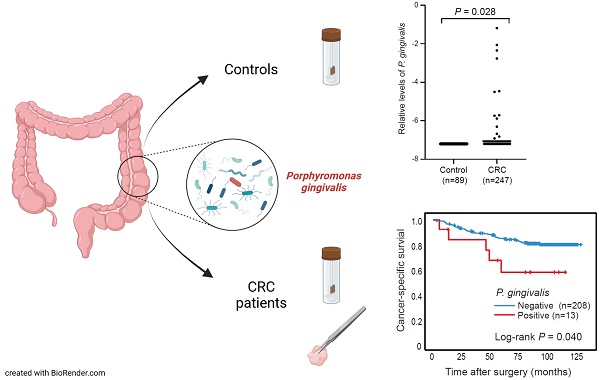
Microbiota dysbiosis may affect both the development and progression of colorectal cancer (CRC). Large metagenomic studies have highlighted specific oral bacteria linked to CRC including Porphyromonas gingivalis. Few studies have however analysed the implications of this bacterium in CRC progression and survival. In this study, we investigated the intestinal presence of P. gingivalis by qPCR in both faecal and mucosal samples from two different patient cohorts, including patients with precancerous dysplasia or CRC, as well as controls. P. gingivalis was detected in 2.6-5.3% of CRC patients and significantly different levels of P. gingivalis were found in faeces of CRC patients compared to controls (P = 0.028). Furthermore, an association was found between the presence of P. gingivalis in faeces and tumour tissue (P < 0.001). Our findings further suggested a potential link between mucosal P. gingivalis and tumours of MSI subtype (P = 0.040). Last but not least, patients with faecal P. gingivalis were found to have a significantly decreased cancer-specific survival (P = 0.040). In conclusion, P. gingivalis could be linked to patients with CRC and to a worse patient prognosis. Further studies are needed to elucidate the role of P. gingivalis in CRC pathogenesis.
Keywords: Porphyromonas gingivalis, colorectal cancer, microbiota, survival
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the second most common cause of cancer death [1]. It is a heterogenous disease, but with a relatively low level of hereditability, which reflects the importance of the environment in development of sporadic CRC.
The colon is highly colonised by microorganisms and harbours around 70% of the human microbiome [2]. The gut microbiota is known to play an important role in mucosal homeostasis, nutritional absorption, immunity, epithelial barrier function as well as carcinogenesis [3]. Recent metagenomic studies have established specific alterations in the gut microbiota associated with CRC [4]. Meta-analyses have further identified a microbial core of enriched bacteria in CRC including bacteria from the oral taxa such as Fusobacterium nucleatum and Parvimonas micra, as well as bacteria from the Porphyromonas genus [5, 6]. A driver/passenger theory has been suggested in CRC progression, where some gut commensal bacteria may be responsible for the overexpression of pro-inflammatory cytokines and the induction of chronic inflammation, which may trigger the development of adenoma followed by adenocarcinoma [7, 8]. Driver bacteria may also directly trigger cancer development by inducing epithelial DNA damage. Succeeding tumourigenesis, the environment is transformed, leading to an overgrowth of opportunistic bacteria. These passenger bacteria may replace the driver bacteria - as they benefit from a growth advantage in the tumour microenvironment - and cause inflammation at a later stage contributing to cancer progression. A good illustration of a potential driver is Escherichia coli, where some strains are capable of releasing the toxin colibactin. This toxin can cause double stranded DNA breaks of epithelial cells which subsequently may lead to tumourigenesis [9]. The pathogenic colibactin-expressing E. coli has been shown to be increased in CRC [10]. Other potential driver/passengers include F. nucleatum and P. micra from the oral microbiota. F. nucleatum has been found interacting with different aspects of CRC progression, in both preclinical and clinical studies (reviewed in [11]), and has been associated with decreased patient survival [12], and resistance to chemotherapy [13]. P. micra is also an interesting bacterium, shown to be associated with tumour immune profiles in CRC and to negatively impact patient survival [14-16].
There are at least three different evolutionary routes described in sporadic colorectal cancer [17]. The traditional pathway occurs in 50% to 70% of the cases and results in the transition from a normal mucosa to an adenoma and further to an adenocarcinoma. This pathway is caused by mutations in tumour oncogenes and suppressor genes (e.g. APC, KRAS, and TP53) and results in chromosomal instability [17]. In the serrated pathway, which occurs in 10% to 20% of the cases, the normal mucosa evolves into sessile serrated lesions (SSL) often with mutations in the BRAF proto-oncogene, followed by the development into an adenocarcinoma through MutL homolog 1 (MLH1) promoter methylation leading to microsatellite instability (MSI). While the traditional pathway occurs more often in the distal colon, the serrated pathway is found mostly in the proximal colon and linked to a good prognosis. Understanding the interactions of the microbiota with these developmental pathways is currently an important objective in colorectal cancer research.
In this study, the aim was to investigate another oral pathogen, P. gingivalis, and its associations with CRC progression and patient survival. P. gingivalis was selected as a putative driver/passenger bacteria in CRC since it has previously been linked to periodontitis, an inflammatory condition in the oral cavity [18], as well as to clinical stages and survival in oral and esophageal cancers [19]. Analyses were conducted to identify P. gingivalis in faecal samples from patients with precancerous dysplasia or CRC using two different patient cohorts. For some of the patients, the presence of P. gingivalis was also analysed in tumour tissue.
Materials and methods
Study patients
The study was based on patients from the Uppsala-Umeå Comprehensive Cancer Consortium (U-CAN) cohort of CRC patients [20], and the Fecal and Endoscopic Colorectal Study in Umeå (FECSU) [21]. The collection of patients for the U-CAN cohort was initiated in 2010 and includes patients diagnosed with CRC with longitudinally collected blood, tissue, faeces, radiological and clinical data. The FECSU cohort consists of patients who underwent colonoscopy at Umeå University hospital between the years 2008-2013, due to gastrointestinal symptoms and includes patients with dysplasia or CRC, but also patients without pathological findings.
For this study, 257 CRC patients (stage I-IV) diagnosed between the years 2010-2014 were included from U-CAN, as well as 135 patients with dysplasia and 39 patients with CRC from the FECSU cohort. Controls were selected from the FECSU cohort and were matched by age and gender from the patients with no neoplastic findings during colonoscopy, as previously described [22]. Inclusion and exclusion criteria, as well as sample collection procedures, for the U-CAN [22] and FECSU [21] cohorts were previously described. For all patients included in this study, faecal samples were collected at time of diagnosis and before start of treatment. Fresh frozen tumour tissue specimens were also collected for CRC patients from the U-CAN cohort (n=115).
The study protocol was approved by the Regional Ethical Review Board in Umeå, Sweden (dnr 2016/219-31 and dnr 08-184M), including the procedure by which the patients gave informed consent.
Detection of P. gingivalis by Quantitative real time PCR
The QIAamp PowerFecal DNA kit (Qiagen, Sollentuna, Sweden) was used to extract gDNA from approximately 0.2 g of stool, according to manufacturer's instructions. Fresh frozen tumour tissues were homogenised using a Precellys 24 homogenizer (Bertin Techologies, Rockville, MD, USA) and gDNA was extracted using the Allprep DNA/RNA/miRNA Universal kit (Qiagen). Double stranded DNA-recovery was measured using the Qubit® dsDNA BR Assay (Invitrogen, Carlsbad, CA, USA) with the Qubit® 2.0 Fluorometer (Invitrogen). Quantitative real time PCR (qPCR) was used to detect P. gingivalis in DNA extracted from faecal samples or fresh frozen tumour tissues using the Quant-StudioTM 6 Flex Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). P. gingivalis was analysed according to previous studies [23, 24], to quantify the gene encoding the small subunit of 16S ribosomal RNA of P. gingivalis. Genomic DNA from P. gingivalis strain 2561 ATCC 33277 (LGC Standards AB, Bora, Sweden) was used as a positive control. The following primers and probes were used: 5′-GCGCTCAACGTTCAGCC-3′ (forward); 5′-CACGAATTCCGCCTGC-3' (reverse) and FAM-CACTGAACTCAAGCCCGGCAGTTTCAA-TAMRA (probe). Reactions were run using the TaqMan Universal PCR Mix (Applied Biosystems, Warrington, UK) with 10 µM of primers and 10 ng of gDNA template in a total volume of 20 µL. The cycle conditions used were: 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of: 95°C for 15 seconds, 60°C for 60 seconds. Data was collected from stable duplicates (standard deviation < 0.5). As an internal criterion, a positive P. gingivalis status was given to samples amplified within 38 cycles. The levels of P. gingivalis were presented as a relative quantification using the 2-ΔCq method. 16SrRNA and the human gene PGT were used as reference for faeces and fresh frozen tissue, respectively, as previously described [22]. Exclusions included depleted samples, low DNA yield, or poor sample quality. After exclusions, analyses were performed on faecal samples from 247 CRC patients from the U-CAN cohort and 89 matched controls (from FECSU), as well as on faecal samples from 128 patients with dysplasia, 38 patients with CRC, and 61 controls from the FECSU cohort. Fresh frozen tumour tissues were analysed from 113 CRC patients. The analyses of P. micra and F. nucleatum in the U-CAN patients were previously described [22].
Statistical methods
IBM SPSS Statistics 28 was used for statistical analyses (SPSS Inc.). The Fischer´s exact test was used for comparisons between categorical variables and the Mann-Whitney U test or Kruskall-Wallis test were used to compare differences in continuous variables between groups. Kaplan-Meier survival analysis was used to estimate cancer-specific survival, with the log-rank test used to compare differences in outcome between groups. Patients were followed from the time of surgery to the time of death or end of follow-up (October 2021). Cancer-specific death was defined as death with known disseminated or recurrent disease. Patients not surgically resected for CRC and patients dying from post-operative complications within 90 days were excluded from survival analyses. Multivariable survival analysis was performed using Cox proportional hazard models. P < 0.05 was used to consider statistical significance.
Results
Significantly different levels of P. gingivalis in faecal samples from CRC patients compared to controls
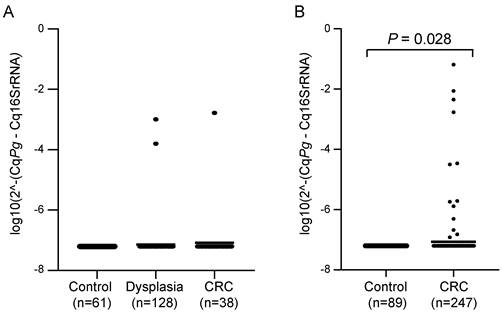
Faecal samples from CRC patients and control individuals (without neoplastic findings) were analysed for the presence of P. gingivalis by qPCR in two different patient cohorts. For the FECSU cohort, two out of 128 patients (1.6%) with dysplasia and one out of 38 patients (2.6%) with CRC were positive for P. gingivalis (Figure 1A). Among the 247 CRC patients from the U-CAN cohort, 13 faecal samples (5.3%) were positive for P. gingivalis, and the levels of P. gingivalis were found to be significantly different in faecal samples from CRC patients compared to controls (P = 0.028) (Figure 1B). No P. gingivalis positive samples were found among the controls. Since so few cases were found positive in the FECSU cohort, we continued our studies focusing on the patients included from the U-CAN cohort.
To assess the presence of P. gingivalis in the colorectal mucosa, we analysed fresh frozen tumour tissues from 113 U-CAN CRC patients. In total, seven samples (6.2%) were positive for mucosal P. gingivalis, and five out of these samples were also positive for the bacterium in faeces. Furthermore, a significant association in distribution of P. gingivalis between these two compartments was found (Table 1).
Cross-tabulation between levels of P. gingivalis in faeces and tumour tissue of CRC patients.
| P. gingivalis in tumour tissue | |||
|---|---|---|---|
| Negative (n=106) | Positive (n=7) | P value | |
| P. gingivalis in faeces, n (%) | < 0.001 | ||
| Negative (n=234) | 102 (98.1) | 2 (1.9) | |
| Positive (n=13) | 4 (44.4) | 5 (55.6) | |
Fisher's exact test was used for comparisons of categorical variables.
Associations between P. gingivalis and clinicopathological and tumour molecular characteristics
We investigated the associations of P. gingivalis in faeces or tumour tissue of U-CAN CRC patients (n=247) to clinical and pathological characteristics (Table 2). We found a slight association of faecal P. gingivalis to patient age, but no significant associations were found with gender or tumour location, stage, grade, or type (mucinous/non-mucinous), neither for faecal nor for mucosal P. gingivalis. Furthermore, P. gingivalis was not significantly associated with perineural or venous invasion. Nevertheless, the presence of P. gingivalis in tumour tissue was associated with a mucinous tumour type (P = 0.045).
Levels of P. gingivalis in faecal samples from patients with dysplasia or CRC versus controls. Scatter plots showing the relative levels of P. gingivalis (Pg) analysed by qPCR in faecal samples from patients with dysplasia or CRC and controls in (A) the FECSU cohort and (B) the U-CAN cohort. Horizontal lines indicate mean relative expression. A Mann-Whitney test was used for the statistical comparison of P. gingivalis levels between CRC patients and controls.
Clinicopathological characteristics of study patients in relation to P. gingivalis in faeces and tumour tissue of CRC patients.
| P. gingivalis in faeces | P. gingivalis in tumour tissue | |||||
|---|---|---|---|---|---|---|
| Negative | Positive | P value | Negative | Positive | P value | |
| Age, n (%) | 0.038 | 0.778 | ||||
| ≤ 59 | 40 (95.2%) | 2 (4.8%) | 17 (94.4%) | 1 (5.6%) | ||
| 60-69 | 90 (97.8%) | 2 (2.2%) | 35 (94.6%) | 2 (5.4%) | ||
| 70-79 | 73 (89.0%) | 9 (11.0%) | 38 (90.5%) | 4 (9.5%) | ||
| ≥ 80 | 31 (100%) | 0 (0%) | 16 (100%) | 0 (0%) | ||
| Gender, n (%) | 0.385 | 0.241 | ||||
| Female | 92 (92.9%) | 7 (7.1%) | 46 (90.2%) | 5 (9.8%) | ||
| Male | 142 (95.9%) | 6 (4.1%) | 60 (96.8%) | 2 (3.2%) | ||
| Location, n (%) | 1.000 | 0.375 | ||||
| Right colon | 49 (94.2%) | 3 (5.8%) | 30 (90.9%) | 3 (9.1%) | ||
| Left colon | 39 (95.1%) | 2 (4.9%) | 19 (90.5%) | 2 (9.5%) | ||
| Rectum | 146 (94.8%) | 8 (5.2%) | 57 (96.6%) | 2 (3.4%) | ||
| Stage, n (%) | 0.953 | 0.164 | ||||
| I | 47 (95.9%) | 2 (4.1%) | 32 (100%) | 0 (0%) | ||
| II | 77 (93.9%) | 5 (6.1%) | 38 (95%) | 2 (5%) | ||
| III | 65 (95.6%) | 3 (4.4%) | 27 (87.1%) | 4 (12.9%) | ||
| IV | 36 (94.7%) | 2 (5.3%) | 9 (100%) | 0 (0%) | ||
| Tumour grade, n (%) | 0.211 | 0.295 | ||||
| High grade | 26 (89.7%) | 3 (10.3%) | 18 (90%) | 2 (10%) | ||
| Low grade | 176 (95.1%) | 9 (4.9%) | 87 (95.6%) | 4 (4.4%) | ||
| Tumour type, n (%) | 0.642 | 0.045 | ||||
| Non-mucinous | 180 (94.7%) | 10 (5.3%) | 91 (96.8%) | 3 (3.2%) | ||
| Mucinous | 24 (92.3%) | 2 (7.7%) | 14 (82.4%) | 3 (17.6%) | ||
| Perineural invasion, n (%) | 0.083 | 0.272 | ||||
| Yes | 28 (87.5%) | 4 (12.5%) | 17 (89.5%) | 2 (10.5%) | ||
| No | 176 (95.7%) | 8 (4.3%) | 88 (95.7%) | 4 (4.3%) | ||
| Venous invasion, n (%) | 0.137 | 0.623 | ||||
| Yes | 41 (89.1%) | 5 (10.9%) | 24 (92.3%) | 2 (7.7%) | ||
| No | 162 (95.9%) | 7 (4.1%) | 81 (95.3%) | 4 (4.7%) | ||
Fisher´s exact test was used for comparisons of categorical variables. Missing cases were present for the following variables in faeces analyses: stage, 10 cases; tumour grade, 33 cases; tumour type and perineural invasion, 31 cases; venous invasion, 32 cases. Missing cases in tumour tissue analyses: stage, 1 case; tumour grade, tumour type, perineural invasion and venous invasion, 2 cases.
We next analysed the presence of P. gingivalis in relation to tumour molecular characteristics. We could not find any significant associations to molecular characteristics for faecal P. gingivalis. However, even though limited by sample size, we did find a significant association between the presence of P. gingivalis in tumour tissue and tumours of MSI subtype (P = 0.040) (Table 3).
Associations between P. gingivalis and other oral CRC-associated bacteria
We further explored the interactions of P. gingivalis with F. nucleatum and P. micra. We found no association of P. gingivalis with these bacteria in faecal samples. However, we observed a significant association between the presence of P. gingivalis and P. micra in tumour tissue (P = 0.005) (Table 4).
Molecular characteristics of study patients in relation to P. gingivalis in faeces and tumour tissue of CRC patients.
| P. gingivalis in faeces | P. gingivalis in tumour tissue | |||||
|---|---|---|---|---|---|---|
| Negative | Positive | P value | Negative | Positive | P value | |
| KRAS status, n (%) | 0.753 | 0.240 | ||||
| Wild-type | 112 (92.6%) | 9 (7.4%) | 62 (91.2%) | 6 (8.8%) | ||
| Mutant | 57 (95%) | 3 (5%) | 44 (97.8%) | 1 (2.2%) | ||
| BRAF status, n (%) | 0.110 | 0.334 | ||||
| Wild-type | 146 (94.8%) | 8 (5.2%) | 89 (94.7%) | 5 (5.3%) | ||
| Mutant | 26 (86.7%) | 4 (13.3%) | 17 (89.5%) | 2 (10.5%) | ||
| MSI status, n (%) | 0.158 | 0.040 | ||||
| MSS | 154 (94.5%) | 9 (5.5%) | 95 (96%) | 4 (4%) | ||
| MSI | 19 (86.4%) | 3 (13.6%) | 11 (78.6%) | 3 (21.4%) | ||
Fisher´s exact test was used for comparisons of categorical variables. Missing cases were present in faeces analyses for the following variables: KRAS status, 66 cases; BRAF status, 63 cases; MSI status, 62 cases.
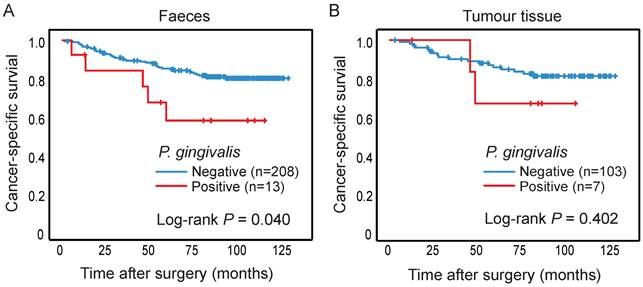
The presence of P. gingivalis in relation to survival of CRC patients. Kaplan-Meier plots of cancer-specific survival in patients negative or positive for P. gingivalis in (A) faeces or (B) tumour tissue. Log-rank tests were used to calculate P values.
Associations between P. gingivalis and other CRC-associated bacteria in faeces and tumour tissue of CRC patients.
| Faeces | Tumour tissue | |||||
|---|---|---|---|---|---|---|
| P. gingivalis | P. gingivalis | |||||
| Negative | Positive | P value | Negative | Positive | P value | |
| F. nucleatum, n (%) | 0.151 | 0.097 | ||||
| Low | 125 (96.9) | 4 (3.1) | 68 (97.1) | 2 (2.9) | ||
| High | 105 (92.1) | 9 (7.9) | 35 (87.5) | 5 (12.5) | ||
| P. micra, n (%) | 0.251 | 0.005 | ||||
| Low | 133 (96.4) | 5 (3.6) | 84 (97.7) | 2 (2.3) | ||
| High | 99 (92.5) | 8 (7.5) | 19 (79.2) | 5 (20.8) | ||
Fischer´s exact tests were used for comparisons of categorical variables. Missing cases were present in faeces analyses for the following variables: F. nucleatum, 4 cases; P. micra, 2 cases. Missing cases in tumour tissue analyses: F. nucleatum, 3 cases; P. micra, 3 cases.
Faecal detection of P. gingivalis is associated with decreased patient survival
The presence of P. gingivalis in faeces and tumour tissue was analysed in relation to cancer-specific patient survival, and patients with P. gingivalis positive faecal samples were found to have a significantly worse prognosis compared to patients with P. gingivalis negative samples (P = 0.040) (Figure 2A). The significance of the prognostic role of faecal P. gingivalis was maintained in a multivariable Cox proportional hazard model including age, gender, and tumor stage (hazard ratio (HR) = 2.90, CI 1.01-8.32, P = 0.047)). No significant difference in survival was found according to P. gingivalis in tumour tissue (Figure 2B).
Discussion
Recent research suggests the involvement of oral microbes in the development and progression of CRC [25]. In this study, we explored the role of the oral bacteria P. gingivalis in CRC, using two different patient cohorts with collected faecal and tumour tissue samples.
We found a significant difference of P. gingivalis faecal levels between CRC patients and non-cancerous controls. P. gingivalis was not detected in the controls, but in some samples from patients with pre-cancerous lesions, suggesting that intestinal presence of P. gingivalis could in some cases be an early event. We further found an association between the presence of faecal and mucosal P. gingivalis. Our findings are in line with previous studies on CRC using both 16SrRNA sequencing and qPCR to identify P. gingivalis [26, 27]. We found a weak association of mucosal P. gingivalis with tumours of MSI subtype. We further found slight associations of P. gingivalis to other oral CRC-associated bacteria in tumour tissue, in particular P. micra. P. gingivalis has previously been related to tumours of CMS1 subtype (including tumours of MSI subtype) together with F. nucleatum and P. micra [28], which is in line with our finding of a putative association between P. gingivalis with these bacteria in CRC. Further larger studies on the relation of P. gingivalis to CRC molecular traits are needed to establish these associations. Interestingly, P. gingivalis has been shown to synergistically promote extra-gastrointestinal infection and to form biofilm together with other oral bacteria in periodontitis [18, 29]. Oral microbiota has also been linked to biofilm formation in CRC [30, 31]. In fact, CRC-associated F. nucleatum has been shown to potentially originate from the oral cavity [32]. Therefore, collecting clinical data about the oral health of the patients would be an important topic for future CRC studies.
The impact of P. gingivalis on cancer-specific patient survival was next investigated. Patients with faecal P. gingivalis - even though few - were found to have a significantly decreased survival compared to patients without faecal P. gingivalis. Our findings were corroborated in a study by Wang et al., where they found that P. gingivalis was enriched in tumour tissue and faecal samples from CRC patients and that its presence was associated with a poor prognosis [27]. Moreover, P. gingivalis has previously been linked to lymph node metastasis and survival in oral and oesophageal cancer [19]. Survival in CRC has also been shown to be impacted by the presence of F. nucleatum and P. micra [12, 14, 16], further supporting a possible interaction between these bacteria in CRC.
Although this study included a relatively large cohort of patients with stool samples, few samples were found positive for P. gingivalis. Studies using larger cohorts to increase the power of statistical tests are required, especially when studying the relation to subgroups of CRC. In this study, we found that when P. gingivalis was identified in faecal samples, it was often present also in tumour tissue, but the precise location within the tumours remains to be explored. Further studies are also needed to address the cause-and-effect relationship of this bacteria in CRC pathogenesis, including mouse models and/or in vitro experiments to explore the molecular mechanisms behind. Few possible mechanisms for P. gingivalis in CRC progression have been described. Using in vitro experiments and mouse models, Okumura et al., reported an increased production of butyrate - a short chain fatty acid reported to enhance tumourigenesis - by P. gingivalis leading to cellular senescence and the onset of CRC tumours [26]. Wang et al., further found that P. gingivalis could promote CRC by NLRP3 inflammasome activation both in vitro and in vivo [27].
In summary, we found that P. gingivalis was associated with CRC and with a worse patient prognosis. A possible interaction between P. gingivalis with other oral CRC-associated bacteria and tumours of MSI subtype was further implied, but these putative interactions need to be further explored using larger patient cohorts. An increased understanding of the pathogenesis of CRC may lead to the identification of potential screening markers, as well as important improvements in personalised medicine.
Abbreviations
CRC: Colorectal cancer; APC: Adenomatous polyposis coli; KRAS: Kirsten rat sarcoma virus; TP53: Tumour protein 53; SSL: Sessile serrated lesion; BRAF: B-Raf; MLH1: MutL Homolog 1; MSI: Microsatellite instability; MSS: Microsatellite stability; U-CAN: Uppsala-Umeå Comprehensive Cancer Consortium; FECSU: Fecal and Endoscopic Colorectal Study in Umeå; PGT: Prostaglandin transporter.
Acknowledgements
The authors thank all the patients who participated in the study. Thanks also to Åsa Stenberg for technical assistance.
Funding
This study was supported by the Swedish Cancer Society, Lion´s Cancer Research Foundation, the County Council of Västerbotten, the Sjöberg Foundation and the Faculty of Medicine Interdisciplinary and innovative research.
Author contributions
Study concept and design: MK, SE, TL, IL, RP, AL; acquisition of data: MK, TL, ALB, CZ, ÅS; data analyses: MK, SE, TL, ALB; interpretation of data: MK, SE, TL, VB, IL, RP, AL; drafting of the manuscript: MK, SE. Critical revision of the manuscript for important intellectual content: MK, SE, TL, VB, ALB, CZ, IL, RP, AL. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Morgan E, Arnold M. et al. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2022;72:338-344
2. Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut Microbiota in Health and Disease. Physiological Reviews. 2010;90:859-904
3. Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur J Immunol. 2015;45:17-31
4. Saus E, Iraola-Guzman S, Willis JR, Brunet-Vega A, Gabaldon T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol Aspects Med. 2019;69:93-106
5. Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z. et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70
6. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679-89
7. Nistal E, Fernandez-Fernandez N, Vivas S, Olcoz JL. Factors Determining Colorectal Cancer: The Role of the Intestinal Microbiota. Front Oncol. 2015;5:220
8. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575-82
9. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107:11537-42
10. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B. et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859-67
11. Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-64
12. Kim Y, Cho NY, Kang GH. Prognostic and clinicopathological significance of Fusobacterium nucleatum in colorectal cancer: a systemic review and meta-analysis. J Pathol Transl Med. 2022;56:144-51
13. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J. et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-63 e16
14. Lowenmark T, Lofgren-Burstrom A, Zingmark C, Ljuslinder I, Dahlberg M, Edin S. et al. Tumour Colonisation of Parvimonas micra Is Associated with Decreased Survival in Colorectal Cancer Patients. Cancers (Basel). 2022Nov30;14(23):5937
15. Löwenmark T, Li X, Löfgren-Burström A, Zingmark C, Ling A, Kellgren TG. et al. Parvimonas micra is associated with tumour immune profiles in molecular subtypes of colorectal cancer. Cancer Immunology, Immunotherapy. 2022Oct;71(10):2565-2575
16. Zhao L, Zhang X, Zhou Y, Fu K, Lau HC, Chun TW. et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene. 2022;41:4200-10
17. Yamagishi H, Kuroda H, Imai Y, Hiraishi H. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer. 2016;35:4
18. Hajishengallis G, Abe T, Maekawa T, Hajishengallis E, Lambris JD. Role of complement in host-microbe homeostasis of the periodontium. Semin Immunol. 2013;25:65-72
19. Kong J, Yuan X, Wang J, Liu Y, Sun W, Gu B. et al. Frequencies of Porphyromonas gingivalis Detection in Oral-Digestive Tract Tumors. Pathol Oncol Res. 2021;27:628942
20. Glimelius B, Melin B, Enblad G, Alafuzoff I, Beskow A, Ahlstrom H. et al. U-CAN: a prospective longitudinal collection of biomaterials and clinical information from adult cancer patients in Sweden. Acta Oncol. 2018;57:187-94
21. Eklof V, Lofgren-Burstrom A, Zingmark C, Edin S, Larsson P, Karling P. et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141:2528-36
22. Lowenmark T, Lofgren-Burstrom A, Zingmark C, Eklof V, Dahlberg M, Wai SN. et al. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci Rep. 2020;10:15250
23. Pignatelli P, Iezzi L, Pennese M, Raimondi P, Cichella A, Bondi D. et al. The Potential of Colonic Tumor Tissue Fusobacterium nucleatum to Predict Staging and Its Interplay with Oral Abundance in Colon Cancer Patients. Cancers (Basel). 2021Mar1;13(5):1032
24. Uraz A, Karaduman B, Isler SC, Gonen S, Cetiner D. Ozone application as adjunctive therapy in chronic periodontitis: Clinical, microbiological and biochemical aspects. J Dent Sci. 2019;14:27-37
25. Koliarakis I, Messaritakis I, Nikolouzakis TK, Hamilos G, Souglakos J, Tsiaoussis J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int J Mol Sci. 2019Aug25;20(17):4146
26. Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y. et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun. 2021;12:5674
27. Wang X, Jia Y, Wen L, Mu W, Wu X, Liu T. et al. Porphyromonas gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 2021;81:2745-59
28. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590
29. Orth RK, O'Brien-Simpson NM, Dashper SG, Reynolds EC. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011;26:229-40
30. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111:18321-6
31. Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J. et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3:34
32. Abed J, Maalouf N, Manson AL, Earl AM, Parhi L, Emgård JEM. et al. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front Cell Infect Microbiol. 2020;10:400
Author contact
![]() Corresponding author: Agnes Ling, Department of Medical Biosciences, Pathology, Building 6M, Umeå University, SE-90185 Umeå, Sweden. Phone: +46(0)907854487; e-mail: agnes.lingse.
Corresponding author: Agnes Ling, Department of Medical Biosciences, Pathology, Building 6M, Umeå University, SE-90185 Umeå, Sweden. Phone: +46(0)907854487; e-mail: agnes.lingse.

 Global reach, higher impact
Global reach, higher impact