3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(15):2867-2877. doi:10.7150/jca.87801 This issue Cite
Review
Tumor Suppressor LncRNA on Chromosome 8p12 (TSLNC8): A Concise Review in Human Malignancies
1. Department of Gastrointestinal Surgery, The Second Affiliated Hospital of Nanchang University, Nanchang 330008, Jiangxi, China.
2. Department of Spleen and Stomach Diseases, Jiujiang Hospital of Traditional Chinese Medicine, Jiujiang 332005, Jiangxi, China.
3. Second School of Clinical Medicine, Nanchang University, Nanchang 330038, Jiangxi, China.
*These two authors contributed equally to this work.
Received 2023-7-5; Accepted 2023-8-28; Published 2023-9-11
Abstract
Tumor Suppressor Long Non-Coding RNA on Chromosome 8p12 (TSLNC8) is an RNA gene that generates a long non-coding RNA transcribed intergenically from both strands. Its significant role in human malignancies attracted significant attention in recent years. Expression analysis of TSLNC8 has been conducted in tissue specimens and cell lines using various techniques, including reverse transcription-quantitative polymerase chain reaction (RT-qPCR), in situ hybridization (ISH), and microarray analysis. Furthermore, functional studies involving the loss and/or gain of TSLNC8 function in cellular and animal models have been carried out. These investigations have highlighted the impact of TSLNC8 on key tumor-related processes, including migration, invasion, and metastasis. Moreover, TSLNC8 has emerged as a regulator capable of modulating critical signaling pathways, such as the Hippo, STAT3, WNT/β-catenin, and MAPK pathways. In this review, we comprehensively synthesize the findings derived from in vitro and in vivo studies, along with analyses conducted on clinical samples, to provide a comprehensive understanding of the multifaceted role of TSLNC8 as a promising tumor biomarker and a potential target for therapeutic interventions.
Keywords: LncRNA, TSLNC8, Malignancy, Tumor biomarker, Therapeutic target
Introduction
Noncoding RNAs (ncRNAs) constitute a substantial portion (~98%) of the human transcriptome and are functionally divided into two distinct types, housekeeping and regulatory ncRNAs [1-3]. Long noncoding RNAs (lncRNAs) represent a key subclass of regulatory ncRNAs that exceed 200 nucleotides in length and lack substantial protein-coding capacity [4, 5]. Emerging as shining star, lncRNAs actively participate in chromatin remodeling, transcriptional, and posttranscriptional events [6, 7]. Through interactions with DNA, RNA, and protein molecules, lncRNAs contribute to intricate regulatory networks that are involved in a wide range of cellular processes and pathological functions [8-11]. Recent evidence continues to accumulate, shedding light on the pivotal roles of lncRNAs as either oncogenes or tumor suppressors, underscoring their potential as therapeutic targets in various disorders, with a particular emphasis on cancer [12-18].
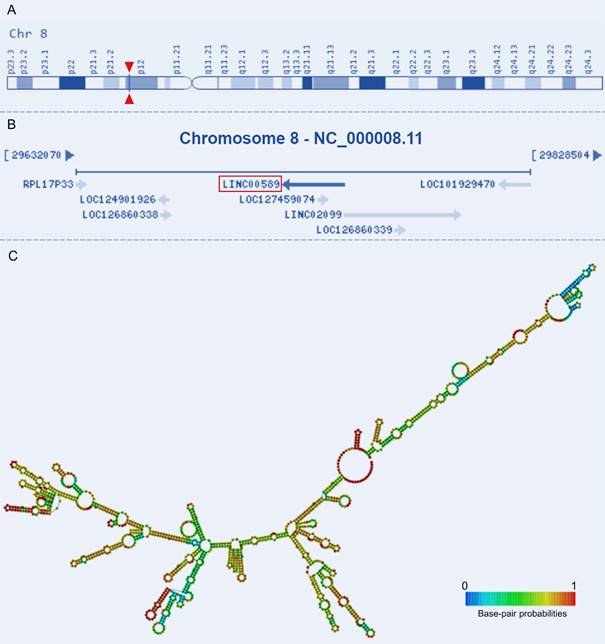
Tumor Suppressor LncRNA on Chromosome 8p12 (TSLNC8), also known as Long Intergenic Non-Protein Coding RNA 589 (LINC00589) or Chromosome 8 Open Reading Frame 75 (C8orf75), is located on Chromosome 8p12 and comprises four exons that are non-overlapping with annotated coding genes (Figure 1A-B). With a total length of 1413 bp, its secondary structure has also been characterized (Figure 1C). This lncRNA has been validated as a non-protein-coding RNA and has emerged as a key player in tumorigenesis[19, 20]. Altered expression of TSLNC8 has been detected in multiple tumorous tissues and cancer cell lines [19-22]. It also participates in a wide range of tumor-related processes and regulates drug resistance and the progression of a variety of human malignancies [19-28]. Given the observed dysregulation of this lncRNA TSLNC8 in diverse malignancies, and its reported significant associations with clinicopathology and survival outcomes [19, 20, 23, 24, 26, 27], TSLNC8 presents a promising prospect as a potential therapeutic target. Therefore, it is imperative to ascertain the molecules and pathways associated with this lncRNA in different tumor contexts. This manuscript provides a comprehensive summary of pertinent studies, categorized into three sections: investigations conducted on cell lines, studies utilizing animal models, and analyses performed on clinical samples.
TSLNC8 in cancers
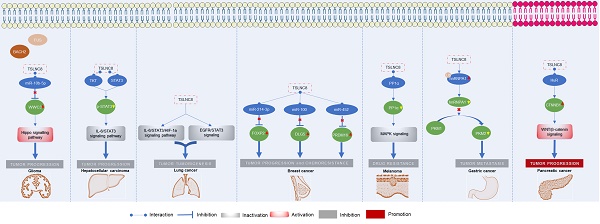
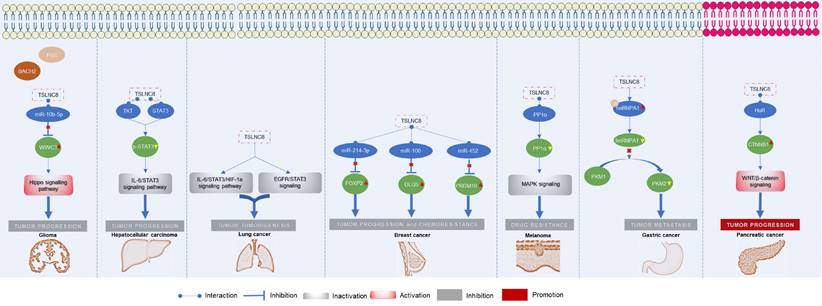
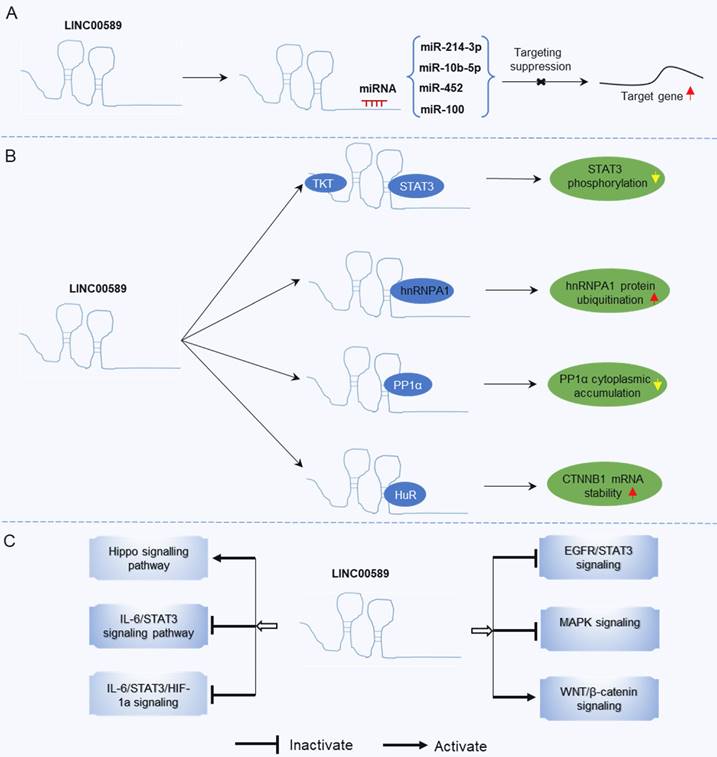
The roles of TSLNC8 have been investigated across multiple cancer types, as depicted in Figure 2. It exhibits diverse functions depending on the specific cancer context. In glioma [20], liver cancer [19], lung cancer [27], breast cancer [21], melanoma [25], and gastric cancer [23], TSLNC8 consistently acts as a tumor suppressor, effectively inhibiting tumor progression and reducing chemoresistance when overexpressed. However, in pancreatic cancer [26], the dysregulated expression of LINC00589 functions as an oncogenic driver, promoting tumor progression and metastasis. Overall, TSLNC8 functions as a regulatory molecule in tumor progression by acting as a competitive endogenous RNA (ceRNA), sequestering specific microRNAs (Figure 3A). Furthermore, it engages in interactions with specific proteins, regulation of target protein phosphorylation, ubiquitination, cytoplasmic accumulation, or stability of target mRNA (Figure 3B). Moreover, TSLNC8 is implicated in multiple signaling pathways, as depicted in Figure 3C. The functional implications of TSLNC8 in vitro and/or in vivo experiments are discussed in detail below.
Genomic view and structure of LINC00589: (A) genomic location extracted from GeneCards database (https://www.genecards.org/cgi-bin/carddisp.pl?gene=LINC00589), (B) genomic context from NCBI database (https://www.ncbi.nlm.nih.gov/gene/619351), (C) minimum free energy secondary structure extracted from RNAfold web server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi?PAGE=3&ID=PEEiuT7Ten), colored by base-pairing probability.
Summary of TSLNC8 expression and roles in tumorigenesis in cancer cell lines.
| Cell expression | In vitro experiment cell lines | Cellular functions | Related molecule/ pathway | Ref. | |||
|---|---|---|---|---|---|---|---|
| Cancer type | Up or down-regulated | Cell lines | Locations | ||||
| Glioma | Downregulated | (U251-MG, SHG-44, BT325, SWO38, CHG-5) vs. a normal astrocyte cell line | - | U251-MG, SWO38, SHG-44, BT325 | cell proliferation, migration, invasion, apoptosis | - | [20] |
| Glioma | Downregulated | (U87, U251) vs. HA cells | Both the cytoplasm and nucleus | U87, U251 | viability, migration, invasion, apoptosis | BACH2, FUS, miR-10b-5p, WWC3, Hippo signaling pathway | [28] |
| Hepatocellular carcinoma | Downregulated | Gradually decreases: Huh-7, SNU-449, Hep3B, Huh-6, HepG2, SMMC-7721, SK-Hep1, HCCLM3, PLC/PRC/5, C3A | Mainly distribution in the nucleus | SMMC-7721, SNU-449, Huh-7 | cell proliferation, invasion, migration | STAT3, TKT, IL-6, p-STAT3-Y705, P-STAT3-S727, IL-6-STAT3 signaling pathway | [19] |
| Non-small cell lung cancer | Downregulated | (A549, H441, H1975) vs. HBE | - | A549 | cell proliferation, migration, invasion, cell apoptosis, autophagy | Beclin-1, p62, ATG14, and LC3-II, IL-6/STAT3/HIF-1a pathway | [22] |
| Lung cancer | Downregulated | (H358, H460, H1975, H1299, H1395, H1650, A549) vs. MRC-5 | - | H1975, H358 | cell proliferation, apoptosis, migration, invasion | EGFR-STAT3 pathway | [27] |
| Breast cancer | Downregulated | (MDA-MB-231, HCC1559, BT549, UACC-812, and MDA-MB-453) vs. NBEC | - | MDAMB-231 | cell proliferation, G1/S phase transition | miR-214-3p, FOXP2 | [21] |
| HER2+ breast cancer | Downregulated | Trastuzumab-resistant cells vs. wild-type cells | Mostly distributed in the cytoplasm | Wild-type cells BT-474, Trastuzumab-resistant cells SKBR3 | cell viability, apoptosis, colony formation, mammosphere formation, trastuzumab resistance, CSC-like properties, and multiple chemoresistance | miR-100, miR-452, DLG5, PRDM16, MUC4 | [24] |
| Pancreatic cancer | Upregulated | (AsPC-1, Capan-2, SW1990, PANC-1, PaCa-2, BxPC-3) vs. HPDE | - | PaCa-2, PANC-1 | cell proliferation, cell invasion | HuR, CTNNB1, WNT/β-catenin signaling pathway | [26] |
| Melanoma | Downregulated | BRAF inhibitor-resistant cell lines vs. BRAF inhibitor-sensitive cells (A357P and SKMEL5) | Mainly in the nucleus | A575P, SKMEL5 | toxicity response, proliferation, apoptosis | PP1α, MAPK signaling | [25] |
| Gastric cancer | Downregulated | (MKN45, MGC-803, AGS, SGC-7901) vs. GES-1 | Predominately localized in the nucleus | MKN45, MGC803, AGS, SGC-7901 | migration, invasion, EMT | hnRNPA1, PKM1, PKM2 | [23] |
In vitro cell line assays
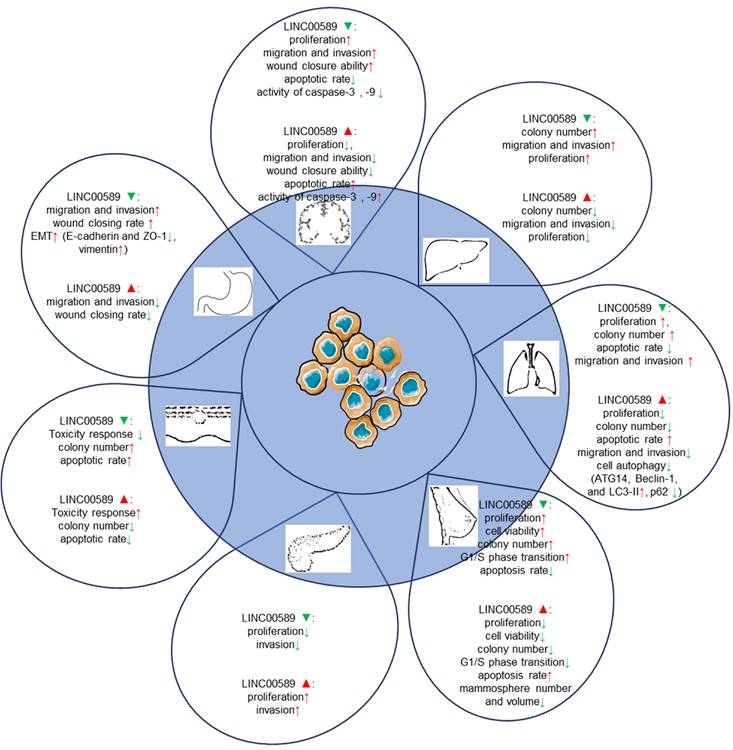
The in vitro studies conducted on TSLNC8 across different cancer types have provided valuable insights into its functional effects. These findings are summarized in Table 1 and illustrated in Figure 4.
In glioma cells, TSLNC8 overexpression resulted in decreased cell proliferation, inhibition of migration and invasion, and an increase in apoptotic rate, while TSLNC8 knockdown exhibited the opposite effects [20, 28]. The suppressive effects of TSLNC8 overexpression are mediated through competitive endogenous RNA (ceRNA) interactions with miR-10b-5p, which attenuates the repression of WWC3 by miR-10b-5p and activates the Hippo signaling pathway [28].
In vitro experiments in HCC cancer cells have revealed that overexpression of
TSLNC8 via lentiviral infection resulted in a marked suppression of colony formation and reduced proliferation rates. Conversely, silencing TSLNC8 accelerated colony formation and increased cell proliferation. TSLNC8 overexpression effectively inhibited the migration and invasion of HCC cells, while its knockdown enhanced these cellular processes. Mechanistically, TSLNC8 physically interacts with TKT and STAT3, leading to the inhibition of STAT3 phosphorylation and transcriptional activity. This interaction ultimately results in the inactivation of the IL-6/STAT3 signaling pathway, thereby contributing to the tumor-suppressive effects of TSLNC8 in HCC.
In lung cancer cells [22, 27], TSLNC8 displayed significant downregulation, whereas its overexpression resulted in the suppression of autophagy and exerted inhibitory effects on cell migration, invasion, and apoptosis promotion. Conversely, TSLNC8 knockdown showed opposite effects. Moreover, TSLNC8 exhibited a remarkable ability to inhibit the aggressive behaviors of lung cancer cells by targeting the IL-6/STAT3/HIF-1a signaling pathway [22]. Additionally, a synergistic effect was observed between TSLNC8 and the EGFR inhibitor osimertinib, effectively suppressing lung cancer tumorigenesis by blocking the EGFR-STAT3 pathway [27].
Functional mechanism of TSLNC8 as a tumor suppressor or oncogene in the initiation and progression of different tumors.
Mechanisms underlying the regulatory role of LINC00589 in tumor progression. (A) LINC00589 functions as a ceRNA, (B) LINC00589 engages in physical interactions with proteins, (C) LINC00589 participates in the modulation of signaling pathways.
In breast cancer [21], up-regulation of TSLNC8 has been shown to decrease the proliferation capacity of breast cancer cells and inhibit the transition from G1 to S phase of the cell cycle. Conversely, TSLNC8 knockdown exhibited the opposite effect. These effects are mediated through the miR-214-3p/FOXP2 axis. In HER2+ breast cancer [24], LINC00589 played a crucial role in enhancing the sensitivity of breast cancer cells to trastuzumab and suppressing anchorage-independent growth. The expression of LINC00589 also reversed cancer stem cell-like properties and reduced chemoresistance in HER2-positive breast cancer. Acting as a ceRNA platform, LINC00589 acts as a sponge for miR-100 and miR-452, thus relieving their suppression of tumor suppressors such as DLG5 and PRDM16. Through this mechanism, LINC00589 exerted multiple inhibitory functions on cancer progression and effectively counteracts drug resistance.
In pancreatic cancer cell lines [26], knockdown of TSLNC8 suppressed cell proliferation and attenuated invasiveness, while overexpression of TSLNC8 increased cell proliferation and enhanced invasion. TSLNC8 interacted with HuR, facilitating HuR's binding to CTNNB1 mRNA and enhancing its stability, ultimately activating the WNT/β-catenin signaling pathway and promoting aggressiveness in pancreatic cancer cells.
Functions of TSLNC8 upregulation and silencing in cell-based assays.
Summary of experiments in murine models to study the roles of LINC00589 in tumor development.
| Cancer type | Animal models | Groups | Experiment phenotypes | Ref. |
|---|---|---|---|---|
| Glioma | Four-week-old athymic nude mice (BALB/c) | U87 and U251 cells (control, stably expressing sh-NC+EV, sh-BACH2, sh-FUS, TSLNC8-OE and sh-BACH2+sh-FUS+TSLNC8-OE) | Tumor volume, percent survival analysis | [28] |
| HCC | Nude mice | SMCC-7721 cells infected with the lentivirus expressing TSLNC8 or the control | Tumor volume, tumor weight, metastatic nodules in the liver, lung, and intestine | [19] |
| Lung cancer | Six-week-old male BALB/c nude (nu/nu) mice | H1975 cells (control, vector, osimertnib + TSLNC8, osimertnib + vector, TSLNC8, osimertnib) | Tumor volume and weight, WB and qPCR of EGFR and phosphorylation of EGFR (Tyr1068) and STAT3 (Tyr705), IHC of EGFR and phosphorylation of STAT3 (Tyr705) | [27] |
| HER2+ breast cancer | Female athymic BALB/c nude mice (4-6 weeks) | TR breast cancer cells (Lv-NC, Lv-LINC00589, miR-NC, miR-100 mimic, or miR-452 mimic) | tumor volume, weight, luciferase activity, qPCR and IHC of DLG5 and PRDM16 | [24] |
| Pancreatic cancer | Nude mice | PaCa-2 cells (Control and TSLNC8-knockdown) | Pulmonary metastatic nodules | [26] |
| Melanoma | 4-week-old female BALB/c nude mice | TSLNC8-overexpressing or vector-transfected cells into nude mice, and treated daily with 40 mg/kg PLX4720 | Tumor weight, tumor volume, WB of p-MEK and p-ERK levels | [25] |
| Gastric cancer | Nude mice | MGC803 cells (control shRNA, sh-LINC00589); PMSNs-control and PMSNs-LINC00589 groups | Peritoneal metastatic nodules, IHC of hnRNPA1, PKM2, Ki67, or CD31 | [23] |
In melanoma [25], TSLNC8 overexpression sensitized cells to the BRAF inhibitor PLX4720, promoting apoptosis and reducing colony formation. TSLNC8 downregulation had the opposite effect, decreasing sensitivity to the inhibitor and increasing colony formation. TSLNC8 achieved its pro-sensitivity effect by binding to PP1α, leading to decreased cytoplasmic accumulation and modulation of MAPK signaling.
In gastric cancer [23], LINC00589 exhibited suppressive effects on migration and invasion of GC cells in vitro. Silencing LINC00589 enhanced the invasion and migration abilities of cancer cells and induced epithelial-mesenchymal transition (EMT). LINC00589 interacted with hnRNPA1 protein, leading to its ubiquitination and degradation, consequently inhibiting PKM2 isoform generation and suppressing carcinogenesis.
In vivo mouse model experiments
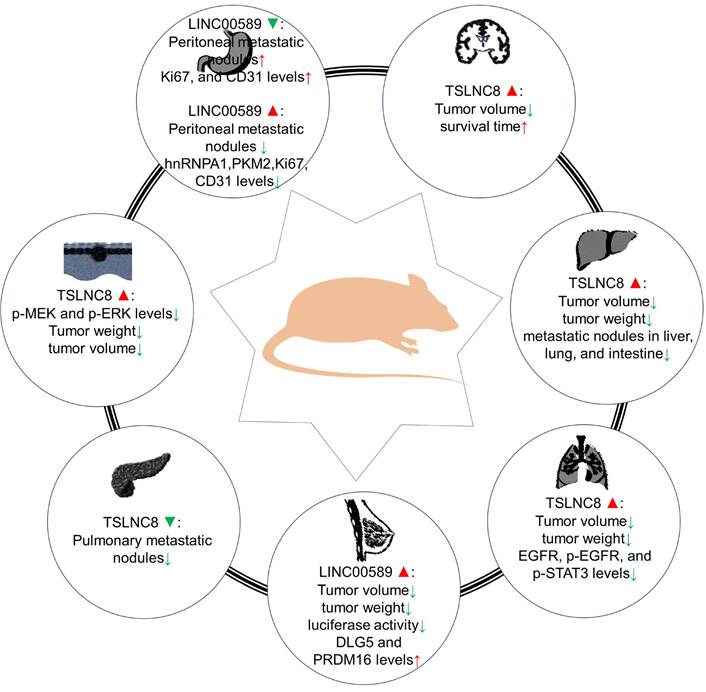
Multiple research teams have investigated the functional implications of LINC00589 up-regulation and/or silencing on tumor development using xenograft models (Table 2 and Figure 5). Similar to in vitro studies, both oncogenic and tumor suppressor role have been reported for LINC00589.
Animal models have provided evidence regarding the impact of LINC00589 modulation on different cancer types. In pancreatic cancer, LINC00589 knockdown reduced pulmonary metastatic nodules [26]. Whereas in glioma models [28], it was demonstrated that knockdown of BACH2 or FUS, overexpression of TSLNC8, or a combination of the three inhibited subcutaneous xenograft growth and prolonged survival in nude mice. In HCC [19], its upregulation was associated with decreased tumor volume, weight, and fewer metastatic nodules in the liver, lung, and intestine. In lung cancer [27], TSLNC8 overexpression led to smaller tumor volume and weight, reduced expression of EGFR, p-EGFR, and p-STAT3 levels, and combination with osimertinib administration effectively suppressed tumor growth, enhancing osimertinib's anti-tumor effects. In HER2+ breast cancer [24], LINC00589 overexpression significantly decreased tumor volume and weight, inhibited luciferase activity, and upregulated DLG5 and PRDM16 protein expressions. Additionally, LINC00589 reversed trastuzumab resistance through miR-100 and miR-452 in breast cancer, as confirmed in xenograft nude mouse models [24]. In melanoma [25], TSLNC8 overexpression diminished tumor growth rate and weight and enhanced the cytotoxic effects of the BRAF inhibitor PLX4720. In gastric cancer [23], LINC00589 knockdown resulted in peritoneal metastatic nodules and decreased Ki67 and CD31 protein levels, while its overexpression reduced the number of peritoneal metastatic nodules and decreased hnRNPA1, PKM2, Ki67, and CD31 levels.
Ex vivo clinical sample studies
Based on the existing studies, TSLNC8 expression levels exhibit diverse implications in different types of cancer. In pancreatic cancer, Chai et al. [26] reported that upregulation of TSLNC8 in cancerous tissues, which was significantly associated with advanced TNM stage, lymph node and distant metastasis, and poorer overall survival (OS). However, in most studies, TSLNC8 was found to be downregulated in cancer samples. In glioma, downregulation of TSLNC8 was linked to larger tumor size, distant metastasis, and higher TNM stage. In hepatocellular carcinoma (HCC), Zhang et al [19] found that TSLNC8 was frequently deleted and downregulated, and lower levels of TSLNC8 RNA were correlated with an increased number of tumor nodules, presence of cancer embolus, poorer differentiation stage, and shorter OS in tumor cases. In lung cancer, the expression level of TSLNC8 was correlated with gender, lymph node metastasis, and TNM stage. In breast cancer, TSLNC8 was downregulated in cancerous tissues compared to adjacent normal tissues. In HER2+ breast cancer, lower expression of LINC00589 was associated with non-response to trastuzumab, advanced TNM stage, shorter survival time and acted as an independent unfavorable prognostic factor for OS. In gastric cancer, TSLNC8 was inversely associated with aggressive pathological features, tumor prognosis, and was an independent prognostic factor. Table 3 presents a detailed summary of the expression of TSLNC8 and its associations with pathological features and prognosis in clinical tumor samples.
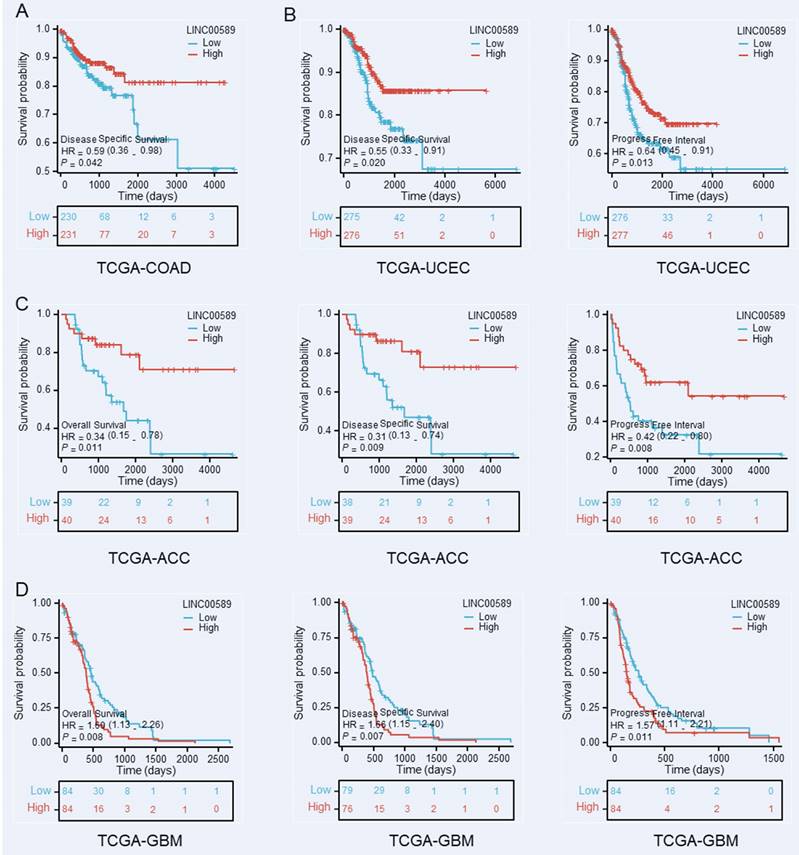
Furthermore, we investigated the relationship between LINC00589 and prognosis in various cancer types using data from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), focusing on OS, disease-specific survival (DSS), and progression-free interval (PFI). Our analysis revealed significant correlations between LINC00589 expression levels and patient prognosis in colorectal adenocarcinoma (COAD), uterine corpus endometrial carcinoma (UCEC), adrenocortical carcinoma (ACC), and glioblastoma multiforme (GBM) (Figure 6). Specifically, low expression of LINC00589 was associated with worse DSS in COAD, shorter DSS and PFI in UCEC, inferior OS, DSS, and PFI in ACC, while better OS, DSS, and PFI in GBM. These findings indicate that the role of LINC00589 may vary depending on the specific cancer type, and its expression levels have the potential to serve as valuable prognostic indicators in certain malignancies.
Roles of TSLNC8 overexpression and/or knockdown in tumorigenesis in mouse xenograft models.
Summary of the expression of TSLNC8 and its associations with pathological features and prognosis in clinical samples.
| Cancer type | Detection method | Expression (Tumor vs. Normal) | Human tissues | Tumor specimens | Significant clinical variables | End-points (analyze methods) | Prognostic biomarker | Ref. |
|---|---|---|---|---|---|---|---|---|
| Glioma | RT-qPCR | Downregulated | 80 paired tumor/adjacent noncancerous tissues | 80 glioma samples | tumor size, distant metastasis, TNM stage | - | - | [20] |
| Glioma | RT-qPCR | Downregulated | 12 for each group (normal brain tissues, grade 1-2, 3-4) | - | - | - | - | [28] |
| HCC | RNA-seq | Downregulated | TCGA (50 paired tumor/ adjacent noncancerous tissues) | - | - | - | - | [19] |
| RT-qPCR | Downregulated | 120 matched tumor/nontumor tissues | 120 HCC samples | cancer embolus, tumorous number, differentiation grade | OS (KM plot) | Favorable | ||
| genomic RT-qPCR | Downregulated | 72 paired tumor/nontumor tissues | - | - | - | - | ||
| Lung cancer | RT-qPCR | Downregulated | 31 pairs of lung cancer tissues and adjacent normal lung tissues | 31 lung cancer samples | gender, lymph node metastasis, TNM stage | - | - | [27] |
| Breast cancer | RT-qPCR | Downregulated | 10 cancer tissues and normal adjacent tissues | - | - | - | - | [21] |
| HER2+ breast cancer | RT-qPCR | Downregulated | Trastuzumab-responding (N= 38) and non-responding (N=33) breast cancer patients | - | - | - | - | [24] |
| ISH | - | - | 92 trastuzumab-treated HER2-positive breast cancer samples | TNM stage | OS (KM plot, Multivariate analysis) | Favorable | ||
| Pancreatic cancer | RT-qPCR | Upregulated | 70 paired tumor/nontumor tissues | 70 pancreatic cancer samples | TNM stage, distant and lymph node metastasis | OS (KM plot) | Unfavorable | [26] |
| Melanoma | RNA-Seq | Downregulated | GEPIA-TCGA dataset (tumor=461, normal =558) | - | - | - | - | [25] |
| RT-qPCR | Downregulated | 8 paired BRAF inhibitors pretreated and treated samples | - | - | - | - | ||
| Gastric cancer | RT-qPCR | Downregulated | 143 paired tumor/adjacent normal tissues | 143 gastric cancer samples | N stage, M stage, TNM stage | OS (KM plot, Multivariate analysis) | Favorable | [23] |
Discussion
Deletion of the short arm of chromosome 8 (8p) is a recurrent genetic abnormality observed across diverse range of cancer types [29, 30]. It is considered one of the most prevalent genetic events associated with oncogenesis. The loss of genetic material on 8p has been identified in numerous malignancies [31-35], including breast cancer [31], colorectal cancer [36, 37], prostate cancer [38], lung cancer [39], bladder cancer [33, 40], and liver cancer [41]. This deletion is implicated in tumor initiation, progression, and clinical outcomes. A recently discovered long non-coding RNA (lncRNA) called TSLNC8, which is located at the 8p12 region, has emerged as a potentially significant contributor in human tumors. This lncRNA has garnered attention for its genomic location within the same chromosomal region where frequent deletions occur in various cancer types.
LINC00589 demonstrates consistent anti-oncogenic properties across various tumor types, except for a single study focusing on pancreatic cancer. However, the precise role of LINC00589 in carcinogenesis remains uncertain, particularly in terms of its tissue-specific functions and whether the findings in pancreatic cancer deviate from its overall tumor-suppressive effects. The primary mechanism underlying the tumor-inhibitory effects of upregulated LINC00589 involves its ability to sequester oncogenic miRNAs. Notably, LINC00589 interacts with multiple miRNAs, such as miR-10b-5p, miR-214-3p, miR-100, and miR-452. Through modulation of these miRNAs, LINC00589 upregulates the expression of tumor suppressor genes such as WWC3, FOXP2, DLG5, and PRDM16. Moreover, LINC00589 interacts physically with proteins involved in the regulation of STAT3 phosphorylation, hnRNPA1 ubiquitination, and PP1α cytoplasmic accumulation. LINC00589 also exerts influence over several signaling pathways, including Hippo, IL-6/STAT3, IL-6/STAT3/HIF-1a, EGFR/STAT3, and MAPK signaling. Additionally, LINC00589 overexpression promotes tumor apoptosis and inhibits metastasis by activating autophagy and suppressing the EMT.
Kaplan-Meier survival curves demonstrate the substantial prognostic significance associated with decreased expression of LINC00589 in COAD (A), UCEC (B), ACC (C), and GBM (D).
LINC00589 has emerged as a critical mediator of drug resistance in multiple cancer types, including lung cancer [27], melanoma [25], and HER2-positive breast cancer [24]. Several researches have demonstrated the role of LINC00589 in regulating chemoresistance and resistance to targeted therapies. Notably, in melanoma [25], the upregulation of LINC00589 has been demonstrated to restore sensitivity to the BRAF inhibitor PLX4720 in resistant cells, offering a potential therapeutic avenue for patients resistant to BRAF inhibitors. Similarly, LINC00589 acts as a modulator, enhancing the efficacy of osimertinib in suppressing the progression of lung cancer [27]. Combinatorial treatment involving the overexpression of TSLNC8 and administration of osimertinib has exhibited substantial inhibition of tumor growth in preclinical models [27]. Furthermore, LINC00589 has been found to sensitize HER2-positive breast cancer cells to trastuzumab and counteract cancer stem cell-like properties, as well as chemoresistance, to various agents including 5-Fluorouracil, doxorubicin, paclitaxel, cisplatin, gemcitabine, and vincristine[24]. These observations suggest that LINC00589 overexpression could potentially lead to a reduction in tumor volume, attenuation of malignant characteristics, and enhanced responsiveness to chemotherapy and targeted therapies. However, the translation of these promising preclinical findings into clinical practice is impeded by obstacles pertaining to safety and bioavailability. Moreover, LINC00589 exhibits intricate interactions with diverse biomolecules, including miRNAs, and plays regulatory effects on multiple signaling pathways such as STAT3 and MAPK, thereby regulating drug resistance. Identification of additional tissue-specific targets will be critical for the development of more targeted therapeutics.
Interestingly, dysregulated expression of LINC00589 has been associated with patient clinical outcomes, indicating its potential as a clinically significant biomarker. Abnormal expression of LINC00589 has shown significant correlations with tumor development, including tumor grade, lymph node and distant metastasis, and tumor stage. Moreover, aberrant LINC00589 expression has been linked to patient survival and disease progression. Despite the availability of extensive data on the prognostic impact of LINC00589, its diagnostic utility, particularly in invasive body fluids such as serum and urine, remains limited. Future studies should focus on investigating the potential of LINC00589 levels to effectively differentiate between cancer patients and healthy individuals.
In summary, TSLNC8 is a versatile long non-coding RNA that plays a pivotal role in tumor development across multiple cancer types. It exerts regulatory control over crucial tumor-related processes and signaling pathways, and its potential as a tumor biomarker is also underscored, with implications for clinical features and prognostic evaluation. Given its substantial impact on carcinogenesis and treatment response, TSLNC8 emerges as a promising candidate for the development of innovative drugs aimed at improving cancer treatment outcomes. These findings strongly support the potential of TSLNC8 as both a tumor biomarker and a therapeutic target. Further investigation into the underlying mechanisms and therapeutic potential of TSLNC8 holds significant promise for advancing cancer treatment strategies in the future.
Abbreviations
LncRNAs: Long noncoding RNAs; TSLNC8: Tumor Suppressor LncRNA on Chromosome 8p12; LINC00589: Long Intergenic Non-Protein Coding RNA 589; C8orf75: Chromosome 8 Open Reading Frame 75; ceRNA: Competitive endogenous RNA; EMT: Epithelial-mesenchymal transition; OS: Overall survival; HCC: Hepatocellular carcinoma; TCGA: The Cancer Genome Atlas; DSS: Disease-specific survival; PFI: Progression-free interval; COAD: Colorectal adenocarcinoma; UCEC: Uterine corpus endometrial carcinoma; ACC: Adrenocortical carcinoma; GBM: Glioblastoma multiforme.
Acknowledgements
Author contributions
HLL conceived, reviewed, and edited reviews; XL, HH, MCL, and HLL retrieved papers and analyzed data. XL, HH, and HLL wrote the manuscript; Final draft read and approved by all authors.
Availability of data and materials
The datasets analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/). And the datasets used and/or analyzed are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Palazzo AF, Koonin EV. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell. 2020;183:1151-61
2. Zhang P, Wu W, Chen Q, Chen M. Non-Coding RNAs and their Integrated Networks. Journal of integrative bioinformatics. 2019;16:20190027
3. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Frontiers in genetics. 2015;6:2
4. Perkel JM. Visiting "noncodarnia". BioTechniques. 2013;54:301 3-4
5. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nature reviews Molecular cell biology. 2023;24:430-47
6. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics, proteomics & bioinformatics. 2016;14:42-54
7. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y. et al. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Frontiers in oncology. 2020;10:598817
8. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature reviews Molecular cell biology. 2021;22:96-118
9. Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics, proteomics & bioinformatics. 2017;15:177-86
10. Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochimica et biophysica acta Gene regulatory mechanisms. 2020;1863:194417
11. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. International journal of molecular sciences. 2018;19:1310
12. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nature reviews Cancer. 2018;18:5-18
13. Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnology & genetic engineering reviews. 2018;34:153-80
14. Li R, Wang X, Zhu C, Wang K. lncRNA PVT1: a novel oncogene in multiple cancers. Cellular & molecular biology letters. 2022;27:84
15. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-7
16. Yang J, Qi M, Fei X, Wang X, Wang K. LncRNA H19: A novel oncogene in multiple cancers. International journal of biological sciences. 2021;17:3188-208
17. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Akbari Dilmaghani N. A review on the role of PTENP1 in human disorders with an especial focus on tumor suppressor role of this lncRNA. Cancer cell international. 2022;22:207
18. Xu J, Wang X, Zhu C, Wang K. A review of current evidence about lncRNA MEG3: A tumor suppressor in multiple cancers. Frontiers in cell and developmental biology. 2022;10:997633
19. Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D. et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology (Baltimore, Md). 2018;67:171-87
20. Chen D, Yu X. Long noncoding RNA TSLNC8 suppresses cell proliferation and metastasis and promotes cell apoptosis in human glioma. Molecular medicine reports. 2018;18:5536-44
21. Qin CX, Yang XQ, Jin GC, Zhan ZY. LncRNA TSLNC8 inhibits proliferation of breast cancer cell through the miR-214-3p/FOXP2 axis. European review for medical and pharmacological sciences. 2019;23:8440-8
22. Fan H, Li J, Wang J, Hu Z. Long Non-Coding RNAs (lncRNAs) Tumor-Suppressive Role of lncRNA on Chromosome 8p12 (TSLNC8) Inhibits Tumor Metastasis and Promotes Apoptosis by Regulating Interleukin 6 (IL-6)/Signal Transducer and Activator of Transcription 3 (STAT3)/Hypoxia-Inducible Factor 1-alpha (HIF-1α) Signaling Pathway in Non-Small Cell Lung Cancer. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:7624-33
23. Wang S, Wo L, Zhang Z, Zhu C, Wang C, Wang Y. et al. Delivery of LINC00589 via mesoporous silica nanoparticles inhibits peritoneal metastasis in gastric cancer. Cancer letters. 2022;549:215916
24. Bai W, Peng H, Zhang J, Zhao Y, Li Z, Feng X. et al. LINC00589-dominated ceRNA networks regulate multiple chemoresistance and cancer stem cell-like properties in HER2(+) breast cancer. NPJ breast cancer. 2022;8:115
25. Han Y, Fang J, Xiao Z, Deng J, Zhang M, Gu L. Downregulation of lncRNA TSLNC8 promotes melanoma resistance to BRAF inhibitor PLX4720 through binding with PP1α to re-activate MAPK signaling. Journal of cancer research and clinical oncology. 2021;147:767-77
26. Chai W, Liu R, Li F, Zhang Z, Lei B. Long noncoding RNA TSLNC8 enhances pancreatic cancer aggressiveness by regulating CTNNB1 expression via association with HuR. Human cell. 2021;34:165-76
27. Zhou SZ, Li H, Wang ZW, Wang MH, Li N, Wang YF. LncRNA TSLNC8 synergizes with EGFR inhibitor osimertinib to inhibit lung cancer tumorigenesis by blocking the EGFR-STAT3 pathway. Cell cycle (Georgetown, Tex). 2020;19:2776-92
28. Yang Y, Liu X, Zheng J, Xue Y, Liu L, Ma J. et al. Interaction of BACH2 with FUS promotes malignant progression of glioma cells via the TSLNC8-miR-10b-5p-WWC3 pathway. Molecular oncology. 2020;14:2936-59
29. Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S. et al. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer research. 1992;52:5368-72
30. Baffa R, Santoro R, Bullrich F, Mandes B, Ishii H, Croce CM. Definition and refinement of chromosome 8p regions of loss of heterozygosity in gastric cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6:1372-7
31. Yaremko ML, Kutza C, Lyzak J, Mick R, Recant WM, Westbrook CA. Loss of heterozygosity from the short arm of chromosome 8 is associated with invasive behavior in breast cancer. Genes, chromosomes & cancer. 1996;16:189-95
32. Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M. et al. Loss of Chromosome 8p Governs Tumor Progression and Drug Response by Altering Lipid Metabolism. Cancer cell. 2016;29:751-66
33. Stoehr R, Wissmann C, Suzuki H, Knuechel R, Krieg RC, Klopocki E. et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Laboratory investigation; a journal of technical methods and pathology. 2004;84:465-78
34. Chen M, Yang Y, Liu Y, Chen C. The Role of Chromosome Deletions in Human Cancers. Advances in experimental medicine and biology. 2018;1044:135-48
35. Ishii H, Baffa R, Numata SI, Murakumo Y, Rattan S, Inoue H. et al. The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3928-33
36. Friedman JB, Brunschwig EB, Platzer P, Wilson K, Markowitz SD. C8orf4 is a transforming growth factor B induced transcript downregulated in metastatic colon cancer. International journal of cancer. 2004;111:72-5
37. Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T. et al. Isolation of a novel gene on 8p21.3-22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes, chromosomes & cancer. 2000;29:9-15
38. Matsuyama H, Pan Y, Oba K, Yoshihiro S, Matsuda K, Hägarth L. et al. Deletions on chromosome 8p22 may predict disease progression as well as pathological staging in prostate cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2001;7:3139-43
39. Nonaka D, Fabbri A, Roz L, Mariani L, Vecchione A, Moore GW. et al. Reduced FEZ1/LZTS1 expression and outcome prediction in lung cancer. Cancer research. 2005;65:1207-12
40. Choi C, Kim MH, Juhng SW, Oh BR. Loss of heterozygosity at chromosome segments 8p22 and 8p11.2-21.1 in transitional-cell carcinoma of the urinary bladder. International journal of cancer. 2000;86:501-5
41. Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C. et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes & development. 2008;22:1439-44
Author contact
![]() Corresponding author: Hongliang Luo, email: ndefy13028edu.cn.
Corresponding author: Hongliang Luo, email: ndefy13028edu.cn.

 Global reach, higher impact
Global reach, higher impact