Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(13):2399-2409. doi:10.7150/jca.86974 This issue Cite
Research Paper
Bioinformatics and Expression Analyses of miR-639, miR-641, miR-1915-3p and miR-3613-3p in Colorectal Cancer Pathogenesis
1. Department of Biology, Faculty of Art and Science, Gaziantep University, 27310, Gaziantep, Turkey.
2. Department of General Surgery, Faculty of Medicine, Gaziantep University, 27310, Gaziantep, Turkey.
Received 2023-6-9; Accepted 2023-7-14; Published 2023-7-31
Abstract
Objectives: MicroRNAs (miRNAs) have important function in cancer development and progression. This study aims to determine the expression levels of miR-639, miR-641, miR-1915-3p, and miR-3613-3p in tissues of colorectal cancer (CRC) patients and the role of these miRNAs in the CRC pathogenesis.
Methods: Tumor and non-tumor tissues were collected from a total of 59 CRC patients. qRT-PCR was used to identify the expressions of miR-639, miR-641, miR-1915-3p and miR-3613-3p. Through bioinformatics analysis, the target genes of miRNAs were identified by using DIANA mirPath v.3. Signaling pathways were generated using KEGG pathway database. Biological pathway, cellular component analysis, and analysis of Protein-Protein Interactions (PPI) Networks were performed using FunRich and STRING database.
Results: Our findings revealed that miR-639, miR-641 and miR-3613-3p were significantly downregulated, and miR-1915-3p was significantly upregulated in tumor tissues compared to non-tumor tissues (p˂0.05). Furthermore, MAPK signaling pathway was the most enriched KEGG pathway regulated by miR-639, miR-641, miR-1915-3p and miR-3613-p. According to the FunRich, it was demonstrated that the targeted genes by miRNAs related to the cellular component and biological pathways such as beta-catenin-TCF7L2, axin-APC-beta-catenin-GSK3B complexes, Arf6 signaling, Class I PI3K signaling, etc. And, by the PPI analysis, it was established that the target genes were clustered on CTNNB1 and KRAS.
Conclusions: These outcomes imply that miR-639, miR-641 and miR-3613-3p have tumor suppressor roles, while miR-1915-3p has an oncogenic role in the pathogenesis of CRC. According to the results of the current study, dysregulated miR-639, miR-641, miR-1915-3p, and miR-3613-3p might contribute to the development of CRC.
Keywords: colorectal carcinoma, expression, microRNA, bioinformatics analysis, biomarker
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and is the second leading cause of cancer-related deaths globally. According to the latest data, it is estimated that approximately 153,000 individuals will be diagnosed with CRC and 52,550 cases will die in 2023. Although many different diagnostic and treatment methods have been used so far, its incidence has been rising steadily, and it is not possible to prevent the progression of cancer in these patients [1, 2]. In most cases, CRC is asymptomatic until it progresses to advanced stages. Thus, early diagnosis and improving screening programs would be critically important to reduce the incidence rates and mortality rates of CRC patients. Therefore, new biomarkers are needed to improve the diagnosis and treatment strategies of patients with CRC.
MicroRNAs (miRNAs) that are approximately 17-25 nucleotides in length are involved in the regulation of gene expression. Through binding to the 3' untranslated regions (3' UTR) of target mRNA molecules, miRNAs either degrade mRNA molecules or prevent the translation of these molecules, thus affecting multiple signaling pathways. miRNAs play a crucial role in various cancer-related biological processes such as development, differentiation, proliferation, cell death, and signal transduction [3]. Due to their essential roles in carcinogenesis, miRNAs are classified as oncomirs or tumor suppressor miRNAs. While oncomirs are highly expressed gene regulators that inhibit the translation of tumor suppressor genes, tumor suppressor miRNAs are effective in the translation of mRNAs with oncogenic properties [4]. Recent studies have revealed the relationship between miRNAs and many types of cancer including CRC [5]. In many of the studies carried out so far, it has been reported that many miRNAs were identified as oncogenic miRNAs in CRC such as miR-17, miR-19a and miR-19b, while several other miRNAs, including miR-30a, miR-198, miR-485-3p and miR-4728-5p were found to play a tumor suppressor role in CRC [6-10]. Taking all into consideration, identification of new miRNA biomarkers and revealing the roles of these miRNAs are critical steps in colorectal carcinogenesis.
Numerous studies have shown that miR-639 is localized on 19p13.12 is dysregulated in different cancer types such as liver [11], tongue squamous cell carcinoma [12], ovary [13], breast [14] and bladder [15]. miR-641 is located on 19q13.2 chromosome region and is expressed at different levels in osteosarcoma [16], lung [17], cervical [18] and gastric [19] cancers. A great number of studies have demonstrated that miR-1915-3p, which is located on the 10p12.31 chromosome, is dysregulated in breast [20], lung [21] and thyroid [22] cancers. In addition, miR-3613-3p (on the 13q14.2 chromosome region) was found to be downregulated in breast [23], melanoma [24] and gastric [25] cancers. Motivated by previous studies, we aimed to determine the expression levels of miR-639, miR-641, miR-1915-3p, and miR-3613-3p in tissues with CRC. Furthermore, using bioinformatics analyses, it was purposed to examine the relationship between the genes targeted by dysregulated miRNAs and colorectal carcinoma, and to perform pathway enrichment, biological pathway, and cellular component analyses of these genes.
Materials and methods
Tissue samples
Between January 2017 and February 2020, we collected 59 pairs of fresh tumor and adjacent non-tumor tissue obtained from CRC patients who underwent surgical operation at Gaziantep University Hospital, Turkey. All patients in this study should meet the following criteria: (1) confirmation of CRC by pathological analysis, (2) no previous history of CRC or other malignant tumors, (3) no radiotherapy or chemotherapy, and (4) no other anti-tumor therapy before surgery. Before collection of the tissues, a written informed consent form was obtained from the participants of this study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the local ethics committee of Gaziantep University, Turkey (Ethical approved numbers: 2019/420 and 2019/421). All tissues were stored in RNAlater (Thermo Fisher Scientific, USA) at -80 ˚C.
RNA extraction and cDNA synthesis
Following the instructions provided by the company, total RNA from fresh tissue specimens was extracted using mirVana ™ miRNA Isolation Kit (Invitrogen, USA). NanoDrop spectrometer was used to assess the quality and the quantity of RNA. The isolated RNA was stored at -80 °C. Thereafter, RNA was reverse transcribed into cDNA using miScript II RT Kit (Qiagen, USA) according to the manufacturer's instructions. The obtained cDNA samples were stored at -20 °C.
Quantitative Real-time PCR (qRT-PCR)
Using 7500 Fast Real-Time PCR system (Applied Biosystems) with miScript SYBR Green PCR Kit (Qiagen, USA), we performed qRT-PCR analysis. While U6 was used as an endogenous control, we analyzed the expression levels of miR-639, miR-641, miR-1915-3p and miR-3613-3p. The reactions were completed in triplicate. The sequences of primers are: miR-639: Forward 5'-ATCGCTGCGGTTGCGAGCGCTGT-3', Revers 5'-CGAGGAAGAAGACGGAAGAAT-3'; miR-641: Forward 5'-AAAGACATAGGATAGAGTCACCTC-3', Revers 5'-CGAGGAAGAAGACGGAAGAAT-3'; miR-1915-3p: Forward 5'-CCCCAGGGCGACGCGGCGGG-3', Revers 5'-CGAGGAAGAAGACGGAAGAAT-3', miR-3613-3p: 5'-ACAAAAAAAAAAGCCCAACCCTTC-3', Revers 5'-CGAGGAAGAAGACGGAAGAAT-3'; U6: Forward 5'-GCTTCGGCAGCACATATACTAAAAT-3'; Revers 5'-CGCTTCACGAATTTGCGTGTCAT-3'.
The 2 -ΔΔCt method was used for real-time PCR data analysis [26]. For this method, we firstly determined fluorescence threshold cycle (Ct) of each sample. Then, ΔΔCt (ΔΔCt= ΔCttumor - ΔCtcontrol) values were calculated using ΔCt (ΔCt = Cttarget - CtU6) values obtained from tumor and healthy tissues. Finally, using 2-ΔΔCt values, fold change values were determined. The high expression level was defined as if those with fold change value above 1 while low expression as if those with fold change value below 1 [10].
Pathway and target gene analysis of miRNAs
DIANA-mirPath that can use predicted miRNA targets is a miRNA pathway analysis online tool. The target genes of miRNAs were identified by using DIANA mirPath v.3 with microT-CDS (v5.0) algorithm. A KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis was accomplished on the genes targeted by miRNAs by using DIANA-mirPath v3.0 [27] and DIANA-TarBase v7.0 [28] databases. Signaling pathways and target genes of miRNAs involved in the formation of CRC carcinogenesis were generated using KEGG pathway database [29].
Functional enrichment analysis
Functional enrichment analysis (FunRich) is a software package and this software is used for functional enrichment and interaction network analysis of genes and proteins [30]. FunRich tool was used for biological pathway and cellular component analyses of genes targeted by miRNAs involved in CRC carcinogenesis, obtained using the DIANA database.
Construction and analysis of protein-protein interactions (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING) database is an online tool that generates direct and indirect connections between protein-protein interaction networks. The interactions between genes targeted by miRNAs were demonstrated with PPI network by using STRING database [31]. A minimum required interaction score 0.4 was set as the cut-off criterion.
Statistical analysis
All statistical analyses were performed using SPSS (version 22.0; IBM Corp.). The data was presented as the mean ± standard deviation. A paired t-test was used to test the difference in miRNA expression in tumor and adjacent non-tumor tissues by comparing ΔCttumor and ΔCtcontrol values. Furthermore, the association between miRNA expression levels and clinicopathological parameters was tested by Pearson chi-square (χ2) and Fishers' Exact tests. p <0.05 was considered a statistically significant level.
Results
Characteristics of patients with CRC
In this study, we used the tumors and adjacent healthy tissues from a total of 59 patients with CRC. While 59.3% of these patients were men, 40.7% of the patients were women. 66.1% of the patients were 55 years old and over (the median age: 55.06±11.89). 62.7% of the samples in the study were collected from the colon tissue and the rest from the rectum tissue. Based on patient medical history, 28.8% of the patients used to smoke, while 8.5% consumed alcohol. Lymph node metastasis was observed in 40.7% of all cases, and distant metastasis was detected in 16.9% of the patients. According to the American Joint Committee on Cancer (AJCC) TNM system, 50.8% of patients were detected in Stage I and II, and 49.2% in Stage III and IV. In terms of histological types, 72.9% of the tissues were adenocarcinoma and 27.1% were mucinous adenocarcinoma. The demographic, clinical and pathological data of the patients with CRC are summarized in Table 1.
Demographic, clinical and pathological features of patients with CRC
| Characteristics | Patients (%) |
|---|---|
| Age (years) | |
| ≥55 | 39 (66.1) |
| ˂55 | 20 (33.9) |
| Gender | |
| Male | 35 (59.3) |
| Female | 24 (40.7) |
| Cigarette smoking | |
| Yes | 17 (28.8) |
| No | 42 (71.2) |
| Alcohol drinking | |
| Yes | 5 (8.5) |
| No | 54 (91.5) |
| Tumor location | |
| Colon | 37 (62.7) |
| Rectum | 22 (37.3) |
| Invasion | |
| T1+T2 | 13 (22) |
| T3+T4 | 46 (78) |
| Neural invasion | |
| Yes | 11 (18.6) |
| No | 48 (81.4) |
| Lymphovascular invasion | |
| Yes | 16 (27.1) |
| No | 43 (72.9) |
| Distant metastasis | |
| M0 | 49 (83.1) |
| M1 | 10 (16.9) |
| Lymph node metastasis | |
| N0 | 35 (59.3) |
| N1+N2 | 24 (40.7) |
| Tumor stage | |
| I-II | 30 (50.8) |
| III-IV | 29 (49.2) |
| Tumor size (cm) | |
| ≤6 | 33 (55.9) |
| >6 | 26 (44.1) |
| Tumor histological type | |
| Adenocarcinoma | 43 (72.9) |
| Mucinous adenocarcinoma | 16 (27.1) |
T1: Tumor invades submucosa, T2: Tumor invades muscularis propria, T3: Tumor invades through the muscularis propria into pericolorectal tissues; M0: No distant metastasis, M1: Metastases in more than one organ/site or the peritoneum; N0: No regional lymph node metastasis, N1: 1-3 pericolic lymph involvement N2:2-4 pericolic lymph involvement or perirectal involvement.
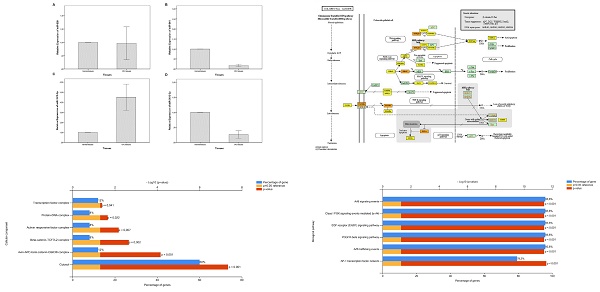
Expression levels of miR-639 (A), miR-641 (B), miR-1915-3p (C) and miR-3613-3p (D) in the tumor and adjacent non-tumor tissues of patients with CRC

Significant differences of expression levels of miR-639, miR-641, miR-1915-3p and miR-3613-3p between tumor and adjacent non-tumor tissues
In the present study, the expression levels of miR-639, miR-641, miR-1915-3p and miR-3613-3p were analyzed in the tumor and adjacent healthy tissues of a total of 59 patients with CRC. When compared to healthy tissues, miR-639 (p=0.000), miR-641 (p=0.000) and miR-3613-3p (p=0.000) (fold change values; 0.20±0.26, 0.17±0.22, and 0.26±0.48, respectively) were significantly downregulated in tumor tissues, whereas miR-1915-3p was significantly upregulated (p=0.032) (fold change value; 4.48±5.01) (Figure 1).
The relationship between expressions of miR-639, miR-1915-3p and miR-3613-3p and clinicopathological features of patients with CRC
Our results showed that the decreased expression of miR-639 was significantly higher compared to the increased miR-639 expression in both colon and rectum tissues of patients (p=0.049). In patients with advanced-stage tumors (Stage III and IV), miR-639 was found to be significantly downregulated (p=0.001). However, we did not observe any significant association between miR-639 expression and other variables (p>0.05). We further did not find a significant relationship between the expressions of both miR-1915-3p and miR-3613-3p and the clinicopathological characteristics of the patients (p>0.05) (Table 2). miR-641 was excluded from these assessments since it was downregulated in all patients.
Bioinformatics analyses
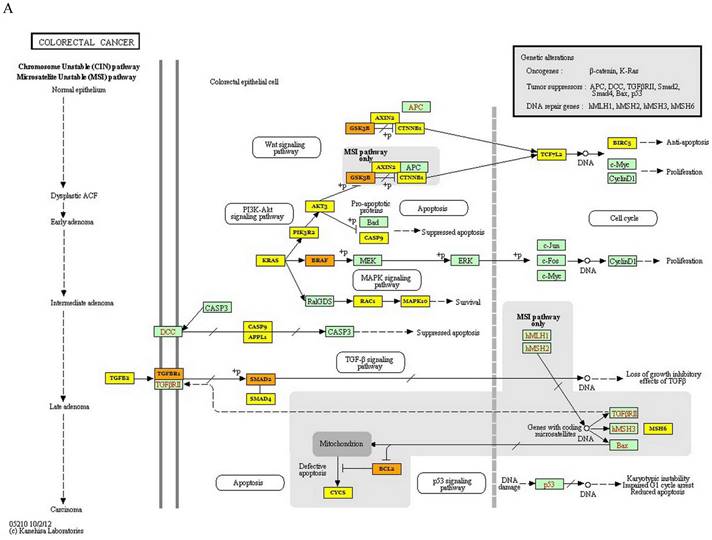
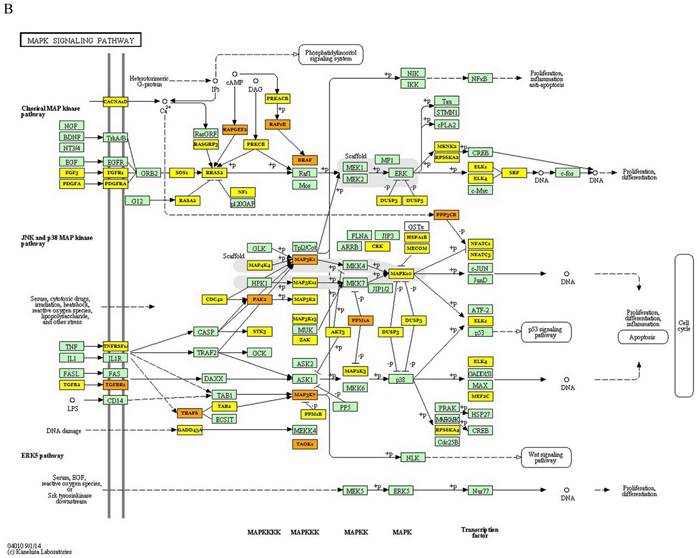
KEGG analysis of 3 miRNAs was performed with DIANA miRpath online tool in this study. The target genes of each miRNA in CRC were determined. As a result of this analysis, it was determined that 24, 7 and 3 genes were targeted by miR-3613-3p, miR-641 and miR-1915-3p, respectively (p values: 4.483447∙10-79, 2.315548∙10-19 and 8.078544∙10-8, respectively). A total of 26 genes were found to be targeted by these 3 miRNAs in CRC (p=0.00808911396543) (Figure 2A). The target gene of miR-639, which is effective in colorectal carcinogenesis, was not found in the DIANA miRpath software. Furthermore, KEGG pathway analysis revealed that MAPK, Wnt, PI3K-Akt, p53, and TGF- signaling pathways were significantly enriched in the pathogenesis of CRC. Additionally, the most enriched KEGG pathway regulated by miR-639, miR-641, miR-1915-3p and miR-3613-p was the MAPK signaling pathway (p-value threshold=0.0022616148417), and TGFB1, BRAF, MAP3K1, MAP3K7, PAK2, TAOK1, PPP3CB, RAPGEF2, TRAF6, CACNA2D1 and RAP1B were among the target genes of the pathway (Figure 2B).
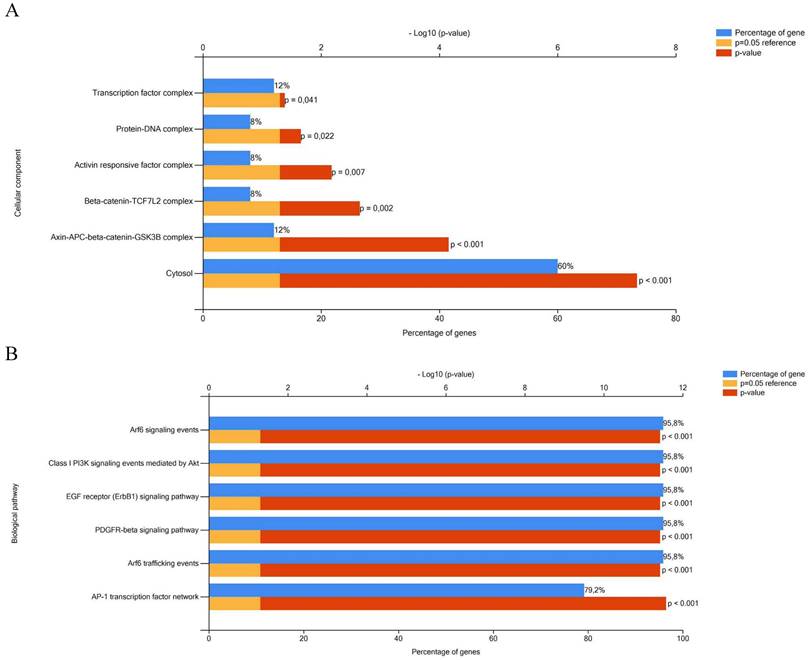
Biological pathway and cellular component analyses of a total of 26 genes targeted by miR-641, miR-1915-3p and miR-3613-3p dysregulated in colorectal carcinogenesis were performed using FunRich enrichment assay. According to the FunRich enrichment analyses, it was seen that these genes related to transcription factor (12%, p=0.041), protein-DNA (8%, p=0.022), activin responsive factor (8%, p=0.007), beta-catenin-TCF7L2 (8%, p=0.022), axin-APC-beta-catenin-GSK3B (12%, p˂0.001) complexes and cytosol (60%, p˂0.001) among the cellular components (Figure 3A). As can be seen in the Figure 3B, these 26 targeted genes were involved in biological pathways such as Arf6 signaling (95.8%, p˂0.001), Class I PI3K signaling (95.8%, p˂0.001), EGF receptor (ErbB1) signaling (95.8%, p˂0.001), PDGFR-beta signaling (95.8%, p˂0.001), Arf6 trafficking (95.8%, p˂0.001) and AP-1 transcription factor network (79.2%, p˂0.001).
The PPI network was comprised of 26 nodes and 136 edges, which were mapped by STRING software (Figure 4). It was observed that the average node degree was 10.5, expected number of edges was 34 and PPI enrichment p-value was < 1.0∙10-16. As a result of the PPI analysis, it was established that these 26 genes were clustered on CTNNB1 and KRAS. This interaction between genes is thought to be effective in the regulation of cell proliferation in CRC.
The association between miRNA expressions and clinicopathological characteristics of CRC patients
| Variables | miR-639 Expression | miR-1915-3p Expression | miR-3613-3p Expression | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | p value | Low | High | p value | Low | High | p value | |
| Age (years) | |||||||||
| ≥55 | 19 | 20 | 0.07 | 13 | 26 | 0.380 | 34 | 5 | 0.653 |
| ˂55 | 17 | 3 | 9 | 11 | 19 | 1 | |||
| Gender | |||||||||
| Male | 12 | 12 | 0.151 | 11 | 13 | 0.261 | 21 | 3 | 0.679 |
| Female | 24 | 11 | 11 | 24 | 32 | 3 | |||
| Cigarette smoking | |||||||||
| Yes | 12 | 5 | 0.338 | 5 | 12 | 0.426 | 15 | 2 | 1.000 |
| No | 24 | 18 | 17 | 25 | 38 | 4 | |||
| Alcohol drinking | |||||||||
| Yes | 4 | 1 | 0.639 | 1 | 4 | 0.641 | 4 | 1 | 0.427 |
| No | 32 | 22 | 21 | 33 | 49 | 5 | |||
| Tumor location | |||||||||
| Colon | 19 | 18 | 0.049* | 12 | 25 | 0.317 | 32 | 5 | 0.396 |
| Rectum | 17 | 5 | 10 | 12 | 21 | 1 | |||
| Invasion | |||||||||
| T1+T2 | 9 | 4 | 0.492 | 5 | 8 | 1.00 | 13 | 0 | 0.322 |
| T3+T4 | 27 | 19 | 17 | 29 | 40 | 6 | |||
| Neural invasion | |||||||||
| Yes | 5 | 6 | 0.310 | 5 | 6 | 0.731 | 9 | 2 | 0.310 |
| No | 31 | 17 | 17 | 31 | 44 | 4 | |||
| Lymphovascular invasion | |||||||||
| Yes | 8 | 8 | 0.290 | 4 | 12 | 0.234 | 14 | 2 | 0.658 |
| No | 28 | 15 | 18 | 25 | 39 | 4 | |||
| Distant metastasis | |||||||||
| M0 | 32 | 17 | 0.166 | 19 | 30 | 0.729 | 44 | 5 | 1.000 |
| M1 | 4 | 6 | 3 | 7 | 9 | 1 | |||
| Lymph node metastasis | |||||||||
| N0 | 23 | 12 | 0.372 | 14 | 21 | 0.603 | 33 | 2 | 0.212 |
| N1+N2 | 13 | 11 | 8 | 16 | 20 | 4 | |||
| Tumor stage | |||||||||
| I-II | 12 | 18 | 0.001* | 12 | 18 | 0.661 | 28 | 2 | 0.424 |
| III-IV | 24 | 5 | 10 | 19 | 25 | 4 | |||
| Tumor size (cm) | |||||||||
| ≤6 | 19 | 14 | 0.541 | 13 | 20 | 0.706 | 30 | 3 | 1.000 |
| >6 | 17 | 9 | 9 | 17 | 23 | 3 | |||
| Tumor histological type | |||||||||
| Adenocarcinoma | 25 | 18 | 0.458 | 14 | 29 | 0.218 | 37 | 6 | 0.176 |
| Mucinous adenocarcinoma | 11 | 5 | 8 | 8 | 16 | 0 | |||
*p <0.05 was considered significant.
Kyoto Encyclopedia of Genes and Genomes pathway of target genes in CRC. A- Signaling pathways and the target genes associated with colorectal carcinogenesis. B- MAPK signaling pathway (the target genes given in the yellow and orange boxes (the genes targeted by 1 and more than 1 miRNAs, respectively) indicate the target genes in the colorectal carcinogenesis and MAPK signaling pathway)
Functional enrichment analysis of the targeted genes by miRNAs in CRC. A- Cellular component analysis. B- Biological pathway analysis
Discussion
CRC is the second leading reason of cancer-related deaths worldwide. Therefore, early diagnosis of the tumor is crucial for the survival period of patients with CRC. In this perspective, the identification of specific and strong molecular biomarkers for the early detection of CRC is highly essential [32]. In recent years, there have been many studies showing that various miRNAs have an oncogene or tumor suppressor role in almost all human malignancies, including CRC. They also have an effect on tumor formation and progression and, therefore, are used as prognostic biomarkers [18-33]. These studies highlight the importance of the dysregulation of miRNAs in the tumor formation process. Therefore, herein, we investigated the expression levels of miR-639, miR-641, miR-1915-3p, and miR-3613-3p in tissues with CRC. Also, in this study, we examined the biological pathways of the target genes of these miRNAs, their effects on colorectal carcinoma, and the interaction network between the proteins encoded by these genes using bioinformatics analysis tools.
In the current study, we found that miR-639 decreased significantly in CRC tissue samples compared to non-tumor tissue samples. In fact, similar results were reported for hepatocellular carcinoma, and miR-639 was shown to act as a tumor suppressor [34]. In a previous study, Xiao et al. (2020) showed that the expression of miR-639 was decreased in liver cancer tissues and this decrease was due to the hypermethylation of the promoter region of miR-639 by DNMT3A. They further reported that miR-639 binds to the 3'-UTR regions of MYST2 and ZEB1 and suppresses the expression of these genes [11]. Wu et al. (2021) demonstrated that expression of miR-639 that targeted FOXC1 was downregulated in ovarian cancer [13]. Moreover, our results reveal a significant relationship between the advanced stages of CRC (stage III and IV) and low miR-639 expression, indicating that miR-639 can be an important biomarker which can be used in the diagnosis and prognosis of CRC. Contrary to our study, miR-639 was previously found to have an oncogenic role in thyroid, bladder and breast cancers [14, 15, 35].
Protein-Protein Interactions networks of the targeted genes by miRNAs in CRC

The present study indicated that the expression of miR-641 was decreased in all tissues with CRC significantly when compared to healthy tissues. Similarly, a previous study reported that expression of miR-641 was downregulated in lung cancer tissues and cell lines and also targets MDM2, which was defined as an oncogene [36]. In another study, miR-641 was also shown to have low expression in cervical cancer and bind directly to the 3'-UTR region of ZEB1, a member of the deltaEF1 family of two-handed zinc-finger factors [18]. Recently, Meng et al. (2020) showed that down-regulation of miR-641 increased cell proliferation in lung adenocarcinoma [17]. Wang et al. (2019) reported that in gastric carcinoma tissues and cells, expression of miR-641 was decreased and miR-641 expression was negatively correlated with lncRNA OPI5-AS1 [19]. Likewise, miR-641 was also downregulated in osteosarcoma tissues and cells [16]. These results also support the findings of the present study in that miR-641 plays a tumor suppressor role in CRC and may be a powerful marker that can be used in the diagnosis of CRC.
Our RT-qPCR results showed that miR-1915-3p was upregulated in tissues with CRC compared to healthy tissues. Similarly, it was shown in a previous study that high expression of miR-1915-3p was associated with infiltrative follicular variants of papillary thyroid cancer and might be a diagnostic factor in thyroid cancer [22]. In another published study, it was indicated that miR‑1915‑3p that targeted DRG2/PBX2, played a role as a silencer of apoptosis in lung cancer, and this miRNA might be a therapeutic target in lung carcinoma [21]. It is also known that miR-1915-3p plays a tumor suppressive role in non-small-cell lung cancer [37]. Guo et al. (2018) showed that miR-1915-3p was upregulated in serum samples of breast cancer patients compared with healthy individuals and that miR-1915-3p might be a diagnostic marker for breast cancer by repression of DUSP3 [20]. These results are compatible with the finding of recent studies that miR-1915-3p, which also plays an oncogenic role, is downregulated in CRC.
miR-3613-3p was among miRNAs that we investigated, and the results herein showed that miR-3613-3p was downregulated in tissues with CRC. In parallel with our outcomes, Yan et al. (2018) reported that the expression of miR-3613-3p was decreased in SW480 and SW620, which are human colon cancer cell lines with different metastatic features [38]. However, miR-3613-3p, defined as a tumor suppressor in gastric cancer, was shown to be downregulated in melanoma cell lines and targeted to CD7 [24, 25]. Chen et al. (2021) indicated that miR-3613-3p plays a tumor suppressor role in the development of breast cancer and miR-3613-3p might be a novel biomarker for breast carcinogenesis [23]. In another study conducted in triple negative breast cancer, it was shown that miR-3613-3p plays a role in suppressing cancer formation [39]. These data also corroborate the conclusion that miR-3613-3p plays a tumor suppressor role and may be a potential biomarker for CRC.
KEGG that contains information of gene regulatory pathways, is an important data source for the modelling of biological systems [29]. In the enriched KEGG pathway analysis, we determined that the signaling pathway in which the target genes of the 3 miRNAs affect together was MAPK. The MAPK is a significant signaling pathway that regulates various biological processes associated with tumor proliferation, development, differentiation, migration and apoptosis, and this signaling pathway plays an important role in CRC carcinogenesis [40]. In the present study, it was determined that BRAF, TAOK1, TRAF6 and PAK2, which were involved in the MAPK signaling pathway, were among the genes targeted by downregulated miR-641 and miR-6313-3p (Figure 2B). Previous studies showed that BRAF was effective in the formation of CRC and played an oncogenic role in this carcinogenesis [41, 42]. Capra et al. (2006) demonstrated that increased expression of TAOK1 was associated with CRC [43]. In the other studies, TRAF6 and PAK2 were upregulated in colon cancer [44, 45].
Furthermore, we concluded that Arf6 signaling, Class I PI3K signaling, EGF receptor (ErbB1) signaling, PDGFR-beta signaling, Arf6 trafficking and AP-1 transcription factor pathways were associated with dysregulated miRNAs. These pathways have an important role in oncogenesis and cancer progression, such as tumor growth, differentiation, invasion, migration, metastasis, apoptosis and poor survival [46-50].
In this study, as a result of PPI analysis, we determined that 26 genes targeted by miRNAs were clustered on CTNNB1 and KRAS. CTNNB1, a transcriptional factor, was upregulated in colorectal cancer tissues [51]. KRAS, known as the oncogene, is regulated by tyrosine kinase receptors and the KRAS protein is an essential part of the MAPK signaling pathway. Previous studies demonstrated that KRAS mutations led to uncontrollable cell growth, cell differentiation, metastasis and angiogenesis in CRC [52]. As we discussed above, although some of the previous studies investigating the relationships of miR-639, miR-641, miR-1915-3p, and miR-3613-3p with different types of cancer have resulted in similar outcomes to our study, there have been also other studies that obtain contrary outcomes. These contradictory results may be mainly related to the heterogeneity of the cancer. However, it is important to highlight that miRNAs target many different mRNA molecules in the genome and some of these target molecules act as oncogenes while some as tumor suppressors. Therefore, it is not a surprising outcome that the same miRNAs play different roles in different cancer types depending on the mRNA molecules they target.
The present study has several limitations. First, we did not perform an experimental study to identify genes targeted by miRNAs in this study. Therefore, it may focus on identification of the target genes of miR-639, miR-641, miR-1915-3p and miR-3613-3p that are effective in CRC in future studies. Second, the sample size of this study may be limited to clearly demonstrating the relationship between clinicopathological characteristics of patients and expression levels of miRNAs. Third, the effects of miRNAs on CRC cell proliferation, invasion, metastasis, and apoptosis were not investigated in vitro. Although the current research has several limitations, the results of this study will guide future studies to reveal the genetic mechanisms that are effective in determining the relationship between colorectal carcinogenesis and miRNAs.
Conclusions
As a result, our findings suggest that miR-639, miR-641, and miR-3613-3p play tumor suppressor roles in CRC, whereas miR-1915-3p plays an oncogenic role. These findings indicate that miR-639, miR-641, miR-1915-3p, and miR-3613-3p can be used as new candidate biomarkers for CRC diagnosis. However, a functional evaluation of the dysregulation of these miRNAs remains to be elucidated in future studies.
Abbreviations
miRNA: microRNA; CRC: colorectal cancer; qRT-PCR: quantitative real time PCR; KEGG: kyoto encyclopedia of genes and genomes; PPI: protein-protein interactions; FunRich: functional enrichment analysis; 3'UTR: 3' untranslated region; Ct: threshold cycle; STRING: search tool for the retrieval of interacting genes.
Acknowledgements
This study was supported by Gaziantep University, Scientific Research Projects Governing Unit (Project Numbers: FEF.DT.19.29 and FEF.ALT.19.28).
Author contributions
All authors contributed to the design of the present study. TG supervised the research, designed and performed experiments, analyzed data, discussed the results, performed revision for important intellectual content and commented on the manuscript. RA performed experiments. AA operated on patients, collected the samples and evaluated clinical findings. All authors have read and approved the final manuscript.
Informed consent
Informed consent was obtained from all individuals included in this study.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the local ethics committee of Gaziantep University, Turkey (approval numbers: 2019/420 and 2019/421).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA: A Cancer Journal for Clinicians. 2023;73:233-54
2. Lv Y, Duanmu J, Fu X, Li T, Jiang Q. Identifying a new microRNA signature as a prognostic biomarker in colon cancer. PLoS One. 2020;15:e0228575
3. Hernández R, Sánchez-Jiménez E, Melguizo C, Prados J, Rama AR. Downregulated microRNAs in the colorectal cancer: diagnostic and therapeutic perspectives. BMB Rep. 2018;51:563-71
4. Smolarz B, Durczyński A, Romanowicz H, Szyłło K, Hogendorf P. miRNAs in Cancer (Review of Literature). Int J Mol Sci. 2022;23:2805
5. Asadi M, Talesh ST, Gjerstorff MF, Shanehbandi D, Baradaran B, Hashemzadeh S. et al. Identification of miRNAs correlating with stage and progression of colorectal cancer. Colorectal Cancer. 2019;8:CRC06
6. Fang L, Li H, Wang L, Hu J, Jin T, Wang J. et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974-87
7. Huang L, Cai JL, Huang PZ, Kang L, Huang MJ, Wang L. et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7:1996-2008
8. Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin D. et al. MicroRNA-30a Mediates Cell Migration and Invasion by Targeting Metadherin in Colorectal Cancer. Technol Cancer Res Treat. 2018;17:1533033818758108
9. Li LX, Lam IH, Liang FF, Yi SP, Ye LF, Wang JT. et al. MiR-198 affects the proliferation and apoptosis of colorectal cancer through regulation of ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:1487-93
10. Gurer T, Aytekin A, Caki E, Gezici S. miR-485-3p and miR-4728-5p as Tumor Suppressors in Pathogenesis of Colorectal Cancer. Molecular Biology. 2022;56:474-88
11. Xiao J, Liu Y, Wu F, Liu R, Xie Y, Yang Q. et al. miR-639 Expression Is Silenced by DNMT3A-Mediated Hypermethylation and Functions as a Tumor Suppressor in Liver Cancer Cells. Mol Ther. 2020;28:587-98
12. Lin Z, Sun L, Chen W, Liu B, Wang Y, Fan S. et al. miR-639 regulates transforming growth factor beta-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting FOXC1. Cancer Sci. 2014;105:1288-98
13. Wu S-G, Zhou P, Chen J-X, Lei J, Hua L, Dong Y. et al. circ-PTK2 (hsa_circ_0008305) regulates the pathogenic processes of ovarian cancer via miR-639 and FOXC1 regulatory cascade. Cancer Cell International. 2021;21:277
14. Ismail A, Abulsoud AI, Mansour OA, Fawzy A. Diagnostic Significance of miR-639 and miR-10b in Βreast Cancer Patients. Meta Gene. 2019;19:155-9
15. Scheffer AR, Holdenrieder S, Kristiansen G, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs in serum: novel biomarkers for patients with bladder cancer? World J Urol. 2014;32:353-8
16. Wu H, Li W, Zhu S, Zhang D, Zhang M. Circular RNA circUBAP2 regulates proliferation and invasion of osteosarcoma cells through miR-641/YAP1 axis. Cancer Cell Int. 2020;20:223
17. Meng F, Zhou Y, Dong B, Dong A, Zhang J. Long non-coding RNA LINC01194 promotes the proliferation, migration and invasion of lung adenocarcinoma cells by targeting miR-641/SETD7 axis. Cancer Cell International. 2020;20:588
18. Yao R, Zheng H, Wu L, Cai P. miRNA-641 inhibits the proliferation, migration, and invasion and induces apoptosis of cervical cancer cells by directly targeting ZEB1. Onco Targets Ther. 2018;11:8965-76
19. Wang LW, Li XB, Liu Z, Zhao LH, Wang Y, Yue L. Long non-coding RNA OIP5-AS1 promotes proliferation of gastric cancer cells by targeting miR-641. Eur Rev Med Pharmacol Sci. 2019;23:10776-84
20. Guo J, Liu C, Wang W, Liu Y, He H, Chen C. et al. Identification of serum miR-1915-3p and miR-455-3p as biomarkers for breast cancer. PLoS One. 2018;13:e0200716
21. Xu C, Li H, Zhang L, Jia T, Duan L, Lu C. MicroRNA-1915-3p prevents the apoptosis of lung cancer cells by downregulating DRG2 and PBX2. Mol Med Rep. 2016;13:505-12
22. Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P. et al. miRNA expression profiling of 'noninvasive follicular thyroid neoplasms with papillary-like nuclear features' compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol. 2017;30:39-51
23. Chen C, Pan Y, Bai L, Chen H, Duan Z, Si Q. et al. MicroRNA-3613-3p functions as a tumor suppressor and represents a novel therapeutic target in breast cancer. Breast Cancer Research. 2021;23:12
24. Gad SA, Ali HEA, Gaballa R, Abdelsalam RM, Zerfaoui M, Ali HI. et al. Targeting CDC7 sensitizes resistance melanoma cells to BRAF(V600E)-specific inhibitor by blocking the CDC7/MCM2-7 pathway. Sci Rep. 2019;9:14197
25. Bibi F, Naseer MI, Alvi SA, Yasir M, Jiman-Fatani AA, Sawan A. et al. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genomics. 2016;17:751
26. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402-8
27. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T. et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460-6
28. Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I. et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153-9
29. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353-d61
30. Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA. et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-601
31. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2018;47:D607-D13
32. Ghareib AF, Mohamed RH, Abd El-Fatah AR, Saadawy SF. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J Gastrointest Cancer. 2020;51:818-23
33. Wang W, He Y, Rui J, Xu MQ. miR-410 acts as an oncogene in colorectal cancer cells by targeting dickkopf-related protein 1 via the Wnt/β-catenin signaling pathway. Oncol Lett. 2019;17:807-14
34. Bai Z, Xia X, Lu J. MicroRNA-639 is Down-Regulated in Hepatocellular Carcinoma Tumor Tissue and Inhibits Proliferation and Migration of Human Hepatocellular Carcinoma Cells Through the KAT7/Wnt/β-Catenin Pathway. Med Sci Monit. 2020;26:e919241
35. Lei ST, Shen F, Chen JW, Feng JH, Cai WS, Shen L. et al. MiR-639 promoted cell proliferation and cell cycle in human thyroid cancer by suppressing CDKN1A expression. Biomed Pharmacother. 2016;84:1834-40
36. Kong Q, Shu N, Li J, Xu N. miR-641 Functions as a Tumor Suppressor by Targeting MDM2 in Human Lung Cancer. Oncol Res. 2018;26:735-41
37. Pan H, Pan Z, Guo F, Meng F, Zu L, Fan Y. et al. MicroRNA-1915-3p inhibits cell migration and invasion by targeting SET in non-small-cell lung cancer. BMC Cancer. 2021;21:1218
38. Yan W, Yang W, Liu Z, Wu G. Characterization of microRNA expression in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). Onco Targets Ther. 2018;11:4701-9
39. Yu Y, Liao H, Xie R, Zhang Y, Zheng R, Chen J. et al. Overexpression of miRNA-3613-3p Enhances the Sensitivity of Triple Negative Breast Cancer to CDK4/6 Inhibitor Palbociclib. Front Oncol. 2020;10:590813
40. Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH. et al. Signaling pathways involved in colorectal cancer progression. Cell & Bioscience. 2019;9:97
41. Sanz-Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol. 2017;28:2648-57
42. Taieb J, Lapeyre-Prost A, Laurent Puig P, Zaanan A. Exploring the best treatment options for BRAF-mutant metastatic colon cancer. Br J Cancer. 2019;121:434-42
43. Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M. et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147-54
44. Sun H, Li X, Fan L, Wu G, Li M, Fang J. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol. 2014;53:195-201
45. Koo KH, Kwon H. MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3. Cell Death & Disease. 2018;9:77
46. Krasinskas AM. EGFR Signaling in Colorectal Carcinoma. Patholog Res Int. 2011;2011:932932
47. Papadatos-Pastos D, Rabbie R, Ross P, Sarker D. The role of the PI3K pathway in colorectal cancer. Crit Rev Oncol Hematol. 2015;94:18-30
48. Manzat Saplacan RM, Balacescu L, Gherman C, Chira RI, Craiu A, Mircea PA. et al. The Role of PDGFs and PDGFRs in Colorectal Cancer. Mediators Inflamm. 2017;2017:4708076
49. Grossmann AH, Zhao H, Jenkins N, Zhu W, Richards JR, Yoo JH. et al. The small GTPase ARF6 regulates protein trafficking to control cellular function during development and in disease. Small GTPases. 2019;10:1-12
50. Wu Z, Nicoll M, Ingham RJ. AP-1 family transcription factors: a diverse family of proteins that regulate varied cellular activities in classical hodgkin lymphoma and ALK+ ALCL. Exp Hematol Oncol. 2021;10:4
51. Ghatak S, Mehrabi SF, Mehdawi LM, Satapathy SR, Sjölander A. Identification of a Novel Five-Gene Signature as a Prognostic and Diagnostic Biomarker in Colorectal Cancers. International Journal of Molecular Sciences. 2022;23:793
52. Marco C, Samantha E, Maria Celeste P, Milo F, Sara De D. Research progress on KRAS mutations in colorectal cancer. Research progress on KRAS mutations in colorectal cancer. 2021;7:26
Author contact
![]() Corresponding author: Adress: Department of Biology, Faculty of Art and Science, Gaziantep University, 27310, Gaziantep, Turkey. Phone: +90 532 505 13 92; E-mail: taytekinedu.tr; turkanaytecom.
Corresponding author: Adress: Department of Biology, Faculty of Art and Science, Gaziantep University, 27310, Gaziantep, Turkey. Phone: +90 532 505 13 92; E-mail: taytekinedu.tr; turkanaytecom.

 Global reach, higher impact
Global reach, higher impact