Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(13):2529-2537. doi:10.7150/jca.86678 This issue Cite
Research Paper
Impact of LINC00673 genetic variants on uterine cervical cancer clinicopathologic characteristics
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
2. Department of Obstetrics and Gynecology, Chi-Mei Foundation Medical Center, Tainan, Taiwan
3. School of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
4. Department of Medicine, College of Medicine, National Taiwan University, Taipei, Taiwan
5. Department of Obstetrics and Gynecology, Chiayi Chang Gung Memorial Hospital Chiayi, Taiwan
6. Department of Nursing, Chang Gung University of Science and Technology, Chiayi Campus, Chiayi, Taiwan
7. School of Medicine, Chung Shan Medical University, Taichung, Taiwan
8. Department of Obstetrics and Gynecology, Changhua Christian Hospital, Changhua, Taiwan
9. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
10. Department of Obstetrics and Gynecology, Chung Shan Medical University Hospital, Taichung, Taiwan
#Equal contribution as first authors
Received 2023-5-31; Accepted 2023-8-6; Published 2023-8-15
Abstract
To date, no study delineates the relationships among the genetic variants of long intergenic noncoding RNA 673 (LINC00673) and uterine cervical carcinogenesis as well as clinicopathological parameters and 5 years survival of cervical cancer patients in Taiwan. Therefore, the involvement of LINC00673 polymorphisms in cervical cancer was investigated. Genotypic frequencies of three LINC00673 polymorphisms rs6501551, rs9914618 and rs11655237 were determined in 199 patients including 115 patients with invasive cancer, 84 with precancerous lesions, and 274 control females using real-time polymerase chain reaction. It revealed that LINC00673 polymorphisms were not found significantly related to development of cervical cancer. Cervical cancer patients with genotypes AG/GG in LINC00673 rs6501551 had more risk to have tumor diameter larger than 4 cm as compared to those with genotype AA (p=0.043). Cervical cancer patients with genotype GG in rs6501551 had worse 5 years survival as compared to those with genotypes AA/AG in multivariate analysis (hazard ratio: 4.70; p=0.097). However, only two patients exhibiting GG were noted, and one had mortality, another had no mortality. In conclusion, larger sample size needs to verify the associations of LINC00673 genetic variants with clinicopathological parameters and patient survival of cervical cancer for Taiwanese females.
Keywords: long intergenic noncoding RNA 673, genetic variants, cervical carcinogenesis, clincopathological parameters, 5 years survival
Introduction
If the transcribed RNA molecules are longer than 200 nucleotides and have restricted or no capability to encode amino acid sequences, they can be defined as long intergenic noncoding RNAs (lncRNAs) [1]. However, there are accumulative evidences supporting that lncRNAs can affect gene presentation via many patterns, such as assembly of chromatin modifying complex, micro RNA sponges, acceleration or repression of gene presentation, increase or inhibition of DNA methylation, implication in cell proliferation, migration, invasiveness, and apoptosis, and thus are related to a variety of biological functions [2-6]. Because they can exhibit oncogene or tumor suppressor roles, they are particularly associated with cancer development [7-10]. The lncRNA Hox transcript antisense intergenic RNA (HOTAIR) has been investigated for oncogenic function [7, 8, 11]; and long noncoding RNAs growth arrest-specific transcript 5 (GAS5) investigated for tumor suppressor function of lncRNAs as well [9, 10, 12].
Long intergenic noncoding RNA 673 (LINC00673) has been found locating on the chromosome17q25.1 [13, 14]. LINC00673 overexpression has been shown in non-small cell lung cancer and displays its oncogenic capacity by increasing cell growth [14]. In contrast, LINC00673 exhibits a tumor suppressive function in pancreatic cancer. Arnes et al. demonstrated that pancreatic cancer cells presented more efficiency in inducing metastatic lesions if LINC00673 expression was reduced [15]. It is a lncRNA among the most epithelial-enriched pancreatic ductal adenocarcinoma related (PDA) lncRNAs. LINC00673 is situated at in a recurrent, focally amplified area in PDA and is associated with a PDA-related single nucleotide polymorphism (SNP).
When there is a different allele present in the shared DNA sequence of a gene between the members of a species or paired chromosomes in a frequency more than 5% of certain population, SNP occurs [16]. When the genetic variants exert a variation on the promoter region, exon or 3'-untranslated district of a gene, they may have an impact on the gene presentation, or alter the encoded amino acids, and thereafter yield the susceptibility to various diseases or cancers [16-18]. A number of SNPs within lncRNA genes have been showed to exert impacts on the expression and sequence of lncRNAs, which are defined as regulatory RNAs without protein-coding capacity, and then influence the development of individual cancer and patient survival [19, 20]. For example, LINC00673 genetic variants were reported to be associated with the risk of numerous cancers including gastric cancer [21], and liver cancer [22]. Moreover, in a meta-analysis study, Zhang et al. concluded that LINC00673 rs11655237 contributed to occurrence of cancer [23]. In the neuroblastoma, the previous studies reported that the LINC00673 rs11655237 polymorphism might be associated with neuroblastoma development and susceptibility [24, 25].
Uterine cervical cancer has been reported to rank the eighth most common incidence and the eighth common mortality among female malignancy on the basis of the data from Health Promotion Administration of the Ministry of Health and Welfare as well as Annual Cancer Registry Report in Taiwan in 2016. However, until now, no research reports the relationships among LINC00673 genetic polymorphisms, and development of cervical cancer as well as clinicopathological parameters and survival of cervical cancer patients in Taiwan. Therefore, the purposes of this study were to investigate the implications of LINC00673 genetic polymorphism in cervical carcinogenesis, progression and 5 years survival of cervical cancer patients.
Materials and Methods
Enrolled female subjects
The retrospective research was conducted by investigating the associations among LINC00673 genetic variants and the occurrence of uterine cervical cancer as well as clinicopathological parameters and 5 years survival of cervical cancer patients. A total of four hundreds and seventy-three Taiwanese females including 199 subjects suffering from uterine cervical neoplasia (115 patients having invasive cancer and 84 having precancerous lesions) and 274 normal control females were enrolled. These patients underwent standard treatment protocols at the Department of Obstetrics and Gynecology in Chung Shan Medical University Hospital in Taichung, Taiwan since February 1994 until February 2015. Two hundred and seventy-four normal females, who participated in general examination at the Outpatient Patient Department and without history of cancer of any sites, were enrolled as the control group of our study in these time. Normal cytological results were diagnosed for them and further confirmed according to the detailed colposcopic results. Chung Shan Medical University Hospital institutional review board approved the research (CSMUH No: CS18208).
Deoxyribonucleic acid (DNA) extraction of blood samples from all subjects for the distributions of LINC00673 genetic polymorphisms
The laboratory technicians obtained the blood samples from all participants via venipuncture. Thereafter, these samples were mixed with ethylenediaminetetraacetic acid, which were previously set in Vacutainer tubes. After completing the above step, the blood specimens were immediately stored at 4℃. Then, the technicians extracted DNA from leukocytes in conforming to the description in previous publication and subsequently dissolved these extracts into pH 7.8 TE buffer [26]. The DNA quality was defined after the measurement of OD260. The OD260/OD280 ratio was determined and the range of 1.8-2.0 accorded with our criteria, and was regarded as pure to prevent its cross reactivity form the existed homologous RNA in the specimens. We ultimately stored the products at -20°C and used them as the templates for the polymerase chain reaction (PCR).
Selection of three LINC00673 genetic variants and genotyping
The selections of three LINC00673 genetic variants, rs6501551, rs9914618 and rs11655237 were defined based on the data of International HapMap Project and previous research [27]. SNP rs11655237, together with rs6501551 and rs9914618 situated at LINC00673 with RegulomeDB score < 3 was defined as the tagSNPs from SNPinfo [27, 28]. The LINC00673 rs9914618 was selected because the LINC00673 rs9914618 polymorphism was associated with progression of oral cancer [29] and hepatocellular carcinoma [30]. The methods of gene polymorphisms determination have been described in using ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and assessing the data with SDS vers. 3.0 software previously [31].
Power and sample size calculations
Based on the hypothesis and our previous study, the frequencies of at least one mutated allele of LINC00673 rs6501551, rs9914618 and rs11655237 were 25.5%, 35.5% and 35.3%, respectively [29]. Assuming a 95% confidence interval, a p value of 0.05, a ratio of cases to healthy controls of 1:1, and at least 90% power to detect a 1.5-fold risk in gene variances of LINC00673, the sample sizes were about 200 case samples for gene polymorphisms of LINC00673.
Statistical analysis
Analysis of variance (ANOVA) with Welch test was performed to assess the age differences among patients with cervical invasive cancer and precancerous lesions, and control females, and then the Games-Howell test was done for post hoc analysis. Hardy-Weinberg equilibrium was performed to assess the genotypic frequencies of rs6501551, rs9914618 and rs11655237 in control females [degree of freedom (d.f.) = 2].
The associations of cervical carcinogenesis with three LINC00673 genetic polymorphisms were assessed by chi-square or Fisher exact tests. Logistic or multinomial logistic regression models were performed to calculate the adjusted odds ratios (AORs) and their 95% confidence intervals (95% CIs) after age adjustment in comparing these associations. Chi-square or Fisher exact tests were also applied to relate the frequencies of three LINC00673 SNPs rs6501551, rs9914618 and rs11655237 with several clinicopathological parameters including clinical stage, pathologic type, cell grading, cervical stromal invasion depth, tumor size, as well as parametrium invasion and vagina invasion, and pelvic lymph node metastasis.
The impacts of LINC00673 genetic polymorphisms and clinicopathological variables on 5 years survival of patients with invasive cervical cancer were checked using Kaplan-Meier model plotting in univariate analysis. The log-rank test was applied to define the statistical significance among them. The influences of LINC00673 genetic variants and above mentioned clinicopathological parameters on 5 years survival of these patients were assessed using Cox proportional hazard model in multivariate analysis in relation to 5 years survival intervals. The SPSS, version 25.0 and WinPepi Software, version 10.0 was applied for checking statistical significance. Hazard ratios (HRs) and their 95% confidence intervals (CIs) were also defined by the SPSS, version 25.0. P < 0.05 was determined to exert a significant difference.
Results
There was significant difference for the age distribution between patients with cervical neoplasm and control females (51.2 ± 13.8 vs. 43.5 ± 10.1, p < 0.001). There was a significant difference among patients with invasive cancer and precancerous lesions of uterine cervix as well as control women based on the Welch test (p < 0.001). Using Games-Howell test as post hoc analysis, the age differences were significant between patients with cervical cancer and patients with precancerous lesions (56.1 ± 12.5 vs. 44.4 ± 12.6, p < 0.001) as well as between cervical cancer patients and control women (56.1 ± 12.5 vs. 43.5 ± 10.1, p < 0.001). But no significant difference was noted for the age distribution between patients with precancerous lesions and control females (44.4 ± 12.6 vs. 43.5 ± 10.1, p = 0.805).
There was no statistical difference in the distribution of LINC00673 genetic variant rs6501551 for genotypes A/A A/G and G/G between patients with cervical neoplasias and control women (p = 0.398). The frequencies of other LINC00673 SNPs, rs9914618 and rs11655237 exhibited no statistical significance between them (p = 0.915 and 0.573, respectively; Table 1). Although age was adjusted, it still did not reveal significant difference of genotypic distributions of LINC00673 SNPs rs6501551, rs9914618 and rs11655237 between patients with cervical neoplasias and control females (p=0.425, 0.829, and 0.543, respectively; Table 1).
Thereafter, patients with cervical neoplasias were reclassified into two subgroups i.e. patients with invasive cervical cancer and those with precancerous lesions, to investigate the relationships among the development of cervical cancer and LINC00673 SNPs. However, it still revealed no significant association among the genotypic distributions of A/A, A/G and G/G in LINC00673 rs6501551 and patients with cervical invasive cancer, and those with precancerous lesion as well as control females (p = 0.560; Table 2). Moreover, no significant associations were also found among development of cervical cancer and LINC00673 rs9914618 and rs11655237 (p = 0.949 and p = 0.584, respectively; Table 2). Even after age adjustment, there were no relationships among the risks of cervical precancerous lesions and invasive cancer and genotypic distributions of these LINC00673 genetic polymorphisms (Table 2).
The relationships among LINC00673 genetic polymorphisms and clinicopathological factors of cervical cancer were further assessed. It showed that cervical cancer patients with genotypes AG/GG in LINC00673 rs6501551 had more risk to have tumor diameter larger than 4 cm as compared to those with AA (p = 0.043; Table 3). Furthermore, cervical cancer patients with allele T in rs11655237 had more risk to have adenocarcinoma histologic type as compared to those with only allele C (p = 0.049; Table 3). However, above associations only reached marginally statistical significances.
Upon univariate analysis, distributions of LINC00673 SNPs rs6501551, rs9914618 and rs11655237 were not found to be correlated with the 5 years survival of cervical cancer patients [p = 0.635, HR = 1.28 (95% CI = 0.46-3.60), AG/GG vs. AA and p = 0.097, HR = 4.70 (95% CI = 0.62-35.42), GG vs. AA/AG in rs6501551; p = 0.970, HR = 0.98 (95% CI = 0.37-2.62), GA/AA vs. GG and p = 0.415, HR = 2.26 (95% CI = 0.30-16.99), AA vs. GG/GA in rs9914618; p = 0.996, HR = 1.00 (95% CI = 0.36-2.80) CT/TT vs. CC and p = 0.418, HR = unavailable (u.a.), TT vs. CC/CT in rs11655237; Table 4]. Whereas, HRs with worse 5 years survival could be found in cervical patients with clinical stage ≥ II (p = 0.011, HR = 3.32, 95% CI = 1.25-8.86), stromal invasion > 10 mm (p = 0.011, HR = 3.83, 95% CI = 1.25-11.77), tumor diameter > 4cm (p = 0.008, HR: 3.69, 95% CI: 1.32-10.37), positive parametrium invasion (p = 0.032, HR = 2.70, 95% CI = 1.05-6.98) and positive lymph node metastasis (p < 0.001, HR = 8.95, 95% CI = 3.19-25.16; Table 4).
Genetic variant frequencies of long intergenic noncoding RNA 673 in Taiwanese females with cervical neoplasias and normal controls
| Genetic variants | Normal controls (n = 274) | Cervical neoplasiasa (n= 199 ) | ORs (95% CIs) | p values | AORs (95% CIs)b | Adjusted p valuesb |
|---|---|---|---|---|---|---|
| rs6501551 | ||||||
| A/Ac | 208 | 148 | 1.00 | 0.398 | 1.00 | 0.425 |
| A/G | 56 | 47 | 1.18 (0.76-1.83) | 0.463 | 1.28 (0.80-2.04) | 0.305 |
| G/G | 2 | 4 | 2.81 (0.51-15.55) | 0.236 | 2.17 (0.38-12.32) | 0.384 |
| A/Ac | 208 | 148 | 1.00 | 1.00 | ||
| A/G & G/G | 58 | 51 | 1.24 (0.80-1.90) | 0.336 | 1.32 (0.83-2.08) | 0.238 |
| A/A & A/Gc | 264 | 195 | 1.00 | 1.00 | ||
| G/G | 2 | 4 | 2.71 (0.49-14.93) | 0.253 | 2.05 (0.36-11.64) | 0.417 |
| HWE values | χ2=0.719 | |||||
| rs9914618 | ||||||
| G/Gc | 164 | 126 | 1.00 | 0.915 | 1.00 | 0.829 |
| G/A | 92 | 65 | 0.92 (0.62-1.36) | 0.676 | 0.88 (0.58-1.33) | 0.544 |
| A/A | 11 | 8 | 0.95 (0.37-2.42) | 0.909 | 1.00 (0.37-2.69) | 0.998 |
| G/Gc | 164 | 126 | 1.00 | 1.00 | ||
| G/A & A/A | 103 | 73 | 0.92 (0.63-1.35) | 0.677 | 0.89 (0.60-1.33) | 0.576 |
| G/G & G/Ac | 256 | 191 | 1.00 | 1.00 | ||
| A/A | 11 | 8 | 0.98 (0.39-2.47) | 0.957 | 1.05 (0.40-2.78) | 0.926 |
| HWE values | χ2=0.181 | |||||
| rs11655237 | ||||||
| C/Cc | 170 | 136 | 1.00 | 0.573 | 1.00 | 0.543 |
| C/T | 88 | 57 | 0.81 (0.54-1.21) | 0.304 | 0.81 (0.53-1.24) | 0.334 |
| T/T | 7 | 6 | 1.07 (0.35-3.26) | 0.903 | 1.29 (0.40-4.20) | 0.670 |
| C/Cc | 170 | 136 | 1.00 | 1.00 | ||
| C/T & T/T | 95 | 63 | 0.83 (0.56-1.23) | 0.346 | 0.84 (0.56-1.27) | 0.416 |
| C/C & C/Tc | 258 | 193 | 1.00 | 1.00 | ||
| T/T | 7 | 6 | 1.15 (0.38-3.46) | 0.809 | 1.38 (0.43-4.45) | 0.589 |
| HWE values | χ2=1.238 |
Statistical analysis: logistic regression model or chi-square or Fisher's tests. aCervical neoplasias consist of precancerous lesions and invasive cancer of the uterine cervix. bThe adjusted p values as well as adjusted odds ratios (AORs) and their 95% confident intervals (95% CIs) were calculated by logistic regression model after age adjustment. cUsed as a reference for comparison to assess the odds ratios of other genotypes.
Genetic variant frequencies of long intergenic noncoding RNA 673 in Taiwanese females with uterine cervical invasive cancer or precancerous lesion and normal controls
| Genetic variants | Normal controls (n =274 ) | Precancerous lesions (n =84 ) | Invasive cancer (n =115) | p values | AORs (95% CIs)a | Ad. p values | AORs (95% CIs)b | Ad. p values |
|---|---|---|---|---|---|---|---|---|
| rs6501551 | ||||||||
| A/Ac | 208 | 61 | 87 | 0.560 | 1.00 | 1.00 | ||
| A/G | 56 | 21 | 26 | 1.29 (0.73-2.31) | 0.382 | 1.27 (0.70-2.30) | 0.434 | |
| G/G | 2 | 2 | 2 | 3.17 (0.43-23.09) | 0.256 | 1.76 (0.23-13.79) | 0.589 | |
| A/Ac | 208 | 61 | 87 | 0.555 | 1.00 | 1.00 | ||
| A/G & G/G | 58 | 23 | 28 | 1.36 (0.78-2.39) | 0.279 | 1.29 (0.72-2.30) | 0.395 | |
| A/A & A/Gc | 264 | 82 | 113 | 0.383 | 1.00 | 1.00 | ||
| G/G | 2 | 2 | 2 | 2.99 (0.41-21.71) | 0.279 | 1.68 (0.22-13.04) | 0.622 | |
| rs9914618 | ||||||||
| G/Gc | 164 | 51 | 75 | 0.949 | 1.00 | 1.00 | ||
| G/A | 92 | 29 | 36 | 1.00 (0.60-1.69) | 0.989 | 0.77 (0.46-1.31) | 0.342 | |
| A/A | 11 | 4 | 4 | 1.17 (0.36-3.84) | 0.794 | 0.85 (0.24-3.07) | 0.803 | |
| G/Gc | 164 | 51 | 75 | 0.744 | 1.00 | 1.00 | ||
| G/A & A/A | 103 | 33 | 40 | 1.02 (0.62-1.69) | 0.933 | 0.78 (0.47-1.30) | 0.344 | |
| G/G & G/Ac | 256 | 80 | 111 | 0.899 | 1.00 | 1.00 | ||
| A/A | 11 | 4 | 4 | 1.17 (0.36-3.78) | 0.793 | 0.93 (0.26-3.31) | 0.908 | |
| rs11655237 | ||||||||
| C/Cc | 170 | 54 | 82 | 0.584 | 1.00 | 1.00 | ||
| C/T | 88 | 28 | 29 | 1.00 (0.59-1.68) | 0.985 | 0.67 (0.39-1.16) | 0.150 | |
| T/T | 7 | 2 | 4 | 0.93 (0.19-4.64) | 0.932 | 1.69 (0.41-6.99) | 0.468 | |
| C/Cc | 170 | 54 | 82 | 0.376 | 1.00 | 1.00 | ||
| C/T & T/T | 95 | 30 | 33 | 0.99 (0.59-1.65) | 0.972 | 0.73 (0.43-1.23) | 0.233 | |
| C/C & C/Tc | 258 | 82 | 111 | 0.925 | 1.00 | 1.00 | ||
| T/T | 7 | 2 | 4 | 0.93 (0.19-4.60) | 0.933 | 1.91 (0.47-7.84) | 0.367 |
aAdjusted p values and adjusted odds ratios with their 95% CIs were calculated using multinomial logistic regression models after age adjustment between patients with uterine cervical precancerous lesions and control females. bAdjusted p values and adjusted odds ratios with their 95% CIs were calculated using multinomial logistic regression models after age adjustment between patients with uterine cervical invasive cancer and control females. cUsed as a reference for comparison to assess the odds ratios of other genotypes. AORs, adjusted odds ratios; 95% CIs, 95% confidence intervals; Ad. p, adjusted p.
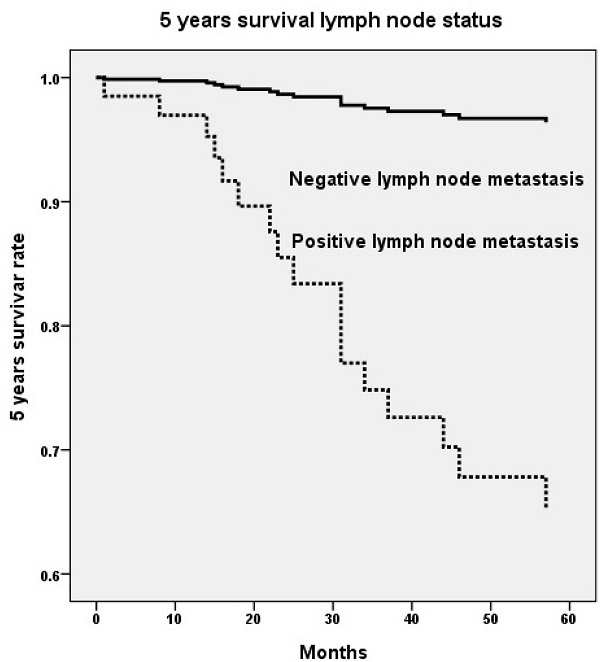
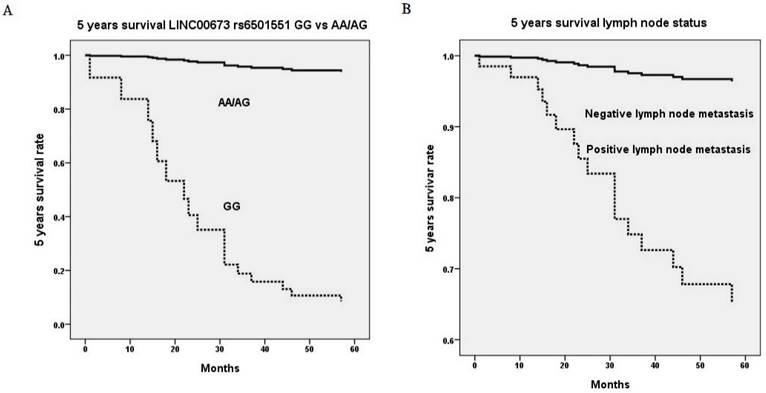
Five years survival rate based on genetic variant of long intergenic noncoding RNA 673 (LINC00673) rs6501551 (p=0.008, HR=38.7, 95% CI=2.55-587.95) (A) and pelvic lymph node status (p=0.002, HR=11.6, 95% CI=2.45-55.06) (B) using Cox proportional hazard model. HR, hazard ratio; 95% CI, 95% confidence interval.
Relationships between genotypic distributions of long intergenic noncoding RNA 673 and clinicopathological parameters of the patients with cervical invasive cancer
| Parametersa | rs6501551 | rs9914618 | rs11655237 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAb AG/GG | AA/AGb GG | GGb GA/AA | GG/GAb AA | CCb CT/TT | CC/CTb TT | |||||||
| Clinical stage | ||||||||||||
| stage Ib | 52 | 18 | 68 | 2 | 44 | 26 | 67 | 3 | 49 | 21 | 68 | 2 |
| ≥ stage II | 34 | 10 | 44 | 0 | 31 | 13 | 43 | 1 | 33 | 11 | 42 | 2 |
| P value | 0.718 | 0.522 | 0.405 | 1.000 | 0.563 | 0.639 | ||||||
| Pathologic type | ||||||||||||
| squamous cell carcinomab | 77 | 25 | 100 | 2 | 67 | 35 | 100 | 2 | 76 | 26 | 99 | 3 |
| adenocarcinoma | 10 | 3 | 13 | 0 | 8 | 5 | 11 | 2 | 6 | 7 | 12 | 1 |
| P value | 1.000 | 1.000 | 0.765 | 0.062 | 0.049* | 0.385 | ||||||
| Cell grading | ||||||||||||
| well (grade 1)b | 13 | 3 | 16 | 0 | 11 | 5 | 15 | 1 | 11 | 5 | 16 | 0 |
| moderate & poor (grades 2/3) | 74 | 25 | 97 | 2 | 64 | 35 | 96 | 3 | 71 | 28 | 95 | 4 |
| P value | 0.758 | 1.000 | 0.749 | 0.455 | 0.774 | 1.000 | ||||||
| Stromal invasion depth | ||||||||||||
| ≤10 mmb | 44 | 13 | 55 | 2 | 40 | 17 | 54 | 3 | 40 | 17 | 56 | 1 |
| >10 mm | 37 | 13 | 50 | 0 | 33 | 17 | 49 | 1 | 37 | 13 | 47 | 3 |
| P value | 0.701 | 0.497 | 0.643 | 0.621 | 0.660 | 0.338 | ||||||
| Tumor diameter | ||||||||||||
| ≤ 4 cmb | 54 | 11 | 63 | 2 | 40 | 25 | 62 | 3 | 46 | 19 | 63 | 2 |
| >4 cm | 32 | 16 | 48 | 0 | 35 | 13 | 47 | 1 | 36 | 12 | 46 | 2 |
| P value | 0.043* | 0.507 | 0.206 | 0.636 | 0.618 | 1.000 | ||||||
| Parametrium | ||||||||||||
| no invasionb | 54 | 18 | 70 | 2 | 46 | 26 | 69 | 3 | 48 | 24 | 69 | 3 |
| invasion | 32 | 9 | 41 | 0 | 29 | 12 | 40 | 1 | 34 | 7 | 40 | 1 |
| P value | 0.715 | 0.534 | 0.459 | 1.000 | 0.063 | 1.000 | ||||||
| Vagina | ||||||||||||
| no invasionb | 56 | 18 | 72 | 2 | 51 | 23 | 71 | 3 | 56 | 18 | 71 | 3 |
| invasion | 30 | 9 | 39 | 0 | 24 | 15 | 38 | 1 | 26 | 13 | 38 | 1 |
| P value | 0.882 | 0.544 | 0.430 | 1.000 | 0.308 | 1.000 | ||||||
| Pelvic lymph node | ||||||||||||
| no metastasisb | 66 | 19 | 83 | 2 | 59 | 26 | 82 | 3 | 60 | 25 | 81 | 4 |
| metastasis | 20 | 8 | 28 | 0 | 16 | 12 | 27 | 1 | 22 | 6 | 28 | 0 |
| P value | 0.503 | 1.000 | 0.233 | 1.000 | 0.412 | 0.570 | ||||||
Statistical analyses: chi-square or Fisher's exact tests. *p<0.05. aClinicopathological data of some cases could not be obtained from the patients with cervical invasive cancer because of incomplete medical charts or records. bAs a reference.
Upon multivariate analysis including all LINC00673 SNPs and significant univariate parameters for analysis, invasive cervical patients with genotype GG in LINC00673 rs6501551 had worse 5 years survival as compared to those with genotypes AA/AG in Taiwanese females (p = 0.008, HR = 38.7, 95% CI = 2.55-587.95; Table 5, Figure 1A). Moreover, other LINC00673 SNPs rs9914618 and rs11655237 were not related to 5 years survival. Moreover, lymph node metastasis was another factor that could predict 5 years survival of cervical cancer patients (p = 0.002, HR = 11.6, 95% CI = 2.45-55.06; Table 5, Figure 1B).
Discussion
Huang et al. found that serum levels of LINC00673 were highest in patients with cervical cancer as compared to those in patients with CIN and normal controls [32]. In addition, increased cell proliferation and cell cycle progression were also reported in SiHa and HeLa cervical cancer cell lines, which exhibited LINC00673 overexpression. Moreover, Shi et al. revealed that LINC00673 overexpression was noted in cervical cancer tissues and was related to poor prognosis in cervical cancer patients [33]. Through negatively modulating miR-126-5p expression and further enhancing PTEN/PI3K/AKT signaling pathway, LINC00673 presents oncogenic function in uterine cervical cancer. In contrast, Wang et al. found that expression of LINC00673 was significantly reduced in cervical cancer tissues than in their normal counterparts [34].
Wang et al. further demonstrated that patients with rs11655237 allele A (T) had significantly lower LINC00673 expression. It has been reported that rs11655237 is situated on exon 4 of LINC00673 [13, 35]. LINC00673 rs11655237 allele A was inferred to be related to increased risk of cervical cancer, possibly via down-regulating LINC00673 expression in cervical tissues [34]. It implies that LINC00673 is a tumor suppressor in cervical cancer and the G > A (C > T) change at rs11655237 probably leads to a target site for miR-1231 binding, which reduces the function of LINC00673 [34-36]. In pancreatic cancer cells, Zheng et al. demonstrated when LINC00673 was bound to miR1231, it was downregulated. Then, PTPN11 was accumulated, and an oncogene IFNAR1 was upregulated, increasing the proliferation of pancreatic cancer cells [35]. In addition, LINC00673 genetic variants were also reported to be associated with the risk of a number of cancers including gastric cancer [21], and liver cancer [22]. Moreover, in a meta-analysis study, Zhang et al. concluded that rs11655237 contributed to occurrence of cancer in all models in Chinese population [23]. As far as our knowledge, no study explores the relationships between LINC00673 SNPs and the development of cervical cancer in Taiwanese females. Therefore, we investigated the involvement of LINC00673 genetic variants in carcinogenesis of uterine cervix. However in the research, three LINC00673 genetic polymorphisms rs6501551, rs9914618 and rs11655237 could not be found to exert significantly different distributions among patients with cervical cancer and those with precancerous lesions and control females.
While correlating LINC00673 genetic variants with clinicopathological parameters, it revealed that cervical cancer patients with genotypes AG/GG in LINC00673 rs6501551 had more risk to exhibit tumor diameter larger than 4 cm as compared to those with AA. Moreover, cervical cancer patients with genotypes CT/TT in rs11655237 had more risk to have adenocarcinoma histologic type as compared to those with CC. But, these associations only reached marginally statistical significances. No association was found between LINC00673 and lymph node metastasis in cervical cancer patients. As far as to our knowledge, no research investigates the involvement of LINC00673 or its SNPs in metastasis potentials of cervical cancer. Considering in relation to clinicopathological variables, Yu et al. demonstrated that LINC00673 was related to invasion and metastasis in tongue squamous cell carcinoma (SCC) [37]. Moreover, Su et al. found that genotype GA/AA in LINC00673 rs9914618 was related to the occurrence of lymphatic spread in oral cancer as compared to GG [29]. They purposed that rs9914618 is located within a CCAAT box and presents as a putative binding motif of nuclear transcription factor Y (NF-Y) [38, 39], a critical transcriptional regulator for many genes overexpression in various cancer types or CCAAT/enhancer binding proteins (C/EBPs) [40, 41], a tumor suppressor in SCC of head and neck. Furthermore, Yuan et al. suggested that the LINC00673 rs9914618 polymorphism may be a promising HCC biomarker, especially in elderly populations [30]. Above diverse findings indicated that LINC00673 genetic variant rs9914618 probably exerted a different expression profile of cancer-associated genes mainly via impaired interactions with NF-Y or C/EBPs.
Univariate analysis of genetic variants of long intergenic noncoding RNA 673 and clinicopathological variables for 5 years survival in cervical cancer patients
| 5 years survival | ||||
|---|---|---|---|---|
| Variablesa | + | - | P value | HR (95% CIs)c |
| rs6501551 | ||||
| AAb | 71 | 13 | 0.635 | 1.00 |
| AG/GG | 22 | 5 | 1.28 (0.46-3.60) | |
| AA/AGb | 92 | 17 | 0.097 | 1.00 |
| GG | 1 | 1 | 4.70 (0.62-35.42) | |
| rs9914618 | ||||
| GGb | 60 | 12 | 0.970 | 1.00 |
| GA/AA | 33 | 6 | 0.98 (0.37-2.62) | |
| GG/GAb | 91 | 17 | 0.415 | 1.00 |
| AA | 2 | 1 | 2.26 (0.30-16.99) | |
| rs11655237 | ||||
| CCb | 65 | 13 | 0.996 | 1.00 |
| CT/TT | 28 | 5 | 1.00 (0.36-2.80) | |
| CC/CTb | 89 | 18 | 0.418 | 1.00 |
| TT | 4 | 0 | u.a. | |
| Clinical stage | ||||
| stage Ib | 60 | 6 | 0.011* | 1.00 |
| ≥ stage II | 32 | 12 | 3.32 (1.25-8.86) | |
| Pathologic type | ||||
| squamous cell carcinomab | 85 | 14 | 0.118 | 1.00 |
| adenocarcinoma | 8 | 4 | 2.36 (0.78-7.18) | |
| Cell grading | ||||
| well (grade 1)b | 12 | 3 | 0.775 | 1.00 |
| moderate & poor (grades 2/3) | 81 | 15 | 0.84 (0.24-2.89) | |
| Stromal invasion depth | ||||
| ≤ 10 mmb | 49 | 4 | 0.011* | 1.00 |
| > 10 mm | 37 | 13 | 3.83 (1.25-11.77) | |
| Tmour diameter | ||||
| ≤ 4 cmb | 56 | 5 | 0.008* | 1.00 |
| > 4 cm | 35 | 13 | 3.69 (1.32-10.37) | |
| Parametrium | ||||
| no invasionb | 61 | 7 | 0.032* | 1.00 |
| invasion | 30 | 11 | 2.70 (1.05-6.98) | |
| Vagina | ||||
| no invasionb | 61 | 9 | 0.115 | 1.00 |
| invasion | 30 | 9 | 2.07 (0.82-5.22) | |
| Pelvic lymph node | ||||
| no metastasisb. | 76 | 5 | < 0.001* | 1.00 |
| metastasis | 15 | 13 | 8.95 (3.19-25.16) | |
Statistical analyses: Kaplan-Meier curve model.
*p < 0.05.
aClinicopathological data of some cases could not be obtained from the patients with cervical invasive cancer because of incomplete records of medical chart.
bAs a reference.
cHR, hazard ratio and 95% CI, 95% confidence interval for long intergenic noncoding RNA 673 genetic polymorphisms rs6501551, rs9914618 and rs11655237 as well as clinicopathological variables, compared to their respective controls.
Survival: +, survival, -, mortality.
Multivariate analysis of genetic variants of long intergenic noncoding RNA 673 and clinicopathological variables for 5 years survival in cervical cancer patients
| 5 years survival | ||
|---|---|---|
| Variables | P value | HR & 95% CIb |
| LINC00673 genetic polymorphisms | ||
| rs6501551 | ||
| AG /GG vs. AAa | 0.537 | 0.64 (0.15-2.67) |
| GG vs. AA/AGa | 0.008 | 38.7 (2.55-587.95) |
| rs9914618 | ||
| GA/AA vs. GGa | 0.412 | 0.62 (0.19-1.97) |
| AA vs. GG/GAa | 0.069 | 10.03 (0.84-120.41) |
| rs11655237 | ||
| CT/TT vs. CCa | 0.204 | 2.04 (0.68-6.10) |
| TT vs. CC/CTa | 0.984 | u.a. |
| Clinicopathological characteristics | ||
| Pelvic lymph node | ||
| metastasis vs. no metastasisa | 0.002* | 11.6 (2.45-55.06) |
Statistical analyses: Cox proportional hazard model.
*p < 0.05.
aAs a comparison reference.
bHR, hazard ratio and 95% CI, 95% confidence interval for LINC00673 genetic polymorphisms rs6501551, rs9914618 and rs11655237, and clinicopathological characteristics, as compared to their respective controls.
LINC00673, long intergenic noncoding RNA 673; u.a., unavailable.
With regard to the relationships LINC00673 SNPs with patient survival, cervical cancer patients with genotype GG in rs6501551 had worse 5 years survival as compared to those with genotypes AA/AG in multivariate analysis in Taiwan. However other LINC00673 genetic variants rs9914618 and rs11655237 were not related to patient survival. Until now, no study reveals the relationship among LINC00673, its SNPs and patient survival in cervical cancer. Whereas, it has been reported that LINC00673 was associated with poor prognosis and enhanced invasion and metastasis in tongue SCC [37]. Moreover, pelvic lymph node metastasis was the only independent factor that could predict 5 years survival among various clinicopathological parameters for cervical cancer patients. As a prognosis predictor in cervical cancer, lymph node metastasis was also corroborated by previous researches [42, 43]. Five years survival rate has been reported declining from 85%-90% in negative pelvic lymph node metastasis down to 30%-50% in positive lymph node with a statistical difference in patients with cervical cancer [44].
As far as our knowledge, this study may be the first research in relating the LINC00673 polymorphisms to various clinicopathological variables and 5 years survival in cervical cancer patients in Taiwan. However, the current study has some weakness. Although cervical cancer patients with genotypes AG/GG in LINC00673 rs6501551 had more risk to have tumor diameter larger than 4 cm as compared to those with genotype AA as well as patients with CT/TT in rs11655237 had more risk to present adenocarcinoma as compared to those with CC, these associations only had marginally statistical difference probably with a relative weak effect of specific SNPs and not significant enough to reach a definite relationship. After adjusting for LINC00673 SNPs and various clinicopathological variables, the statistical significances might disappear. Although cervical cancer patients with genotype GG in rs6501551 had worse 5 years survival as compared to those with genotypes AA/AG in multivariate analysis, only two patients exhibiting GG were noted, and one had mortality, another had no mortality. Moreover, the study was a hospital-based cohort research and occurrence of selection bias was inevitably possible, and external validity might be limited. These problems may be resolved by enlarging the sample size in the future study. In addition, whether the LINC00673 genetic variants affect LINC00673 expression or change the linkage to its interacting proteins or microRNAs should be delineated and need further explorations.
Acknowledgements
This study was supported by research grants from Chung Shan Medical University and Chi-Mei Foundation Medical Center (CMCSMU11101). This study was also supported by Chung Shan Medical University Hospital (CSH-2023-D-009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-1789
2. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641
3. Caceres-Duran MA, Ribeiro-Dos-Santos A, Vidal AF. Roles and Mechanisms of the Long Noncoding RNAs in Cervical Cancer. Int J Mol Sci. 2020 21
4. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159
5. Su SC, Reiter RJ, Hsiao HY, Chung WH, Yang SF. Functional Interaction between Melatonin Signaling and Noncoding RNAs. Trends Endocrinol Metab. 2018;29:435-445
6. Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY. et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915
7. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458-464
8. Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H. et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126-1136
9. Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G. et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159
10. Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial-Mesenchymal Transition in Osteosarcoma by Regulating the miR-221/ARHI Pathway. J Cell Biochem. 2017;118:4772-4781
11. Weng SL, Wu WJ, Hsiao YH, Yang SF, Hsu CF, Wang PH. Significant association of long non-coding RNAs HOTAIR genetic polymorphisms with cancer recurrence and patient survival in patients with uterine cervical cancer. Int J Med Sci. 2018;15:1312-1319
12. Weng SL, Ng SC, Lee YC, Hsiao YH, Hsu CF, Yang SF. et al. The relationships of genetic polymorphisms of the long noncoding RNA growth arrest-specific transcript 5 with uterine cervical cancer. Int J Med Sci. 2020;17:1187-1195
13. Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M. et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911-916
14. Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X. et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558-25575
15. Arnes L, Liu Z, Wang J, Maurer C, Sagalovskiy I, Sanchez-Martin M. et al. Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma. Gut. 2019;68:499-511
16. Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3-22
17. Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS. et al. 3'UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. 2017;7:4466
18. Su CW, Chien MH, Lin CW, Chen MK, Chow JM, Chuang CY. et al. Associations of genetic variations of the endothelial nitric oxide synthase gene and environmental carcinogens with oral cancer susceptibility and development. Nitric Oxide. 2018;79:1-7
19. Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7-16
20. Lv Z, Xu Q, Yuan Y. A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Mutat Res Rev Mutat Res. 2017;771:1-14
21. Zhao K, Zhang R, Li T, Xiong Z. Functional variants of lncRNA LINC00673 and gastric cancer susceptibility: a case-control study in a Chinese population. Cancer Manag Res. 2019;11:3861-3868
22. Yang T, Li J, Wen Y, Tan T, Yang J, Pan J. et al. LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front Genet. 2019;10:506
23. Zhang H, Wu B, Liang K, Ke L, Ma X, Luo C. et al. Association between the LINC00673 rs11655237 C> T polymorphisms with cancer risk in the Chinese population: A meta-analysis. Medicine (Baltimore). 2022;101:e30353
24. Zhang Z, Chang Y, Jia W, Zhang J, Zhang R, Zhu J. et al. LINC00673 rs11655237 C>T confers neuroblastoma susceptibility in Chinese population. Biosci Rep. 2018 38
25. Li Y, Zhuo ZJ, Zhou H, Liu J, Liu Z, Zhang J. et al. Additional data support the role of LINC00673 rs11655237 C>T in the development of neuroblastoma. Aging (Albany NY). 2019;11:2369-2377
26. Lee CY, Ng SC, Hsiao YH, Yang SF, Hsu CF, Wang PH. Impact of the Receptor for Advanced Glycation End Products Genetic Polymorphisms on the Progression in Uterine Cervical Cancer. J Cancer. 2018;9:3886-3893
27. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790-1797
28. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600-605
29. Su SC, Lin CW, Ju PC, Chang LC, Chuang CY, Liu YF. et al. Association of LINC00673 Genetic Variants with Progression of Oral Cancer. J Pers Med. 2021 11
30. Yuan LT, Yang YC, Lee HL, Shih PC, Chen LH, Tang CH. et al. Genetic Polymorphisms of lncRNA LINC00673 as Predictors of Hepatocellular Carcinoma Progression in an Elderly Population. Int J Mol Sci. 2022 23
31. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. Int J Med Sci. 2019;16:1424-1429
32. Huang SK, Ni RX, Wang WJ, Wang D, Zhao M, Lei CZ. et al. Overexpression of LINC00673 Promotes the Proliferation of Cervical Cancer Cells. Front Oncol. 2021;11:669739
33. Shi WJ, Liu H, Ge YF, Wu D, Tan YJ, Shen YC. et al. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine. 2020;136:155286
34. Wang Y, Luo T. LINC00673 rs11655237 Polymorphism Is Associated with Increased Risk of Cervical Cancer in a Chinese Population. Cancer Control. 2018;25:1073274818803942
35. Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J. et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747-757
36. Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429-3431
37. Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-16632
38. Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15-27
39. Dolfini D, Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20:676-685
40. Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561-575
41. Bennett KL, Hackanson B, Smith LT, Morrison CD, Lang JC, Schuller DE. et al. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67:4657-4664
42. Kamura T, Tsukamoto N, Tsuruchi N, Saito T, Matsuyama T, Akazawa K. et al. Multivariate analysis of the histopathologic prognostic factors of cervical cancer in patients undergoing radical hysterectomy. Cancer. 1992;69:181-186
43. Choi KH, Kim JY, Lee DS, Lee YH, Lee SW, Sung S. et al. Clinical impact of boost irradiation to pelvic lymph node in uterine cervical cancer treated with definitive chemoradiotherapy. Medicine (Baltimore). 2018;97:e0517
44. Montz FJ, Holschneider CH, Solh S, Schuricht LC, Monk BJ. Small bowel obstruction following radical hysterectomy: risk factors, incidence, and operative findings. Gynecol Oncol. 1994;53:114-120
Author contact
![]() Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: wang082160com
Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: wang082160com

 Global reach, higher impact
Global reach, higher impact