3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(15):2771-2783. doi:10.7150/jca.85640 This issue Cite
Research Paper
CSNK1G2-AS1 promotes metastasis, colony formation and serves as a biomarker in testicular germ cell tumor cells
1. NHC Key Laboratory of Human Stem Cell and Reproductive Engineering, Institute of Reproductive and Stem Cell Engineering, School of Basic Medical Science, Central South University, Changsha, 410078, People's Republic of China.
2. Hunan Cancer Hospital, the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410078, People's Republic of China.
3. Clinical Research Center for Reproduction and Genetics in Hunan Province, Reproductive and Genetic Hospital of CITIC-Xiangya, Changsha, 410078, People's Republic of China.
# These authors equally contributed to this work.
Received 2023-4-26; Accepted 2023-7-9; Published 2023-9-4
Abstract
Background/Aim: Some long non-coding RNAs (lncRNAs) have been found to significantly participate in the progression of TGCTs. In comparison to the normal testis, the TGCT tissues showed significantly decreased CSNK1G2-AS1 expression, however, its effect on TGCTs and its mechanism are still unclear. The aim of this study is to investigate the effect of CSNK1G2-AS1 on TGCTs and explore the mechanism underlying its effect on TGCTs.
Materials and Methods: In this study, to evaluate the expression of CSNK1G2-AS1 in tissue samples from TGCTs, the UCSC and GEPIA databases were applied and qRT-PCR was conducted. The Kaplan-Meier Plotter was applied to analyze the correlation between CSNK1G2-AS1 methylation levels and the prognosis of TGCTs patients. The assays of MTS, clone formation, transwell, and flow cytometry were performed to investigate the effect of CSNK1G2-AS1 overexpression on the proliferation, metastasis, and apoptosis of TGCT cells, respectively. Finally, western blotting was conducted to determine the expressions of the proteins associated with EMT and AKT.
Results: Our study first found that, compared to the normal testis, TGCTs tissue showed significantly decreased CSNK1G2-AS1 expression, and hypomethylation of CSNK1G2-AS1 was significantly correlated with a better prognosis with TGCTs patients. In vitro, we found that overexpression of CSNK1G2-AS1 dramatically promoted the clone formation, invasion, and migration of TGCT cells, but inhibited apoptosis. And CSNK1G2-AS1 overexpression significantly decreased the expression of EMT-associated proteins ZO-1 but increased the expression and phosphorylation of AKT.
Conclusions: CSNK1G2-AS1 may play an essential role in the pathogenesis and metastasis of TGCTs through the EMT- and AKT-mediated signal pathways.
Keywords: Testicular germ cell tumors (TGCTs), long noncoding RNA (lncRNA), metastasis, EMT, AKT
Introduction
Testicular germ cell tumors (TGCTs) are the most common solid tumors in adolescent and male young adults [1]. Over the past few decades, TGCTs have continued to increase in the majority of the population [2]. By 2030, it is estimated that there will be 65,827 new cases of testicular cancer worldwide (10,561 more cases than in 2012) [3]. TGCTs are roughly classified into seminomas and non-seminomas, of which non-seminomas include undifferentiated embryonic carcinoma or differentiated teratoma, yolk sac tumor, and choriocarcinoma [4, 5]. Mixed TGCTs are common and may appear in any form. The clinical management of TGCTs can be effectively treated by monitoring, surgery, chemotherapy, radiotherapy, or combining them [6]. Although the 5-year relative survival rates for seminoma and non-seminoma patients are above 90% [7], there is still a risk existed. According to statistics, approximately 15% to 20% of patients with the disseminated disease will relapse, especially in the late stage (2 years after remission), with a poor prognosis [8]. Metastasis and cisplatin resistance are presently the two major causes of death in most TGCT patients [9]. The Serum tumor markers, such as α-fetoprotein (AFP), β-subunit of chorionic gonadotropin (β-hCG), and lactate dehydrogenase (LDH), can be used for the diagnosis of testicular tumors and the monitoring of disease recurrence [10]. However, false positive or negative results can always arise [11, 12]. The genes driving the tumorigenesis or metastasis of TGCTs have not been identified so far, and these genes may be potential therapeutic targets [13]. Despite recent efforts to identify other susceptible genetic/epigenetic lesions, no effective molecular markers are available for screening, diagnosis, or prognosis [14]. Therefore, identifying the potential mechanism underlying TGCTs and novel biomarkers for target therapies are great importance and urgently needed.
Recently, increasing amounts of studies have shown that the epigenetic regulations are closely connected to the pathogenesis of TGCTs [15]. LncRNAs are considered a class of nonprotein-coding transcripts with a minimum length of 200 bases. In recent years, lncRNAs have been increasingly investigated in various human diseases, like cancer [16-18]. Through regulating miRNAs, chromatin remodelling, and histone modification, lncRNAs significantly participated in the regulation of cell growth, apoptosis, metastasis, and angiogenesis in malignant tumors [19]. Alterations of lncRNA expression might be closely associated with the tumorigenesis of TGCTs [20, 21]. For example, the HOXA transcript at the distal tip (HOTTIP) was a noncoding RNA with 3764 nucleotides, and significantly participated in the promotion of cell proliferation through competitive binding of miR-128-3p in testicular embryonic carcinoma [22]. LncRNA Narcolepsy candidate-region 1 C (NLC1-C) was found to be down-regulated in testicular embryonal carcinoma cells, and exerted a tumor-promoting role in testicular cancer [23]. Therefore, understanding the function of lncRNA molecules in tumor is conducive to identify new diagnostic biomarkers or therapeutic targets for TGCTs [20, 21].
LncRNA casein kinase 1 gamma 2 antisense RNA 1 (CSNK1G2-AS1), also known as chromosome 19 open reading frame 34 (C19orf34), is an RNA with 1,313 kb in 19p13 and highly expressed in the testis. CSNK1G2-AS1 is the antisense RNA of CSNK1G2, located on the complementary sequence of intron 1 of CSNK1G2. CSNK1G2 is a major negative regulator of necroptosis, and the CSNK1G2 knockout mice showed premature aging of their testis [24]. In our present study, we analyzed the expression of CSNK1G2-AS1 in TGCTs and its relationship with the prognosis of TGCT patients using the Gene Expression Profiling Interactive Analysis (GEPIA) database [25] (http://gepia.cancer-pku.cn/) and University of California Santa Cruz (UCSC) Xena database, respectively. We found that compared to the normal testis tissues, CSNK1G2-AS1 was significantly down-regulated in TGCT tissues. However, its role in TGCTs and the mechanism underlying the role are still unclear.
In our study, we reported for the first time that CSNK1G2-AS1 was down-regulated in TGCTs and the hypomethylation of CSNK1G2-AS1 was significantly associated with a better prognosis in patients with TGCTs. We hypothesized that CSNK1G2-AS1 is involved in tumorigenesis and progression of TGCTs. Then we confirmed for the first time the expression of CSNK1G2-AS1 in testicular germ cell tumor tissues and cells. The purpose of this study was to investigate the function of CSNK1G2-AS1 in the development of TGCTs, and identify additional therapeutic targets and molecular markers for the clinical treatment and diagnosis of TGCTs.
Materials and methods
Collection of patient testicular tissue samples
Eleven TGCT tissue samples and ten adjacent normal testicular tissue samples were collected from the Department of Urology, the Affiliated Cancer Hospital, Xiangya School of Medicine, Central South University for qRT-PCR detection. This study was approved by the Ethics Committee of Central South University (2021KYKS-46). The informed consent was signed by each patient enrolled in this study. All tissues were confirmed by histopathological examination from the Pathology Department of Hunan Cancer Hospital. Fresh tissues were collected and frozen in liquid nitrogen for storage at -196°C.
Analysis of CSNK1G2-AS1 expression in TGCTs and the total testicular cell population
The expression of CSNK1G2-AS1 in various organs was analyzed using the UCSC database [26] (http://genome.ucsc.edu/). The expression of CSNK1G2-AS1 in TGCTs and normal testicular tissues was evaluated using GEPIA database [25] (http://gepia.cancer-pku.cn/). The RNA sequencing data of 13 tissue samples from TGCT patients and 4 paraneoplastic tissue samples (in our previous study [27]) were also applied for the analysis of the CSNK1G2-AS1 expression. To investigate the expression level of CSNK1G2-AS1 in the total testicular cell population, we retrieved the single cell RNA sequencing data of Human Testis Cell Atlas from the Gene Expression Omnibus (GEO) database (accession number: GSE120508) (https://humantestisatlas.shinyapps.io/humantestisatlas1/) [28].
Evaluation of CSNK1G2-AS1 methylation in TGCTs and prognosis of TGCT patients
The CpG islands around CSNK1G2-AS1 were profiled by the UCSC Genome online tool (https://genome.ucsc.edu/) [26], The data of CSNK1G2-AS1 methylation and prognosis of patients with TGCTs were downloaded from the UCSC Xena database (https://xena.ucsc.edu/) [29]. The relationship of the CSNK1G2-AS1 methylation with the prognosis of patients with TGCTs was analyzed using the Kaplan-Meier Plotter (https://kmplot.com/analysis/) [30]. Progression-free survival (PFS), defined as the time it takes after treatment to disease progression or death [31, 32]. Disease-free survival (DFS), defined as the time after treatment during which no disease is found.
RNA isolation and qRT-PCR detection
According to the manufacturer's instructions, total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen, Waltham, MA, USA). The quality and purity of each RNA sample were measured by Agilent2100 (Agilent, Wilmington, DE, USA). The cDNA was synthesized from 1 µg of extracted RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). The cDNA product was diluted fivefold with enzyme-free water, and then the LightCycler 480 SYBR Green I Master (Roche) was operated as instructed by the manufacturer. β-actin was used as the internal control and all experiments were repeated three times indenpently. Amplification was carried out using the following thermocycling conditions: initial denaturation at 95°C for 5 min, followed by 45 cycles of 95°C for 10 s and 60°C for 10 s, and final extension at 72°C for 10 s. The LightCycler 480 software (Roche) was used to analyze threshold cycle (CT) values, and the 2-∆∆CT method was used to evaluate the relative gene expression [33]. The sequences of the primers for qRT-PCR detection were listed below: CSNK1G2-AS1 Forward: GACAGCCGGCCTCTGAGAAC, Reverse: TCCACTTTGCTTTTGTTGATGGCGT; β-actin Forward: TTCCAGCCTTCCTTCCTGGG, Reverse: TTGCGCTCAGGAGGAGCAAT.
Cell culture and cell transfection
The NCCIT human malignant teratoma cells were purchased from the American Type Culture Collection (ATCC, VA, USA). The TCam-2 human testicular seminoma cells were obtained from Dr. Yuxin Tang [34, 35]. NCCIT and TCam-2 cells were cultured in RPMI 1640 (GIBCO, Grand Island, NY, USA) or Dulbecco's Modified Eagle's Modified (DMEM, GIBCO) medium [36-38] supplemented with 10% fetal bovine serum (FBS, GIBCO), 100 U/mL penicillin and 100 μg/mL streptomycin (GIBCO) respectively, in an incubator containing 5% CO2 at 37°C. The cells at logarithmic phage were collected and transferred to a 6-well plate (5.0 × 105 cells/well) and transfected at 70% confluence. As previously described [27], cells were divided into two groups: the cells transfected with 2.5 µg of the pcDNA3.1(+) vector and the cells transfected with 2.5 µg of the CSNK1G2-AS1 pcDNA3.1(+) vector. Cell transfection was performed using lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. Cells were collected 48 h after transfection for subsequent experiments. The cDNA of CSNK1G2-AS1 was PCR-amplified and subcloned into the HindIII and XhoI sites of the pcDNA3.1(+) vector. The CSNK1G2-AS1 overexpression plasmid was designed and synthesized by Bochu Biotechnology Co., Ltd (Changsha, China).
MTS assay
The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent (Promega) was used to detect cell proliferation. TCam-2 and NCCIT cells were cultured in a 96-well plate with 5 × 103 cells/mL (200 μL/well) for 6 h as day 0 and then transfected with indicated plasmids at 24, 48, 72, 96, and 120 h (day 1, day 2, day 3, day 4, day 5, respectively). Then 20 μL MTS was added and incubated for 4 h at 37°C. The absorbance at 492 nm was determined by the enzyme immunoassay analyzer (Thermo Fisher Scientific, Waltham, MA, USA). Each experiment was repeated three times.
Plate colony formation assay
The transfected cells were inoculated into a 6-well plate at a density of 400 cells/well and cultured in an incubator at 37°C and 5% CO2 for eight days until most single cells could be observed under a microscope to become clones containing 50 cells. The cells were then fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet (Sigma, St. Louis, MO, USA) for 15 min. After dyeing, a photo was taken to calculate the number of clones using Adobe Photoshop CS5. Data from three independent experiments were expressed as mean ± standard deviation (S.D.).
Flow cytometric cell apoptosis assay
After 48 h of transfection, cells were digested into single cell with 0.05% trypsin (GIBCO), washed with pre-cooled PBS (Gibco) three times, and treated with an Annexin V FITC Apoptosis Kit (BD Pharmingen; San Diego, CA, USA) according to the manufacturer's instructions. The Accuri C6 flow cytometer (BD) (FACScan) was employed for detection of apoptosis. Finally, FlowJo VX 10 (Tree Star; Ashland, OR, USA) was applied to analyze the apoptosis results. Each experiment was performed in triplicate.
Transwell cell migration and invasion assays
Transwell migration and invasion assays were performed using 8.0 μm Transwell Permeable
Support (353097) (Corning Inc., Corning, NY, USA). TCam-2 and NCCIT cells were digested into single cell and resuspended with serum-free substrate after 48 h of transfection. Cells were diluted to a concentration of 100,000/mL, 200 μL cells were added to the upper chamber which was precoated with Matrigel Matrix (BD) [27], and 800 μL 15% FBS-contained medium was added to the lower chamber. The 24-well plates were placed in an incubator at 37°C and with 5% CO2 for 48 h. The chamber was then washed twice with PBS, fixed with 4% paraformaldehyde on the upper and lower walls for 30 min. After another washing with PBS, the fixed cells were stained with 0.1% crystal violet (Sigma) for 15 min. Then the cells on the upper surface of the chamber that did not pass through the chamber were rinsed with clean water and gently removed with a sterile cotton swab. Finally, cells were observed and photographed under an inverted microscope and counted in five randomly selected fields. The experiment described above was repeated three times.
Western blotting
Cells were collected and lysed using radioimmunoprecipitation assay (RIPA) lysis buffer (CoWin Biosciences, Beijing, China) according to the manufacturer's instructions to obtain the protein. The Bicinchoninic Acid (BCA) Protein Quantification Kit (Thermo Fisher Scientific) was used for protein quantification, and the samples were adjusted to the same concentration. After mixing with 5× loading buffer (CoWin Biosciences), the protein samples were boiled at at 100°C for 8 min, and then the protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (SDS-PAGE) and transferred to the polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking with 5% fatty-free milk for 1 h, the corresponding primary antibodies were incubated with the PVDF membrane overnight at 4°C. Next, After 3-time washing using Tris Buffered Saline with Tween-20 (TBST), the PVDF membrane was incubated with the secondary antibody at 37°C for 1 h. After 3-time washing with TBST, the protein bands were visualized by fluorography using an enhanced chemiluminescence system (Millipore). Primary antibodies used in this study are as follows: anti-glyceraldehyde-3-phosphate dehydrogenase antibody (GAPDH) (CW0100) (1:1,000 dilution; CoWin Biosciences), anti-zonula occludens-1 (ZO-1) antibody (#8193) (1:1,000 dilution; Cell Signaling Technology), anti-AKT antibody (10176-2-AP) (1:1,000 dilution; ProteinTech), and anti-Phospho-AKT (Ser473) (P-AKT) antibody (66444-1-Ig) (1:1,000 dilution; ProteinTech). Secondary antibodies used are as follows: goat anti-mouse IgG (CW0102) (1:2,000 dilution; CoWin Biosciences) or goat anti-rabbit IgG (CW0103) (1:2,000 dilution; CoWin Biosciences). GAPDH was used as an internal reference. Densitometric analysis of the bands was performed using ImageJ 1.52a software (National Institutes of Health, Bethesda, MD, USA) for protein quantification.
Statistical analysis
The data were represented as means ± standard deviation (S.D.) and each experiment was repeated at least three times independently. The results were analyzed using GraphPad Prism software (Version 5.0; La Jolla, CA, USA). The Student's t test was used to calculate the difference between the two groups. Significant differences between multiple data sets were calculated using one-way analysis of variance (ANOVA). P < 0.05 was deemed statistically significant. PFS and DFS were calculated using a log-rank test.
Results
In the present study, we tried to explore the function of CSNK1G2-AS1 in TGCTs based on a series of bioinformatic analysis and in vitro experiments. We found that CSNK1G2-AS1 had a lower expression in TGCTs and that hypomethylation of CSNK1G2-AS1 was significantly associated with a better prognosis of patients with TGCTs. Further analysis showed that CSNK1G2-AS1 could promote the migration and invasion of TGCT cells and regulate the expression of AKT and the proteins associated with EMT.
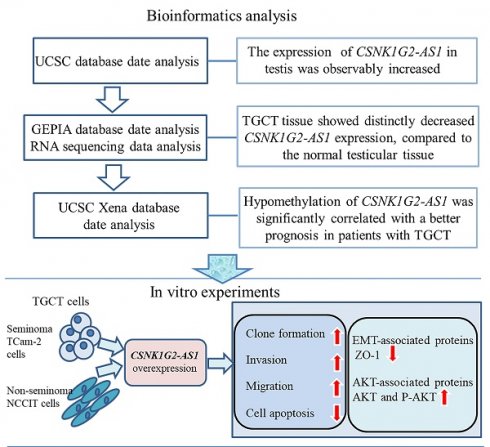
Expression of CSNK1G2-AS1 in tumor tissues. (A) The UCSC database analysis showed that CSNK1G2-AS1 is highly expressed in testis. (B) Representative volcano plots showing that CSNK1G2-AS1 is significantly down-regulated in TGCT tissues. (C) Representive results from the analysis of GEPIA database showed that the expression of CSNK1G2-AS1 in TGCT tissues was significantly lower than that of normal testis. TPM = transcribed per million. (D) Differential expression of CSNK1G2-AS1 in seminomas and non-seminomas versus normal testis from the GEPIA database. TPM = transcribed per million. (E) qRT-PCR was used to verify the expression of CSNK1G2-AS1 in TGCTs and normal testis. 'T' means TGCT samples, 'N' means normal testis. *P < 0.05, *** P < 0.001.
Decreased CSNK1G2-AS1 expression in TGCT tissues and significant association between the CSNK1G2-AS1 hypomethylation and the better prognosis
The levels of CSNK1G2-AS1 expression in different tissues and organs were compared using UCSC database [26]. The results showed significantly increased expression of CSNK1G2-AS1 in the testis (Fig. 1A). In our previous study, to explore the function of lncRNAs in TGCTs, we conducted RNA sequencing and constructed lncRNA expression profiles using 13 TGCT tissues and 4 paraneoplastic tissues [27], and observed that CSNK1G2-AS1 was significantly down-regulated in TGCTs (Fig. 1B). To verify the reliability of differential expression of CSNK1G2-AS1, we analyzed its expression in TGCTs using the GEPIA database [25]. The results showed that CSNK1G2-AS1 was distinctly down-regulated in different types of TGCTs (seminoma and non-seminoma) (Fig. 1C-D). Subsequently, ten adjacent normal testicular tissue samples and eleven TGCT samples were employed, and the decreased CSNK1G2-AS1 expression in TGCTs was confirmed using qRT-PCR detection (Fig. 1E). These results revealed that compared with the normal testis, TGCT tissues exhibited significantly decreased CSNK1G2-AS1 expression.
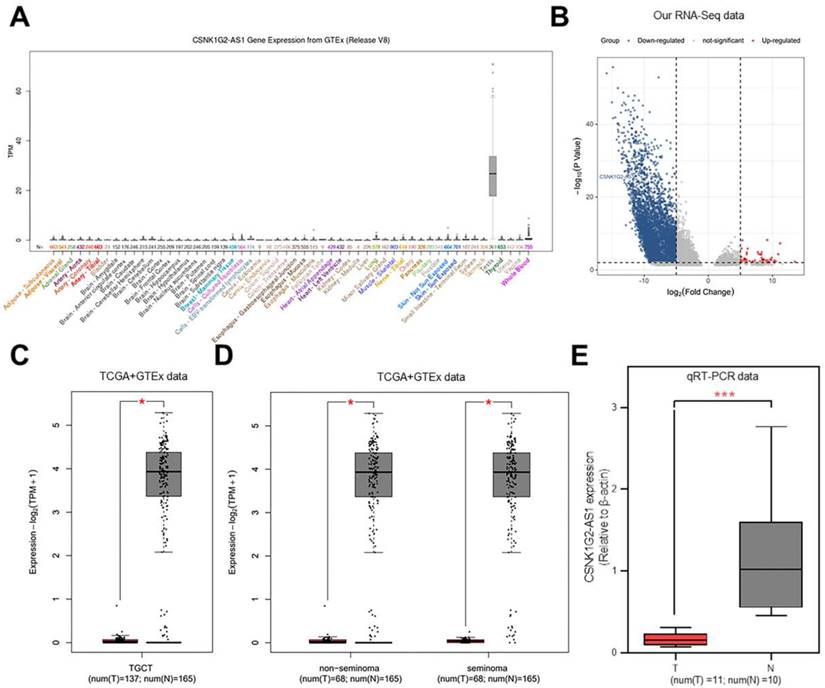
The CpG islands around CSNK1G2-AS1 were profiled using the UCSC Xena online tool (Fig. 2A), and there are 156 samples in TCGA Testicular Cancer (TGCT) (17 datasets) were selected. The correlation analysis between the methylation levels of CSNK1G2-AS1 and the prognosis of TGCT patients showed that patients with hypermethylation of CSNK1G2-AS1 had significantly lower DFS (Fig. 2B) and PFS (Fig. 2C) than patients with hypomethylation. Finally, we plotted the time-dependent receiver operating characteristic (ROC) curves of DFS and PFS. The Area Under Curves (AUC) at 1 and 2 years of DFS were 0.66 and 0.71, respectively (Fig. 2D). The AUC at 1 and 2 years of PFS were 0.62 and 0.65, respectively (Fig. 2E). Therefore, these results suggested that the low methylation level of CSNK1G2-AS1 may be a potential prognostic marker for a good prognosis in TGCT patients.
Methylation level of CSNK1G2-AS1 in TGCTs and the prognosis of patients with TGCTs. (A) The CpG islands around CSNK1G2-AS1 were profiled using the UCSC Xena online tool. (B-C) TGCT data from the TCGA TGCT cohort were used to analyze the correlation between the level of CSNK1G2-AS1 methylation and the prognosis of patients with TGCTs. (D) Representative ROC curves showing the predictive sensitivity and specificity of the DFS at 1 and 2 years. (E) Representative ROC curves showing the predictive sensitivity and specificity of PFS at 1 and 2 years.
Overexpression of CSNK1G2-AS1 increased clone formation but did not affect cell proliferation
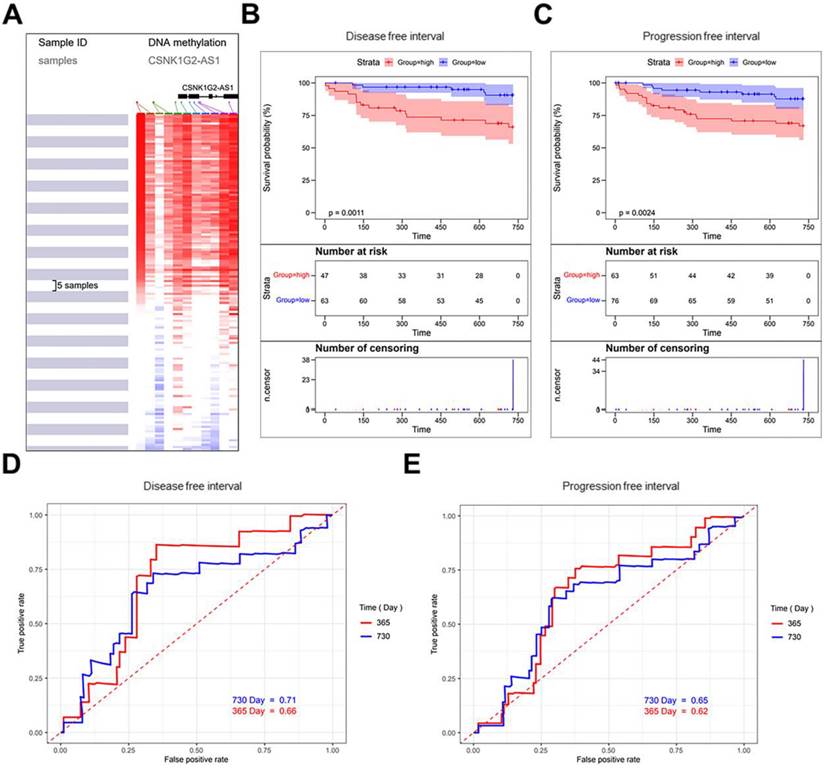
Because CSNK1G2-AS1 expression was decreased in TGCT tissues, we first tested whether overexpression of CSNK1G2-AS1 had an effect on TGCT cell proliferation. The effect of CSNK1G2-AS1 overexpression in TCam-2 and NCCIT cells was determined by qRT-PCR (Fig. 3A-B), then the proliferation was evaluated by the MTS assay. According to the MTS assay, we found that no significant difference was observed between the cells with CSNK1G2-AS1 overexpression or not (Fig. 3C-D). However, significantly high clonal formation was observed in the cells with CSNK1G2-AS1 overexpression compared to the negative control (P < 0.05, Fig. 3E-F).
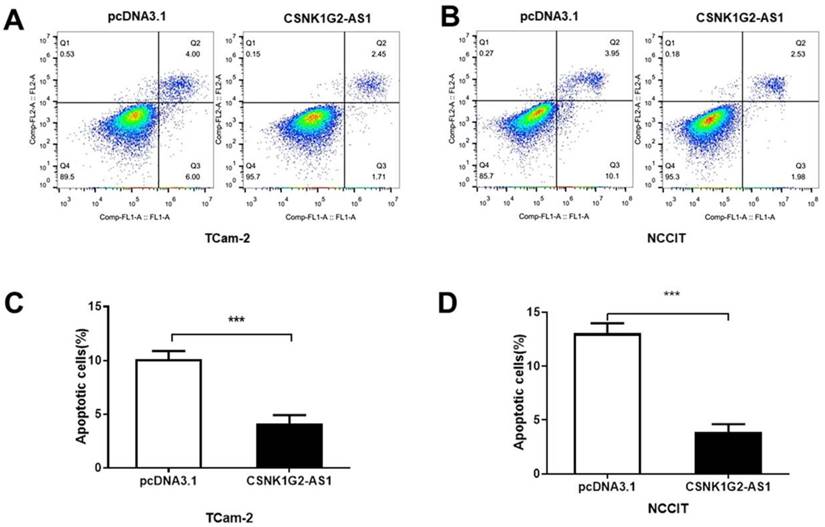
CSNK1G2-AS1 overexpression inhibited the apoptosis of TGCT cells
To elucidate the influence of CSNK1G2-AS1 on the regulation of TGCT cell apoptosis, Annexin V/PI staining and flow cytometry were performed. As shown in Figure 4, the percentages of early and late apoptotic TCam-2 and NCCIT cells were obviously reduced by CSNK1G2-AS1 overexpression compared to the negative control (P < 0.05, Fig. 4A-D). These results showed that overexpression of CSNK1G2-AS1 could inhibit the apoptosis of TGCT cells.
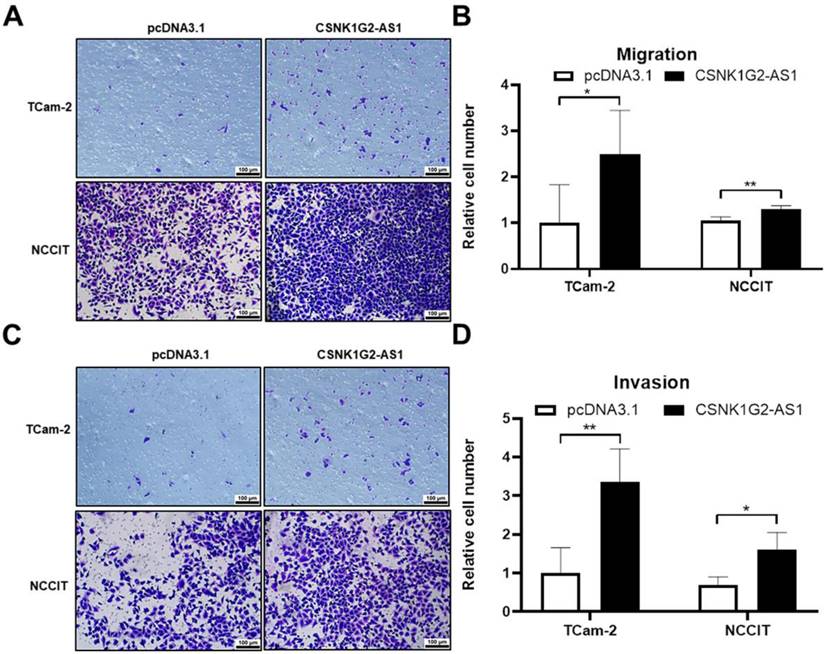
CSNK1G2-AS1 positively regulated the migration and invasion of TGCT cells
After orchiectomy, the probability of recurrence and metastasis in patients with stage I seminoma and patients with non-seminoma was 15-20% and ~30%, according to the European Guidelines for Urological Testicular Cancer [39]. Therefore, to explore the relationship between CSNK1G2-AS1 overexpression and the migrative and invasive abilities of TGCT cells, we performed transwell assays. We found that 48 h after overexpression, the TGCT cells exhibited significantly enhanced migrative (Fig. 5A-B) and invasive (Fig. 5C-D) abilities compared to negative control (P < 0.05). These results indicated that CSNK1G2-AS1 plays a promoting role in the migration and invasion of TGCT cells, which warrants further investigation.
CSNK1G2-AS1 promotes the clonal formation of TGCT cells. (A-B) The overexpression of CSNK1G2-AS1 in TCam-2 and NCCIT cells was verified by qRT-PCR. (C-D) MTS showed that CSNK1G2-AS1 overexpression did not affect TCam-2 and NCCIT proliferation. (E) Colony formation assays showed that CSNK1G2-AS1 overexpression promotes colony formation activity of TGCT cells. (F) Overexpression of CSNK1G2-AS1 increased the relative number of TGCT cell clones. Data represent the mean ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001.
Overexpression of CSNK1G2-AS1 inhibits TGCT cell apoptosis. (A-B) Flow cytometry was used to detect cell apoptosis of TGCT cells overexpressed with CSNK1G2-AS1 and pcDNA3.1 (+). (C-D) Compared to the pcDNA3.1 (+) group, the apoptosis rate of TGCT cells overexpressing CSNK1G2-AS1 was significantly reduced. *** P < 0.001.
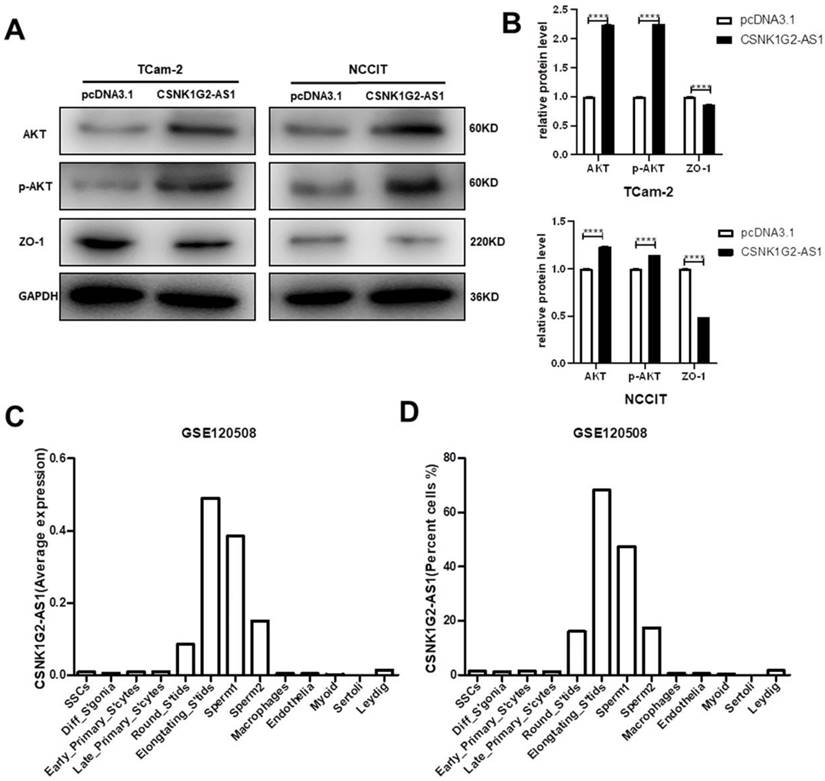
CSNK1G2-AS1 promoted the development of TGCT cells by affecting the signaling pathways mediated by EMT and AKT
Epithelial-mesenchymal transformation (EMT) is a biological process in which epithelial cells are transformed into phenotypic mesenchymal cells by a specific procedure [40]. It plays an important role in cancer metastasis. To find out whether CSNK1G2-AS1 could regulate the metastasis and invasion of TGCT cells through EMT signaling pathway, the proteins associated with EMT were detected by Western blot. We found that in TGCT cells, CSNK1G2-AS1 overexpression dramatically enhanced the expression of the EMT-related protein ZO-1 (Fig. 6A-B). Due to the involvement of the AKT-related signaling pathway in the progression of TGCTs by regulating cell proliferation and migration [41, 42], we also detected the proteins related to the AKT signaling pathway and found that CSNK1G2-AS1 overexpression also significantly enhanced the expression and phosphorylation of AKT. These results indicated that CSNK1G2-AS1 could promote the development of TGCT cells by influencing the signaling pathways related to EMT and AKT (Fig. 6A-B).
Expression of CSNK1G2-AS1 in the total testicular cell population
The results verified by in vitro experiments appear to be inconsistent with bioinformatic analysis. The two main cell groups in normal testis include germ cells and somatic cells. The germ cells include a few undifferentiated spermatogonia and differentiated spermatogonia, and a large number of spermatocytes and spermatids, especially meiotic divisions and late spermatids [43, 44]. The average expression (Fig. 6C) and cell percentage (Fig. 6D) of CSNK1G2-AS1 in germ cells suggested that in the spermatogenesis process, CSNK1G2-AS1 highly expressed in later stages of spermatogenesis (round spermatids, elongated spermatids, and sperm), but low in the proliferative stage of spermatogonial stem cells and meiophase.
Discussion
Recently, the imbalance of lncRNAs in various cancers has attracted much more attentions. Many studies have shown that lncRNAs play a crucial role in the physiological and pathological changes of cancers [45, 46]. Moreover, some studies have also shown that lncRNAs are significantly associated with the pathogenesis and development of TGCTs. For example, the removal of SPRY4-IT1 lncRNA can inhibit the growth of reproductive testicular tumors by decreasing cell survival, proliferation, migration, and invasion [47]. LncRNA H19 knockdown in TCam-2/CDDP cells can help tumor cells survive by affecting cell activity reduction, cell cycle arrest, increase apoptosis, and decrease invasion [48]. However, whether other lncRNAs also affect the tumorigenesis and progression of TGCTs was completely unknown. In this study, our results about the expression of CSNK1G2-AS1 were consistent with the analysis from the GEPIA database described previously. Comparing with the normal testis, the testicular germ cell tumor tissues exhibited significantly decreased CSNK1G2-AS1 expression, suggesting the potential involvement of CSNK1G2-AS1 in the tumorigenesis and development of testicular tumors. Meanwhile, in TGCTs, low CSNK1G2-AS1 methylation indicated high DFS, indicating the association of CSNK1G2-AS1 with the prognosis of TGCT, while its role in TGCTs remains unclear. Therefore, we conducted an in vitro study to overexpress CSNK1G2-AS1 in TCam-2 and NCCIT cells to investigate the role of CSNK1G2-AS1 in TGCT cells. Through in vitro experiments, we found that overexpression of CSNK1G2-AS1 did not affect the proliferation of TGCT cells but promoted clonal formation, invasion, and migration, as well as inhibited the apoptosis of TCam-2 and NCCIT cells. Therefore, CSNK1G2-AS1 could promote the development of TGCTs by increasing the metastasis and survival rate of TGCT cells in vitro. Next, we explored the signaling pathway which involved in the promotion of CSNK1G2-AS1 on the development of TGCTs.
CSNK1G2-AS1 promotes metastasis and invasion of TGCT cells. (A) Transwell assay found that overexpression of CSNK1G2-AS1 promotes TGCT cells metastasis compared to pcDNA3.1 (+) group. (B) The relative number of migrating cells in the overexpression of CSNK1G2-AS1 group was increased. (C) The transwell assay found that CSNK1G2-AS1 overexpression promoted invasion of TGCT cells compared to negative control. (D) The relative number of invasive TGCT cells with CSNK1G2-AS1 overexpression was increased. *P < 0.05, **P < 0.01. Scale bar = 100 μm.
Effects of CSNK1G2-AS1 on EMT, AKT signaling pathway related proteins, and expression levels of CSNK1G2-AS1 in the total testicular cell population. (A) Western blot assays detected the expression levels of AKT, p-AKT and ZO-1 in TCam-2 and NCCIT cells. (B) Relative expression levels of AKT, p-AKT and ZO-1 in TCam-2 and NCCIT cells. GAPDH was used as a loading control. (C-D) Average expression and cell percentage of CSNK1G2-AS1 in the human testis from the Gene Expression Omnibus (GEO) database (accession number: GSE120508). ****P < 0.0001.
Metastasis is an important reason for the occurrence, development, and poor prognosis of tumors [49]. The metastasis and recurrence of TGCTs are some of the most intractable problems in a clinical setting. We found that CSNK1G2-AS1 could promote TGCT cells migration and invasion, which further proved that CSK1G2-AS1 can promote TGCTs development. The EMT is a developmental process that enables stationary epithelial cells to gain the ability to migrate and invade independent cells [50] and is associated with various tumor functions, including tumor initiation, malignant progression, tumor stem cell formation, tumor cell migration, infiltration into the blood, metastasis, and therapeutic resistance [51]. An increasing amount of recent studies have shown that tumor metastasis may be significantly related to the EMT signaling pathway. For example, Lei et al. discovered that overexpression of miR-145 could reverse the EMT phenotype of thyroid cancer cells and reduce the efficiency of cell metastasis [52]. Li et al. indicated that Emodin inhibits EMT and invasion of pancreatic cancer by upregulating microRNA-1271 [53]. In this study, we detected proteins related to the EMT signaling pathway and found that after overexpression of CSNK1G2-AS1, the expression of ZO-1 decreased. Studies have shown that CAPS1 could promote CRC cell metastasis in vitro by inducing EMT, including reduced expression of the epithelial markers E-cadherin and ZO-1 [54]. Zhang et al. indicated that decreased expression of ZO-1 is related to the metastasis of liver cancer [55]. Therefore, CSNK1G2-AS1 may promote TGCT cells migration and invasion by regulating the EMT signaling pathway.
Many cellular processes, including cell growth, survival, apoptosis, angiogenesis, metabolism, and protein synthesis, can be regulated by AKT [56]. Numerous studies have shown that AKT expression is increased in most cancers [57]. In the cancers with increased AKT expression, cytoplasmic P21 expression was also elevated, contributing to tumor progression, chemotherapeutic resistance, and poor prognosis [58]. AKT phosphorylation leads to the activation of several substrates, such as BAD, caspase, and forkhead transcription factors, which could impair apoptosis and enhance survival [59]. In this study, we found that the expression and the phosphorylation of AKT significantly increased after the overexpression of CSNK1G2-AS1 in TGCT cells. Therefore, CSNK1G2-AS1 may inhibit TGCT cells apoptosis through the AKT signaling pathway. Meanwhile, studies have shown that activation of AKT could promote the EMT phenotype of seminoma and enhance seminoma invasion [38]. These results suggested that CSNK1G2-AS1 may promote the development of TGCT cells through regulating the EMT and AKT related signaling pathways.
Our in vitro experiments showed the role and the possible mechanism of CSNK1G2-AS1 in promoting TGCT cells development. However, the down-regulation of CSNK1G2-AS1 in TGCTs and the correlation between CSNK1G2-AS1 and the prognosis of TGCT patients suggested that CSNK1G2-AS1 may resemble a tumor suppressor gene in TGCTs, contrary to the results of the cell experiments in vitro. From an objective point of view, the results of in vitro studies may not accurately represent the situation in vivo. Cancer development and progression are closely related to changes of the surrounding stroma. Cancer cells can functionally shape their microenvironment by secreting various cytokines, chemokines, and other factors [60]. Tumor growth, metastasis, and response to treatment are also associated with the complexity of the tumor microenvironment (TME), which is an “ecological niche” that stimulates cancer progression. The dynamic changes that occur in TME could cause selection of tumor cell variants, which may promote genomic instability [61-63]. The in vitro experiments of tumor cells lose the signals maintained or assisted in vivo, and lose contact with other cells or molecules, which may lead to inconsistent experimental results between in vitro and in vivo experiments. For example, arsenic trioxide (ATO) has opposite effects on the nuclear factor erythroid-derived 2 like 2 (Nrf2) pathway in oral squamous cell carcinoma in vitro and in vivo [64]. Furthermore, the role of RASSF5 in cancer is controversial, Liao et al. explained how RASSF5 can activate MST1/2 and suppress cancer in vivo but inhibit MST1/2 in vitro [65]. However, there is no suitable in vivo model for human TGCTs and no animal model capable of forming precursor cell carcinomas in situ observed in humans [66].
TGCTs originating from CIS can traditionally be divided into seminoma and non-seminoma, each accounting for approximately 50% [67]. Seminoma consists of transformed germ cells that closely resemble primordial germ cells (PGCs)/gonocyte, and NSE can be composed of cells with typical pluripotency of PGCs/gonocytes [68]. Therefore, the gene expression profile of TGCTs is similar to PGCs/gonocytes [69]. Due to the limitations of sample acquisition, in the preliminary study of the expression characteristics of lncRNAs in TGCTs, the control group used was paracancerous testicular tissue. Spermatogenesis is a complex and dynamic cellular differentiation process. We found that in the spermatogenesis process, CSNK1G2-AS1 expression level was high in the later stages of spermatogenesis but low in the proliferative stage of spermatogonial stem cells and meiophase. Perhaps PGCs/gonocytes would be a more appropriate control for the study of genetic/epigenetic lesions in TGCTs. The expression level of CSNK1G2-AS1 in TGCTs compared to PGCs/gonocytes was still unknown, whether CSNK1G2-AS1 was expressed at the wrong developmental node to promote the tumorigenesis and metastasis of TGCTs needs further investigation.
This study is the first to confirm the role of CSNK1G2-AS1 in tumors and its role in TGCTs. Although we have studied the effect of CSNK1G2-AS1 on TGCT cells, this study also has some limitations. Firstly, due to the difficulty for the collection of patient samples, the sample size of this study was not very sufficient. Secondly, we should further study how CSNK1G2-AS1 affects the AKT and EMT signaling pathways and how the AKT and EMT signaling pathways affect the development of TGCT cells. Finally, we must determine the target genes of CSNK1G2-AS1 in TGCTs to further study how they affect the tumorigenesis and prognosis of TGCT. The current findings suggest that CSNK1G2-AS1 can be used as a biomarker for testicular germ cell tumor cells. On the basis of improving relevant studies, it is expected to develop inhibitors of CSNK1G2-AS1 to inhibit the migration of testicular germ tumor cells, and develop new therapeutic drugs to improve TGCT treatment.
Conclusion
In summary, the role of CSNK1G2-AS1 in TGCTs was described for the first time in this study, and its molecular mechanism was preliminarily discussed. In vitro studies have shown that CSNK1G2-AS1 could suppress TGCT cells apoptosis but promote colony formation, migration, and invasion of TGCT cells through the AKT and EMT signaling pathways, suggesting that CSNK1G2-AS1 may play a critical role in the development and metastasis of TGCT cells.
Abbreviations
TGCTs: Testicular germ cell tumors; CSNK1G2-AS1: Casein kinase 1 gamma 2 antisense RNA 1; GEPIA: Gene Expression Profiling Interactive Analysis; ANOVA: One-way analysis of variance; AUC: Area Under Curves; EMT: Epithelial-mesenchymal transformation; PGCs: Primordial germ cells; DFS: Disease-free survival; PFS: Progression-free survival; ROC: Receiver operating characteristic; MTS: 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
Acknowledgements
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31472054), the National Key Research and Development Program of China (No. 2016YFC1000600), and the Fundamental Research Funds for Central Universities of Central South University (No. 2021zzts0322).
Author contributions
Hao Bo and Liqing Fan conceived, designed, and performed the experiments. Guangmin Liu and Zhizhong Liu collected clinical samples. Ruixue Li and Fang Zhu performed the in vitro experiments. Ruixue Li and Qianyin Zhou contributed to figure layout, data analysis, and manuscript drafting. Hao Bo and Qianyin Zhou proofed the manuscript. Liqing Fan revised and confirmed the final manuscript. All authors contributed to the article and approved the submitted version.
Ethical approval
This study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University (2021KYKS-46).
Statement of human and animal rights
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Declaration of Helsinki of 1964 and its subsequent amendments or comparable ethical standards.
Statement of informed consent
All patients provided their informed written consent to collect tissue samples.
Data accessibility
All data generated or analyzed during this study are included in the published article.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Carouge D, Blanc V, Knoblaugh SE, Hunter RJ, Davidson NO, Nadeau JH. Parent-of-origin effects of A1CF and AGO2 on testicular germ-cell tumors, testicular abnormalities, and fertilization bias. Proc Natl Acad Sci U S A. 2016;113:E5425-33
2. Das MK, Evensen HSF, Furu K, Haugen TB. miRNA-302s may act as oncogenes in human testicular germ cell tumours. Scientific reports. 2019;9:9189
3. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
4. Chieffi P, De Martino M, Esposito F. New Anti-Cancer Strategies in Testicular Germ Cell Tumors. Recent Pat Anticancer Drug Discov. 2019;14:53-9
5. Batool A, Liu XM, Zhang CL, Hao CF, Chen SR, Liu YX. Recent advances in the regulation of testicular germ cell tumors by microRNAs. Front Biosci (Landmark Ed). 2019;24:765-76
6. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69:7-34
7. Ylonen O, Jyrkkio S, Pukkala E, Syvanen K, Bostrom PJ. Time trends and occupational variation in the incidence of testicular cancer in the Nordic countries. BJU Int. 2018;122:384-93
8. Kollmannsberger C, Tandstad T, Bedard PL, Cohn-Cedermark G, Chung PW, Jewett MA. et al. Patterns of relapse in patients with clinical stage I testicular cancer managed with active surveillance. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:51-7
9. Lobo J, Constancio V, Leite-Silva P, Guimaraes R, Cantante M, Braga I. et al. Differential methylation EPIC analysis discloses cisplatin-resistance related hypermethylation and tumor-specific heterogeneity within matched primary and metastatic testicular germ cell tumor patient tissue samples. Clin Epigenetics. 2021;13:70
10. Khan O, Protheroe A. Testis cancer. Postgrad Med J. 2007;83:624-32
11. Batool A, Karimi N, Wu XN, Chen SR, Liu YX. Testicular germ cell tumor: a comprehensive review. Cell Mol Life Sci. 2019;76:1713-27
12. Dieckmann KP, Simonsen-Richter H, Kulejewski M, Anheuser P, Zecha H, Isbarn H. et al. Serum Tumour Markers in Testicular Germ Cell Tumours: Frequencies of Elevated Levels and Extents of Marker Elevation Are Significantly Associated with Clinical Parameters and with Response to Treatment. Biomed Res Int. 2019;2019:5030349
13. Lobo J, Gillis AJM, Jeronimo C, Henrique R, Looijenga LHJ. Human Germ Cell Tumors are Developmental Cancers: Impact of Epigenetics on Pathobiology and Clinic. Int J Mol Sci. 2019 20
14. Costa AL, Lobo J, Jerónimo C, Henrique R. The epigenetics of testicular germ cell tumors: looking for novel disease biomarkers. Epigenomics. 2017;9:155-69
15. Looijenga LH, Van Agthoven T, Biermann K. Development of malignant germ cells - the genvironmental hypothesis. Int J Dev Biol. 2013;57:241-53
16. Saha P, Verma S, Pathak RU, Mishra RK. Long Noncoding RNAs in Mammalian Development and Diseases. Advances in experimental medicine and biology. 2017;1008:155-98
17. Zhu C, Xie Y, Li Q, Zhang Z, Chen J, Zhang K. et al. CPSF6-mediated XBP1 3'UTR shortening attenuates cisplatin-induced ER stress and elevates chemo-resistance in lung adenocarcinoma. Drug Resist Updat. 2023;68:100933
18. Jiang M, Qi F, Zhang K, Zhang X, Ma J, Xia S. et al. MARCKSL1-2 reverses docetaxel-resistance of lung adenocarcinoma cells by recruiting SUZ12 to suppress HDAC1 and elevate miR-200b. Mol Cancer. 2022;21:150
19. Wu L, Zhu X, Song Z, Chen D, Guo M, Liang J. et al. Long Non-Coding RNA HOXA-AS2 Enhances The Malignant Biological Behaviors In Glioma By Epigenetically Regulating RND3 Expression. OncoTargets and therapy. 2019;12:9407-19
20. Barchi M, Bielli P, Dolci S, Rossi P, Grimaldi P. Non-Coding RNAs and Splicing Activity in Testicular Germ Cell Tumors. Life (Basel). 2021 11
21. Bresesti C, Vezzoli V, Cangiano B, Bonomi M. Long Non-Coding RNAs: Role in Testicular Cancers. Front Oncol. 2021;11:605606
22. Su Y, Zhou LL, Zhang YQ, Ni LY. Long noncoding RNA HOTTIP is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Mol Genet Genomic Med. 2019;7:e870
23. Kawashima M, Tamiya G, Oka A, Hohjoh H, Juji T, Ebisawa T. et al. Genomewide association analysis of human narcolepsy and a new resistance gene. Am J Hum Genet. 2006;79:252-63
24. Sziva RE, Acs J, Tokes AM, Korsos-Novak A, Nadasy GL, Acs N. et al. Accurate Quantitative Histomorphometric-Mathematical Image Analysis Methodology of Rodent Testicular Tissue and Its Possible Future Research Perspectives in Andrology and Reproductive Medicine. Life (Basel). 2022 12
25. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
26. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM. et al. The human genome browser at UCSC. Genome Res. 2002;12:996-1006
27. Bo H, Zhu F, Liu Z, Deng Q, Liu G, Li R. et al. Integrated analysis of high-throughput sequencing data reveals the key role of LINC00467 in the invasion and metastasis of testicular germ cell tumors. Cell Death Discov. 2021;7:206
28. Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X. et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141-57
29. Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675-8
30. Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101-9
31. Zhang JJ, Sun Z, Yuan H, Wang M. Alternatives to the Kaplan-Meier estimator of progression-free survival. Int J Biostat. 2020;17:99-115
32. Halabi S, Vogelzang NJ, Ou SS, Owzar K, Archer L, Small EJ. Progression-free survival as a predictor of overall survival in men with castrate-resistant prostate cancer. J Clin Oncol. 2009;27:2766-71
33. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8
34. Peng D, Wei J, Gan Y, Yang J, Jiang X, Kitazawa R. et al. Testis developmental related gene 1 regulates the chemosensitivity of seminoma TCam-2 cells to cisplatin via autophagy. J Cell Mol Med. 2019;23:7773-84
35. Gan Y, Wang Y, Tan Z, Zhou J, Kitazawa R, Jiang X. et al. TDRG1 regulates chemosensitivity of seminoma TCam-2 cells to cisplatin via PI3K/Akt/mTOR signaling pathway and mitochondria-mediated apoptotic pathway. Cancer Biol Ther. 2016;17:741-50
36. Cui Y, Miao C, Liu S, Tang J, Zhang J, Bu H. et al. Clusterin suppresses invasion and metastasis of testicular seminoma by upregulating COL15a1. Mol Ther Nucleic Acids. 2021;26:1336-50
37. Batool A, Chen SR, Liu YX. Distinct Metabolic Features of Seminoma and Embryonal Carcinoma Revealed by Combined Transcriptome and Metabolome Analyses. J Proteome Res. 2019;18:1819-26
38. Chen Y, Lu J, Xia L, Xue D, Yu X, Shen D. et al. Testicular orphan receptor 4 promotes tumor progression and implies poor survival through AKT3 regulation in seminoma. Cancer Sci. 2018;109:384-94
39. Albers P, Albrecht W, Algaba F, Bokemeyer C, Cohn-Cedermark G, Fizazi K. et al. Guidelines on Testicular Cancer: 2015 Update. Eur Urol. 2015;68:1054-68
40. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-8
41. Das MK, Furu K, Evensen HF, Haugen OP, Haugen TB. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Sci Rep. 2018;8:2462
42. Yang NQ, Luo XJ, Zhang J, Wang GM, Guo JM. Crosstalk between Meg3 and miR-1297 regulates growth of testicular germ cell tumor through PTEN/PI3K/AKT pathway. Am J Transl Res. 2016;8:1091-9
43. Di Persio S, Tekath T, Siebert-Kuss LM, Cremers JF, Wistuba J, Li X. et al. Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis. Cell Rep Med. 2021;2:100395
44. Guo J, Nie X, Giebler M, Mlcochova H, Wang Y, Grow EJ. et al. The Dynamic Transcriptional Cell Atlas of Testis Development during Human Puberty. Cell Stem Cell. 2020;26:262-76 e4
45. Fan D, Qiu B, Yang XJ, Tang HL, Peng SJ, Yang P. et al. LncRNA SNHG8 promotes cell migration and invasion in breast cancer cell through miR-634/ZBTB20 axis. European review for medical and pharmacological sciences. 2020;24:11639-49
46. Liu X, Peng D, Cao Y, Zhu Y, Yin J, Zhang G. et al. Upregulated lncRNA DLX6-AS1 underpins hepatocellular carcinoma progression via the miR-513c/Cul4A/ANXA10 axis. Cancer gene therapy. 2020
47. Das MK, Furu K, Evensen HF, Haugen Ø P, Haugen TB. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Scientific reports. 2018;8:2462
48. Wei J, Gan Y, Peng D, Jiang X, Kitazawa R, Xiang Y. et al. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer medicine. 2018;7:6247-57
49. Wu X, Gao S, Wang L, Bu T, Wu S, Zhou L. et al. Role of laminin and collagen chains in human spermatogenesis - Insights from studies in rodents and scRNA-Seq transcriptome profiling. Semin Cell Dev Biol. 2022;121:125-32
50. Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Molecular oncology. 2017;11:28-39
51. Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends in cell biology. 2019;29:212-26
52. Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta biochimica et biophysica Sinica. 2017;49:588-97
53. Li N, Wang C, Zhang P, You S. Emodin inhibits pancreatic cancer EMT and invasion by up-regulating microRNA-1271. Molecular medicine reports. 2018;18:3366-74
54. Zhao GX, Xu YY, Weng SQ, Zhang S, Chen Y, Shen XZ. et al. CAPS1 promotes colorectal cancer metastasis via Snail mediated epithelial mesenchymal transformation. Oncogene. 2019;38:4574-89
55. Zhang X, Wang L, Zhang H, Tu F, Qiang Y, Nie C. Decreased expression of ZO-1 is associated with tumor metastases in liver cancer. Oncol Lett. 2019;17:1859-64
56. Neri LM, Cani A, Martelli AM, Simioni C, Junghanss C, Tabellini G. et al. Targeting the PI3K/Akt/mTOR signaling pathway in B-precursor acute lymphoblastic leukemia and its therapeutic potential. Leukemia. 2014;28:739-48
57. Brown JS, Banerji U. Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol Ther. 2017;172:101-15
58. Kumar A, Purohit R. Cancer associated E17K mutation causes rapid conformational drift in AKT1 pleckstrin homology (PH) domain. PloS one. 2013;8:e64364
59. Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA repair. 2016;42:63-71
60. Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557-66
61. Jarosz-Biej M, Smolarczyk R, Cichon T, Kulach N. Tumor Microenvironment as A "Game Changer" in Cancer Radiotherapy. Int J Mol Sci. 2019 20
62. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
63. Mantovani A, Ponzetta A, Inforzato A, Jaillon S. Innate immunity, inflammation and tumour progression: double-edged swords. J Intern Med. 2019;285:524-32
64. Zhang X, Su Y, Zhang M, Sun Z. Opposite effects of arsenic trioxide on the Nrf2 pathway in oral squamous cell carcinoma in vitro and in vivo. Cancer Lett. 2012;318:93-8
65. Liao TJ, Tsai CJ, Jang H, Fushman D, Nussinov R. RASSF5: An MST activator and tumor suppressor in vivo but opposite in vitro. Curr Opin Struct Biol. 2016;41:217-24
66. Olesen IA, Sonne SB, Hoei-Hansen CE, Rajpert-De Meyts E, Skakkebaek NE. Environment, testicular dysgenesis and carcinoma in situ testis. Best Pract Res Clin Endocrinol Metab. 2007;21:462-78
67. Kristensen DM, Sonne SB, Ottesen AM, Perrett RM, Nielsen JE, Almstrup K. et al. Origin of pluripotent germ cell tumours: the role of microenvironment during embryonic development. Mol Cell Endocrinol. 2008;288:111-8
68. Chieffi P, Chieffi S. Molecular biomarkers as potential targets for therapeutic strategies in human testicular germ cell tumors: an overview. J Cell Physiol. 2013;228:1641-6
69. Almstrup K, Hoei-Hansen CE, Wirkner U, Blake J, Schwager C, Ansorge W. et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res. 2004;64:4736-43
Author contact
![]() Corresponding authors: Hao Bo; Liqing Fan, Email: 1172881652com; liqingfanedu.cn.
Corresponding authors: Hao Bo; Liqing Fan, Email: 1172881652com; liqingfanedu.cn.

 Global reach, higher impact
Global reach, higher impact