3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(3):714-728. doi:10.7150/jca.91033 This issue Cite
Review
A concise review of the regulatory, diagnostic, and prognostic implications of HOXB-AS3 in tumors
1. Department of Gastrointestinal Surgery, The Second Affiliated Hospital of Nanchang University, Nanchang 330000, Jiangxi, China.
2. Department of Traditional Chinese Medicine, Jiujiang Hospital of Traditional Chinese Medicine, Jiujiang 332007, Jiangxi, China.
3. Nanchang University Queen Mary School, Nanchang 330038, Jiangxi, China.
4. The Second Clinical Medical College, Nanchang University, Nanchang 330038, Jiangxi, China.
# These two authors contributed equally to this work.
Received 2023-10-12; Accepted 2023-11-28; Published 2024-1-1
Abstract
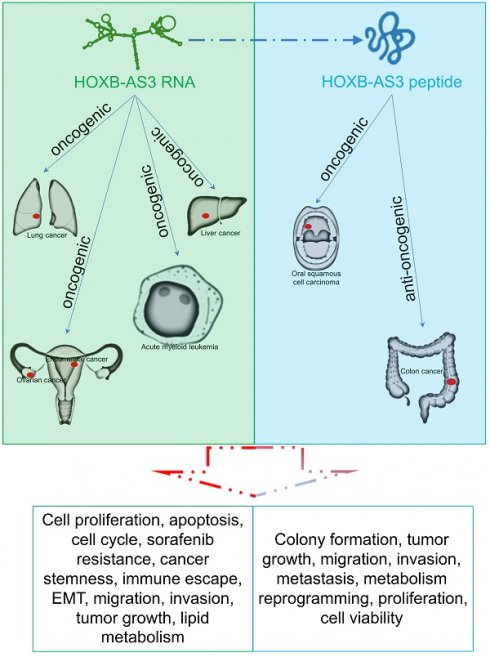
Recent studies have reported that HOXB-AS3 (HOXB Cluster Antisense RNA 3) is an intriguing molecule with dual functionality as a long noncoding RNA (lncRNA) and putative coding peptide in tumorigenesis and progression. The significant expression alterations of HOXB-AS3 were detected in diverse cancer types and closely correlated with clinical stage and patient survival. Furthermore, HOXB-AS3 was involved in a spectrum of biological processes in solid tumors and hematological malignancies, such as stemness, lipid metabolism, migration, invasion, and tumor growth. This review comprehensively analyzes its clinical relevance for diagnosis and prognosis across human tumors and summarizes its functional role and regulatory mechanisms in different malignant tumors, including liver cancer, acute myeloid leukemia, ovarian cancer, lung cancer, endometrial carcinoma, colon cancer, and oral squamous cell carcinoma. Overall, HOXB-AS3 emerges as a promising biomarker and novel therapeutic target in multiple human tumors.
Keywords: HOXB-AS3, human tumors, clinical significance, molecular target, biological functions, regulatory mechanisms
Introduction
Cancer, a leading cause of global mortality, continues to be a significant public health concern. According to recent data, in 2020, there were approximately 19.3 million new cancer cases and 10 million deaths worldwide [1, 2]. This represents a substantial increase from the reported figures in 1990. In China, there were an estimated 4.82 million new cancer cases and 3.21 million cancer-related deaths in 2022 [3]. Similarly, in the United States, it is projected that there were around 1.92 million new cancer cases and 0.61 million deaths in 2022 [4]. The rising incidence and mortality rates of cancer highlight the urgent need for innovative approaches in diagnosis, prognosis, and treatment.
Extensive research efforts have been dedicated to the identification of biomarkers and molecular targets for cancer therapy [5-9]. Among these, the exploration of long non-coding RNAs (lncRNAs) has garnered significant attention [10, 11]. LncRNAs are a type of RNA with a length exceeding 200 nucleotides that do not encode proteins or have limited protein-coding ability [12]. They can be broadly categorized as sense, antisense, intronic, bidirectional, or intergenic, depending on their relationship with neighboring protein-coding genes [12, 13]. In recent years, lncRNAs have gained increasing importance in cancer research, as considerable evidence [14-18] suggests their involvement in tumor initiation and progression by influencing various cellular processes, including cell cycle, survival, tumor invasion, and metastasis through multiple mechanisms at the transcriptional, post-transcriptional, or epigenetic levels. Importantly, lncRNAs have demonstrated promise as potential diagnostic and prognostic markers, as well as therapeutic targets in various types of cancer [19-22].
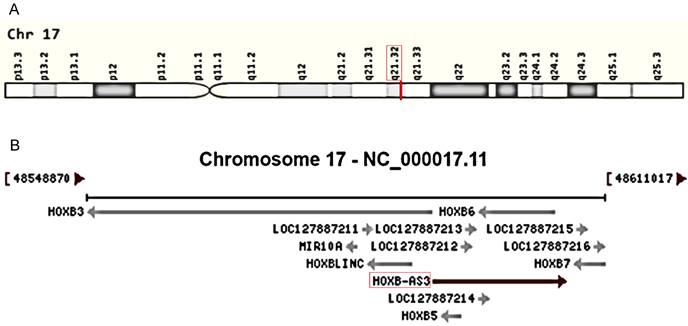
HOX genes, encompassing the HOXA, HOXB, HOXC, and HOXD gene clusters, play a pivotal role in tumorigenesis and serve as valuable drug targets in disease treatment [23-26]. Recently, there has been an increasing focus on lncRNAs associated with these genes in various aspects of cancer, including HOXA-AS3 [27-31], HOTTIP [32-35], and HOTAIR [36-39]. Among them, HOXB-AS3 (HOXB Cluster Antisense RNA 3) has emerged as a novel antisense lncRNA transcribed from the human chromosome 17q21.32 (Figure 1A), overlapping with the HOXB5 and HOXB6 genes on the sense strands (Figure 1B). Since its discovery, HOXB-AS3 has been implicated in cancers. Initially, HOXB-AS3, which is annotated as a lncRNA, was found actually encodes a small peptide that is reported to participate in suppressing colon cancer growth [40], however, subsequent studies primarily focused on the RNA level and revealed tumor-promoting properties of HOXB-AS3 RNA [41-47]. For instance, in liver cancer [47], lncRNA HOXB-AS3 is significantly upregulated in cancerous samples, particularly in the advanced stage. This upregulation enhances tumor cell proliferation while inhibiting apoptosis. Similarly, in endometrial carcinoma (EC) [42], both cancer tissues and EC cell lines exhibit overexpression of lncRNA HOXB-AS3, and lncRNA HOXB-AS3 correlates with survival rates in EC patients. HOXB-AS3 overexpression could promote EC progression [42]. Furthermore, research also unveiled the regulatory role of HOXB-AS3 on tumor development, involving multiple pathways [43, 44, 47, 48]. HOXB-AS3 demonstrates promising potential as an oncological biomarker and therapeutic target in human malignant disorders.
In this review, we systematically searched PubMed and Google Scholar using terms related to HOXB-AS3 and manually screened relevant full-text studies of human malignancies. By synthesizing the results of these studies and combining multiple online databases, we systematically analyzed the aberrant expression of HOXB-AS3 and its clinical relevance in the prognostic and diagnostic value in different human tumors and also outlined the current understanding of the biological functions and regulatory mechanisms of the HOXB-AS3 lncRNA and its encoded micro-peptides in multiple malignant tumors.
Promising Clinical Biomarker in Human Cancer
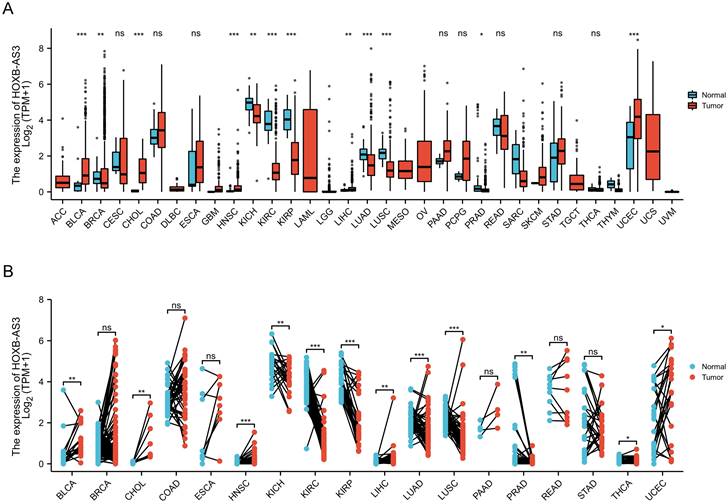
Recent research studies have shown the aberrant expression of HOXB-AS3 in tumor tissues and cancer cell lines, and significant correlations have been established between the dysregulation of HOXB-AS3 and specific clinicopathological characteristics, along with the prognosis of patients across multiple types of tumors, as presented in Table 1. In this section, we aim to provide a comprehensive overview and investigation into the changes observed in the expression of HOXB-AS3 across human malignancies. Additionally, we explore the relationship between HOXB-AS3 and patient prognoses, such as overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI), and discuss its potential as a valuable diagnostic biomarker for different tumor types.
Genomic view for HOXB-AS3 gene for genomic location (A) extracted from GeneCards database (https://www.genecards.org/cgi-bin/carddisp.pl?gene=HOXB-AS3&keywords=HOXB-AS3), and genomic context (B) from NCBI database (https://www.ncbi.nlm.nih.gov/gene/404266).
Expression of HOXB-AS3 in tissues and cells and its relationship with clinical characteristics and survival in malignancies.
| Tumor type | RNA or protein | Expression level | Models | Clinical characteristic | Survival indicator | Prognosis with high expression | Biomarker | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Cell line | Main subcellular location | ||||||||
| Hepatoma | RNA | Up | Human tissues | HepG, PLC, Hep3B, LM3 | Nucleus (Hep3B cells) | TNM stage | - | - | - | [47] |
| Liver cancer | RNA | Up | Human tissues | - | - | - | - | - | - | [49] |
| Acute myeloid leukemia | RNA | Up | Human tissues, TCGA | OCI/AML3, TF-1 | Cytoplasm (OCI/AML3 cells) | Sex, age, PLT, cytogenetic risk group S, relapse rate, somatic gene mutations | Overall survival, Relapse free survival (human) | Adverse | Prognostic | [46] |
| NPM1-mutated acute myeloid leukemia | RNA | Up | Human tissues | Kasumi-1, KG-1a, K-562, MV-4-11, THP-1, MOLM-13, OCI-AML3 | Nuclear (OCI-AML3 cells) | - | Overall survival (murine PDX model) | Adverse | Prognostic | [45] |
| Acute myeloid leukemia | RNA | Up | Human tissues, GSE dataset, TARGET, TCGA | HS-5, THP-1, leukemic stem cells (isolated from THP-1 cells) | - | - | Overall survival (human and murine model) | Adverse | Prognostic | [50] |
| Epithelial ovarian cancer | RNA | Up | Patient specimens, TCGA, GEPIA | HEY, SKOV3, OVCAR3, IOSE80 | - | Histological grade, FIGO stage, lymph node metastasis | Overall survival (human) | Adverse | Prognostic | [44] |
| Epithelial ovarian cancer | RNA | Up | Human sample, TCGA | - | - | Disease-free status and overall survival status | Disease-free survival, Overall survival (human) | Adverse | Prognostic | [41] |
| Lung cancer | RNA | Up | Human sample | H1795, SPC-A1, H460, A549, 16HBE | - | Differentiation, TNM | - | - | - | [43] |
| Endometrial carcinoma | RNA | Up | Human sample, TCGA | HEC-1-A, HEC-1-B, Ishikawa, ESC | - | - | Overall survival (human) | - | Prognostic | [42] |
| Uterine corpus endometrial carcinoma | RNA | Up | - | hEM15A, HEC1A, | - | - | - | - | - | [51] |
| Endometrioid carcinoma | RNA | Up | Human tissues, GSE dataset, TCGA | Ishikawa, AN3-CA, HEC-1A, RL95-2 | - | - | - | - | Diagnostic | [48] |
| Colon cancer | RNA | Down | Human sample, GEO dataset | SW480, SW620, HCT116, HCT116high | - | - | - | - | - | [40] |
| Micro‑peptide | Down | Human sample | SW480, SW620, HCT116, HCT116high | Nucleus (SW480, SW620) | Clinical stage | Overall survival (human) | Favorable | Prognostic | ||
| Oral squamous cell carcinoma | RNA | Up | Human sample, TCGA | - | - | TNM stage | Overall survival (human) | Adverse | Prognostic | [52] |
| Micro‑peptide | Up | Human sample | - | - | - | - | - | - | ||
Gene Expression Across Human Cancers
In recent studies, aberrant expression of the HOXB-AS3 RNA has been found in liver cancer [47, 49], lung cancer [43], ovarian cancer [41, 44], endometrial cancer [42, 48, 51], and acute myeloid leukemia [45, 46, 50]. And at the protein expression level, abnormal expression of the HOXB-AS3 micro-peptide has been reported in oral squamous cell carcinoma [52] and colon cancer [40] (Table 1).
The Interactive Bodymap in GEPIA 2 (http://gepia2.cancer-pku.cn/) displayed the expression profile of the HOXB-AS3 RNA transcript across tumor and normal tissues (Figure 2A). Among the tumor samples, KICH exhibited the highest RNA transcript expression (Figure 2B), while among the paired normal tissues, the normal tissues adjacent to READ showed the highest RNA transcript expression (Figure 2C). To compare the expression levels between tumor and normal samples, the HOXB-AS3 expression levels were assessed in pan-cancers, as well as in paired normal and tumor samples, using UCSC XENA (https://xenabrowser.net/datapages/) (Figure 3A and B). The results demonstrated that HOXB-AS3 was significantly up-regulated in a number of malignancies, such as BLCA, CHOL, HNSC, LIHC, and UCEC. Conversely, it was noticeably downregulated in several types of tumors, such as KICH, KIRC, KIRP, and PRAD. The differential expression patterns observed across various cancer types suggest that HOXB-AS3 expression levels may hold significant clinical value in predicting disease onset and progression.
Prognostic and Diagnostic Potential
The clinical value of HOXB-AS3 as a predictive biomarker has been reported in multiple studies (Table 1). HOXB-AS3 expression levels have been found to correlate with clinical features across several cancers (Table 1). For instance, in epithelial ovarian cancer, overexpression of HOXB-AS3 RNA was distinctly associated with higher histological grade, advanced FIGO stage and lymph node metastasis [44]. Similarly, in lung cancer, the expression level of HOXB-AS3 RNA in tissues was closely linked to tumor differentiation and TNM stage [43]. Furthermore, investigations have explored the relationship between HOXB-AS3 expression and patient prognosis in various tumors (Table 1). In acute myeloid leukemia [46], epithelial ovarian cancer [44], endometrial carcinoma [42], and oral squamous cell carcinoma [52], higher expression of HOXB-AS3 RNA indicated a poorer prognosis. Conversely, in colon cancer, higher expression of the HOXB-AS3 micro‑peptide was associated with longer survival time [40].
Comprehensive Expression Profile of HOXB-AS3 Isoforms Across Human Tissues. Analysis of HOXB-AS3 gene expression encompassing all identified isoforms, utilizing data from GEPIA 2: Interactive Bodymap delineates the median expression of HOXB-AS3 in tumor and normal samples (A). Bar plots provide a detailed gene expression profile across tumor samples (B) and corresponding normal tissues (C). The acronyms used in this figure are listed in Supplementary Table 1 for reference.
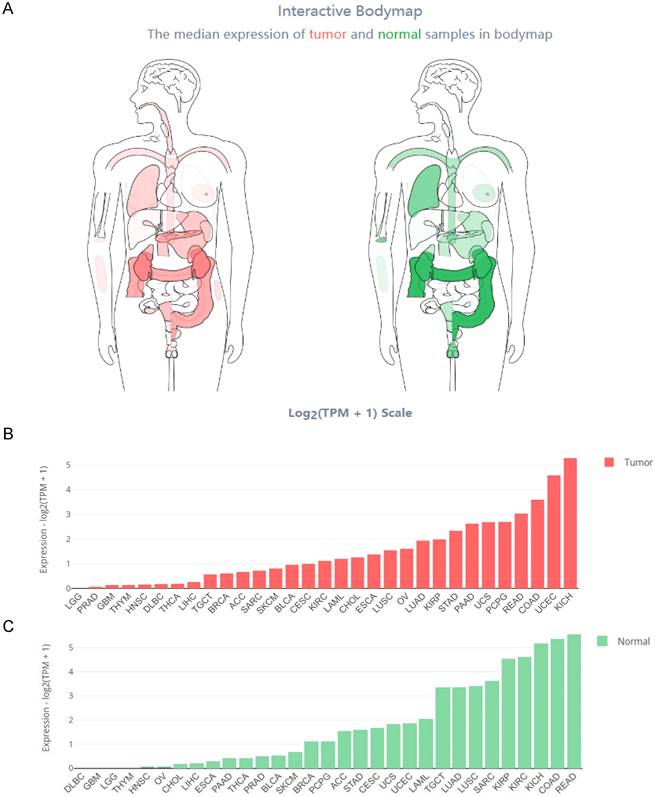
The relative expression level of HOXB-AS3 assessed in pan-cancers (A), as well as in paired normal and tumor samples (B). The acronyms used in this figure are listed in Supplementary Table 1 for reference.
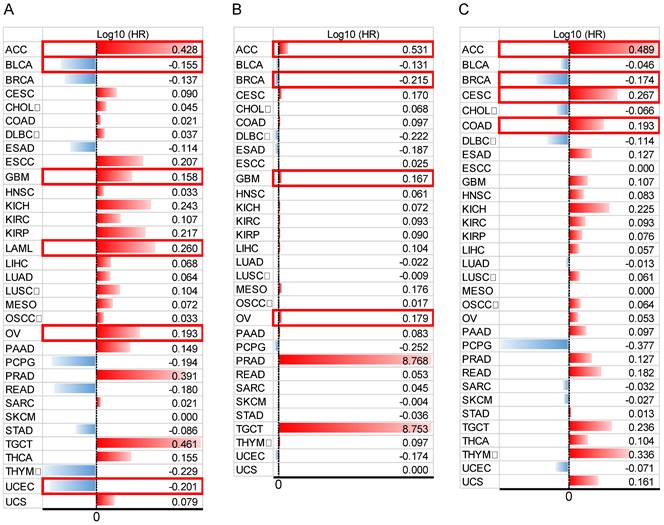
In addition, to comprehensively evaluate the prognostic significance of HOXB-AS3 in pan-cancer, we conducted an extensive analysis using publicly available datasets from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) (Figure 4A - C). Regarding OS (Figure 4A), we observed that HOXB-AS3 overexpression was associated with an unfavorable prognosis in ACC, GBM, LAML, and OV. Conversely, it acted as a favorable predictor in BLCA and UCEC. In terms of DSS (Figure 4B), high expression of HOXB-AS3 was linked to an unfavorable outcome in ACC, GBM, and OV, while it served as a favorable predictor in BRCA.
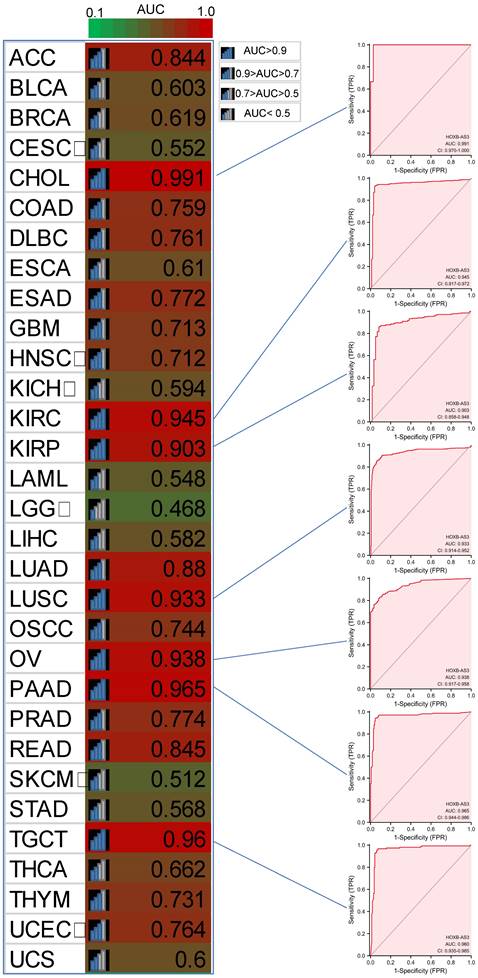
Furthermore, in PFI analysis (Figure 4C), HOXB-AS3 overexpression was indicative of an unfavorable prognosis in ACC, CESC, and COAD. Conversely, it acted as a favorable predictive factor in BRCA. Furthermore, receiver operating characteristic (ROC) curve analyses were also performed (Figure 5), and interestingly, it revealed that HOXB-AS3 expression could serve as a potent diagnostic biomarker in multiple cancer types, particularly in CHOL, KIRC, KIRP, LUSC, OV, PAAD, and TGCT with an area under the curve (AUC) exceeding 0.9. All above results indicated that HOXB-AS3 holds promise as both a diagnostic and prognostic predictor in diverse malignancies.
Role and Mechanisms of HOXB-AS3 in Cancer
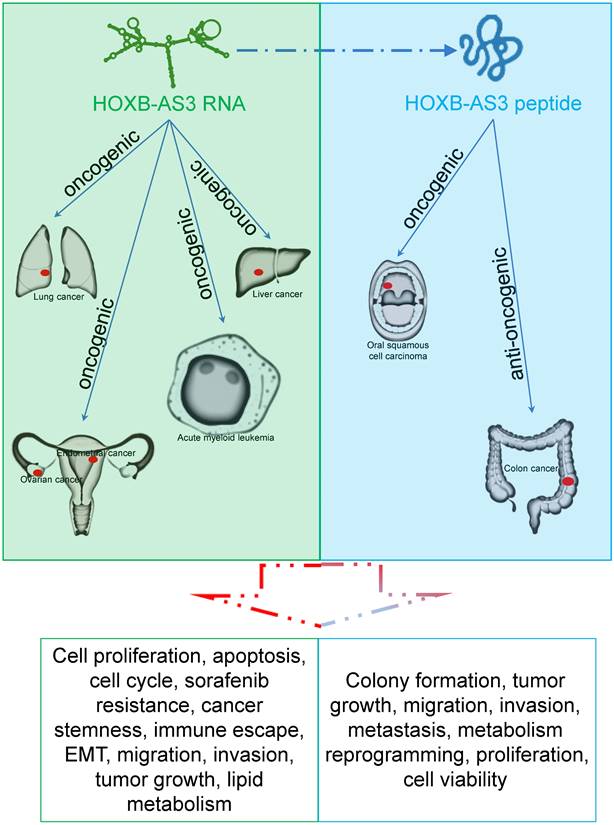
HOXB-AS3 RNA and its encoded micro-peptide have both been reported to play crucial roles in regulating a wide range of biological functions that impact tumor initiation and progression. These functions include cell proliferation, viability, apoptosis, cell cycle, migration, invasion, metastasis, tumor growth, epithelial-mesenchymal transition (EMT), stemness, immune escape, and lipid metabolic reprogramming (Table 2 and Figure 6). Considering these findings, HOXB-AS3 holds promise as a novel therapeutic target for human cancers.
HOXB-AS3 lncRNA: The Noncoding Dimension
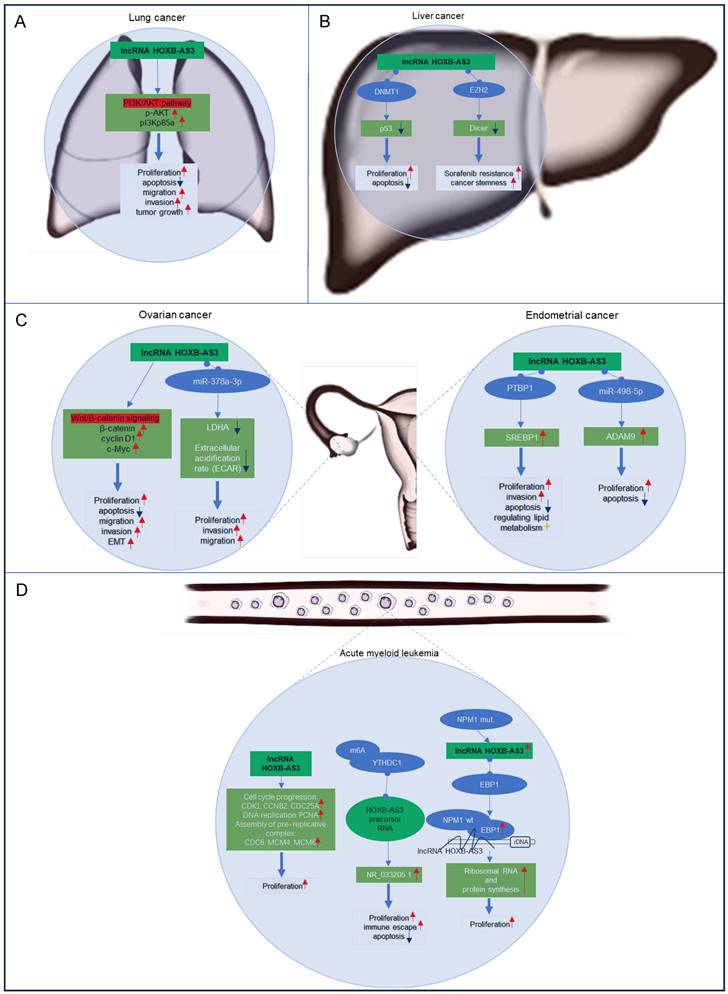
Currently most studies have focused on the functional significance of lncRNA HOXB-AS3 across various cancer types, encompassing lung cancer [43], liver cancer [47, 49], ovarian cancer [41, 44], endometrial cancer [42, 48, 51], and acute myeloid leukemia [45, 46, 50]. The regulatory mechanisms of lncRNA HOXB-AS3 in these tumors are delineated in Figure 7. This section aims to provide an overview of the distinct regulatory mechanisms attributed to lncRNA HOXB-AS3 in different tumor contexts.
Lung Cancer
Lung cancer ranks as the primary cause of tumor-related mortality globally. Increased expression of HOXB-AS3 RNA has been observed in lung cancer tissues and cell lines, exhibiting a noteworthy association with tumor differentiation and TNM stage [43]. Functionally, in vitro investigations have revealed that inhibition of HOXB-AS3 leads to suppressed cell proliferation, colony formation, induced apoptosis, and reduced migration and invasion capabilities [43]. Additionally, in vivo experiments have validated the involvement of HOXB-AS3 in promoting tumor growth through the activation of the PI3K-AKT pathway [43]. These findings highlight the potential of targeting HOXB-AS3 as a promising therapeutic approach for lung cancer.
Liver Cancer
Liver cancer is a highly prevalent and aggressive malignancy. The expression of HOXB-AS3 is significantly upregulated in liver cancer tissues compared to adjacent normal tissues, showing a positive correlation with tumor stage [47]. Functional studies have revealed that interfering with HOXB-AS3 expression affects hepatoma cell proliferation, apoptosis, cancer stemness, and sorafenib resistance [47, 49]. Mechanistically, HOXB-AS3 binds to DNMT1, leading to the downregulation of the tumor suppressor gene p53 and promoting tumor cell proliferation [47]. Additionally, HOXB-AS3 epigenetically suppresses dicer expression by recruiting EZH2, resulting in stem-like cell properties and increased resistance to sorafenib [49]. These findings stressed the role of HOXB-AS3 in hepatocellular carcinoma progression and its potential as a therapeutic target for improving treatment outcomes.
Data bars showing overall survival (OS) (A), disease-specific survival (DSS) (B), and progression-free interval (PFI) (C) in pan-cancer for HOXB-AS3. The horizontal bars represent the data, while the corresponding log10 (hazard ratio) (HR) values are presented on the far right. The cut-off value was determined using the median of HOXB-AS3 expression. Statistical significance was established at a p-value of < 0.05 and is marked with a red outer box. The acronyms used in this figure are listed in Supplementary Table 1 for reference.
Diagnostic ROC curves for HOXB-AS3 expression in normal and cancer tissues using UCSC XENA datasets. The acronyms used in this figure are listed in Supplementary Table 1 for reference.
The role and regulatory mechanism of HOXB-AS3 RNA and micro-peptide in human malignancies.
| Level | Cancer type | Role | Experiments | In vitro | In vivo | Functions | Related molecule/signal | Ref. |
|---|---|---|---|---|---|---|---|---|
| RNA | Hepatoma | Oncogenic | In vitro | Hep3B, LM3 | - | Cell proliferation, apoptosis, cell cycle | p53, DNMT1 | [47] |
| Liver cancer | Oncogenic | In vitro and in vivo | Huh7, Huh7/SR | xenograft mouse model (SCID mice) | Sorafenib resistance, cancer stemness | EZH2, Dicer, H3K27me3, SOX2, OCT4 | [49] | |
| Acute myeloid leukemia | Oncogenic | In vitro | OCI/AML3, TF-1 | - | Cell proliferation | CDK1, CCNB2, CDC25A, PCNA, CDC6, MCM4, MCM6 | [46] | |
| NPM1-mutated acute myeloid leukemia | Oncogenic | In vitro and in vivo | Leukemic stem cells | Human AML patient-derived xenografts (NSG mice) | Proliferation | EBP1, NPM1 | [45] | |
| Acute myeloid leukemia | Oncogenic | In vitro and in vivo | THP-1 cell, leukemic stem cells | Xenograft mouse model (NSG mice) | Proliferation, apoptosis, immune escape | YTHDC1, NR_033205.1 | [50] | |
| Epithelial ovarian cancer | Oncogenic | In vitro | SKOV-3, OVCAR-3 | - | Cell proliferation, migration, invasion, EMT, apoptosis | β-catenin, cyclin D1, c-Myc, Wnt/β-catenin signaling | [44] | |
| Epithelial ovarian cancer | Oncogenic | In vitro | SKOV3, A2780 | - | Proliferation, invasion, migration | LDHA, miR-378a-3p | [41] | |
| Lung cancer | Oncogenic | In vitro and in vivo | A549, H1975 | Xenograft mouse model (BALB/c nude mice) | Cell proliferation, apoptosis, cell cycle, migration, invasion, tumor growth | p-AKT, pI3Kp85a, PI3K/AKT pathway | [43] | |
| Endometrial carcinoma | Oncogenic | In vitro | HEC1A, Ishikawa cells | - | Cell proliferation, apoptosis | ADAM9, miR-498-5p | [42] | |
| Uterine corpus endometrial carcinoma | Oncogenic | In vitro | HEC1A cells | - | Proliferation, migration | - | [51] | |
| Endometrioid carcinoma | Oncogenic | In vitro | Ishikawa, AN3-CA, HEC-1A, RL95-2 | - | Proliferation, invasion, cell cycle, apoptosis, lipid metabolism | PTBP1, SREBP1 | [48] | |
| Micro-peptide | Colon cancer | Anti-oncogenic | In vitro and in vivo | SW480, HTC-116, SW620 | Xenograft mouse model (NOD-SCID mice) | Colony formation, tumor growth, migration, invasion, metastasis, metabolic reprogramming | hnRNPA1, PKM2, PKM1 | [40] |
| Oral squamous cell carcinoma | Oncogenic | In vitro | Cal-27, UM2 | - | Proliferation, viability | IGF2BP2, c-Myc | [52] |
Ovarian and Endometrial Carcinomas
Ovarian cancer and endometrial carcinoma are the two most common gynecologic tumors, and the elevated expression of lncRNA HOXB-AS3 has been observed in both cancer specimens and cancer cell lines associated with these two cancers [41, 42, 44, 48]. In ovarian cancer, elevated levels of HOXB-AS3 indicated an adverse prognosis [41, 44], and HOXB-AS3 overexpression is linked to higher histological grade, advanced FIGO stage, and lymph node metastasis in patients [44]. And HOXB-AS3 plays a critical role in ovarian cancer tumorigenesis by activating the Wnt/β-catenin signaling pathway [44] or acting as a miR-378a-3p sponge [41]. These molecular mechanisms impact essential cellular processes, including proliferation, apoptosis, migration, invasion, and EMT, underscoring its oncogenic role in epithelial ovarian cancer progression [41, 44]. Similarly, in endometrial carcinoma, HOXB-AS3 has been shown to play an oncogenic role [42, 48, 51]. The lncRNA HOXB-AS3 serves as a sponge for miR-498-5p or interacts with PTBP1 protein to regulate downstream target genes, which further regulate lipid metabolism and tumor-related biological behaviors, such as cell proliferation, apoptosis, and invasion, to promote tumor progression [42, 48].
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is the predominant form of acute leukemia in adults [53, 54], characterized by the uncontrolled proliferation of abnormal and immature cells in the blood system, disrupting the normal production of blood cells.
In AML, an upregulation of HOXB-AS3 expression has been observed specifically in leukemic stem cells (LSCs), which play a crucial role in leukemogenesis [55-57]. Patients with elevated levels of HOXB-AS3 expression have shown a significant decrease in overall survival and relapse-free survival rates compared to those with lower expression levels [46]. HOXB-AS3 is implicated in promoting enhanced cell proliferation by upregulating key genes involved in cell cycle progression, DNA replication, and pre-replicative complex assembly [46]. Notably, a distinctive mechanism involves the overexpression of the spliceosome NR_033205.1 of HOXB-AS3, facilitated by the RNA-binding protein YTHDC1 through m6A modification of the HOXB-AS3 precursor RNA [50]. This aberrant overexpression contributes to proliferation, inhibits apoptosis, and promotes self-renewal of LSCs, exacerbating the leukemic phenotype [50].
The main role of HOXB-AS3 RNA and its encoded micro-peptide in the occurrence and development of multiple tumors.
In the context of AML cases harboring nucleophosmin 1 (NPM1) mutations [45], HOXB-AS3 assumes a regulatory role in the proliferative capacity of blasts, which are undifferentiated leukemia cells. It accomplishes this through its interaction with ErbB3-binding protein 1 (EBP1), guiding EBP1 to the ribosomal DNA locus [45]. This intricate mechanism modulates ribosomal RNA transcription and de novo protein synthesis, pivotal processes for the maintenance of cellular functions and malignant growth in AML.
Regulatory mechanisms of lncRNA HOXB‑AS3 in lung cancer (A), liver cancer (B), ovarian cancer and endometrial carcinoma (C), and acute myeloid leukemia (D).
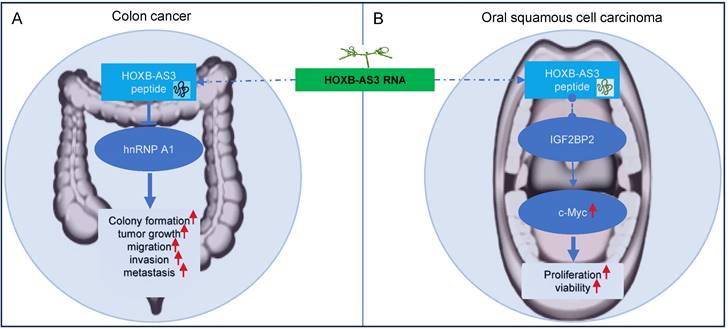
Regulatory mechanisms of HOXB‑AS3 micro-peptide in colon cancer (A) and oral squamous cell carcinoma (B).
HOXB-AS3 Beyond RNA: The Micro-Peptide Dimension
HOXB-AS3 was initially identified as a lncRNA gene in Homo sapiens. However, ribosome profiling studies have revealed that HOXB-AS3 RNA associates with ribosomes [58], hinting at its potential to encode a protein or a small peptide. An open reading frame (ORF) within HOXB-AS3, conserved across primates, has been shown to encode a 53-amino acid micro-peptide absent in other species and lacking known protein homology, indicating a unique functional entity encoded by this lncRNA. Huang et al. [40] confirmed that the presence of this HOXB-AS3 micro-peptide in various cancer cell lines, including colon, breast, ovarian, and nasopharyngeal cancer cells. It's noteworthy that the dysregulated expression of the HOXB-AS3 micro-peptide, rather than the lncRNA itself, has been reported to play a crucial role in modulating tumor growth and cell proliferation in colon and oral cancers [40, 52] (Figure 8).
Experimental validations have further solidified the coding potential of HOXB-AS3 [40]. Constructs designed to express HOXB-AS3 ORF (Open Reading Frame) fusion proteins with GFP and Flag tags confirmed peptide expression, dependent on the intact start codon of the HOXB-AS3 ORF. Western blot analyses using anti-GFP antibodies substantiated the expression of these fusion proteins in transfected cells.
The specificity of the peptide's expression was corroborated by the development of antibodies against the HOXB-AS3 peptide itself, which detected the peptide in cancer cell lines, thus underscoring its endogenous presence [40]. Most compellingly, the use of a translation-blocking antisense oligonucleotide specific to HOXB-AS3, followed by mass spectrometry, confirmed the peptide's independent translation from the HOXB-AS3 RNA [40], decisively establishing its existence beyond theoretical prediction. These findings necessitate a re-evaluation of HOXB-AS3's role in cancer biology, suggesting a novel functional dimension beyond its RNA origins.
Gaining a comprehensive understanding of the role and regulatory mechanisms of the HOXB-AS3 micro-peptide holds promise for developing effective treatment strategies and exploring its potential clinical applications. This multi-dimensional approach could pave the way for novel therapeutic and diagnostic avenues, harnessing the hitherto unexplored coding capacity of lncRNAs.
Colon Cancer and Oral Squamous Cell Carcinoma
Colon cancer and oral squamous cell carcinoma (OSCC) are prevalent cancers with significant global impact [2, 59]. They rank as the 3rd and 12th most common malignancies worldwide, respectively [2, 60]. In colorectal cancer (CRC), approximately 60% of patients are affected by metastasis, resulting in poor prognoses characterized by five-year survival rates below satisfactory levels [61, 62]. Similarly, OSCC presents a concerning scenario, with an overall five-year survival rate in 2020 falling below 50% [63] [52].
In colon cancer [40], the HOXB-AS3 micro-peptide displays aberrant expression patterns, with down-regulated levels observed in tumor tissues compared to adjacent normal tissues. Low levels of HOXB-AS3 peptide are associated with a poor prognosis for CRC patients [40]. Notably, it is the HOXB-AS3 micro-peptide, rather than the RNA, that acts as a tumor suppressor, exerting functional roles in critical cellular processes like cell proliferation, migration, and invasion, thereby inhibiting tumor growth and metastasis in colon cancer [40]. Mechanistically, the HOXB-AS3 peptide suppresses CRC growth by inhibiting hnRNP A1-mediated PKM splicing, consequently preventing PKM2 formation and reprogramming of glucose metabolism [40].
In OSCC [52], the HOXB-AS3 micro-peptide has been reported to exhibit increased expression in tumor tissues. Interestingly, the HOXB-AS3 protein, not the mRNA, exerts its oncogenic function in OSCC tumorigenesis [52]. It primarily influences cell proliferation and viability. Regulatory mechanisms involving the HOXB-AS3 micro-peptide in OSCC include its direct binding with IGF2BP2, which enhances the stability of c-Myc mRNA, and leads to increased expression of c-Myc [52]. The c-myc transcription factor is known for its broad range of functions, affecting cellular activities such as differentiation, proliferation, and metabolism [64-68]. c-Myc functions as an oncogene in numerous cancers, including OSCC [69, 70].
Conclusion and Future Perspectives
In recent years, significant advancements in next-generation sequencing technology have led to the identification of numerous tumor-related lncRNAs [71-74]. These lncRNAs have emerged as potential oncogenes or tumor suppressor genes, playing crucial regulatory roles in tumor development processes [11, 75-77]. The extensive body of evidence highlights the importance of lncRNAs in unraveling the intricate mechanisms underlying cancer progression.
One such lncRNA, HOXB-AS3, has garnered significant attention in the field of tumorigenesis recently. LncRNA HOXB-AS3 has been found to be upregulated in various human cancers, including solid and hematological tumors [41, 43, 45, 48, 50]. And lncRNA HOXB-AS3 levels have shown significant correlations with some clinicopathological features and prognosis, such as TNM stage, tumor metastasis status, and overall survival. In-depth analyses using data from TCGA and UCSC XENA databases further indicate that lncRNA HOXB-AS3 holds promise as both a prognostic and diagnostic biomarker for diverse malignancies.
Moreover, HOXB-AS3 has emerged as a promising therapeutic target in cancer treatment. This lncRNA engages in diverse interactions with molecules, functioning as a multifaceted scaffold that intricately regulates gene expression and cellular dynamics. It plays a role in regulating gene expression at the transcriptional level through interactions with key proteins such as DNMT1 [47], EZH2 [49], and PTBP1 [48]. Additionally, lncRNA HOXB-AS3 contribute to tumor progression by participating in several signaling pathways, including p53 signaling [47], PI3K-AKT pathway [43], Wnt/β-catenin signaling [44], and lipid metabolism pathways [48], underscoring its pivotal role in maintaining cellular equilibrium and influencing tumor growth. And HOXB-AS3's involvement extends to crucial tumor-related processes like cell proliferation, migration, and invasion. Targeting its expression through methods like small interfering RNA (siRNA) or small hairpin RNA (shRNA) has yielded encouraging outcomes. For instance, siRNA-mediated interference in hepatoma cells curtails proliferation, induces apoptosis, and leads to cell cycle arrest [47]. In ovarian cancer [41, 44], suppressing HOXB-AS3 hampers cell proliferation and impedes migration and invasion. Similar siRNA-based approaches in endometrial carcinoma [42] and acute myeloid leukemia [50] demonstrate reduced cancer cell proliferation and enhanced apoptosis. Moreover, in lung cancer [43], shRNA-mediated inhibition of HOXB-AS3 curbs cell proliferation, migration, and invasion, triggers apoptosis, and diminishes tumorigenicity in mouse models. These findings collectively suggest that HOXB-AS3 may be a viable target for drug development. Additionally, HOXB-AS3 has been identified as a significant player in sorafenib resistance [49], cancer stemness [49, 50], and immune escape [50], offering new therapeutic avenues for cancer patients.
Intriguingly, although initially categorized as a lncRNA, debates have arisen regarding HOXB-AS3's potential translation into a micro-peptide [40, 52]. Reports have presented varying effects, suggesting oncogenic and tumor-suppressive roles in colon [40] and oral cancers [52], respectively. Additionally, lower levels of the HOXB-AS3 peptide have been linked to more advanced clinical stages and a poorer prognosis for patients with CRC [40]. These findings raise the possibility that the HOXB-AS3 peptide could serve as a prognostic biomarker and therapeutic target in certain tumor types.
Despite the progress in HOXB-AS3 research, the field is still fraught with challenges and open questions. A primary uncertainty is whether HOXB-AS3 encodes micro-peptides across various human tumors and the extent of aberrant HOXB-AS3 micro-peptide expression, including their potential prognostic and diagnostic implications for diverse tumor types. Addressing these gaps necessitates further study. Moreover, it is vital to unravel the specific mechanisms through which HOXB-AS3 operates in different tumor contexts, whether via RNA-mediated pathways or through the actions of its encoded proteins. Thorough validation with in vitro and in vivo studies is essential to establish definitive evidence. Secondly, the utility of HOXB-AS3 as a diagnostic marker remain to be further explored. Our review suggests that HOXB-AS3 expression in tissues may serve as a promising diagnostic predictor to distinguish between tumor and normal samples across a range of malignancies. Nonetheless, the verification of the ROC curve is preliminary and based on the available dataset. It is required for broader clinical investigations for validation. To date, the detection of HOXB-AS3 in body fluids like plasma, and its feasibility as a non-invasive, and cost-effective tumor detection method remains unreported. However, the literature does report other lncRNAs, such as LINK-A [78-81] and HOTAIR [82-84], which are found to be significantly expressed in the blood of patients with diverse tumors and proposed as a non-invasive diagnostic marker with substantial diagnostic potential in diverse tumorigenic conditions. This highlights the imperative for increased focus and research into the potential of HOXB-AS3 as a tumor biomarker in these readily accessible biofluids. Lastly, the prognostic value of HOXB-AS3 is currently grounded in retrospective clinical samples. Prospective studies or clinical trials, notably those that are large-scale and multi-center, encompassing diverse patient groups, are crucial to delve deeper into the prognostic significance of HOXB-AS3. These future research efforts will be instrumental in enriching our collective understanding of HOXB-AS3 and its clinical relevance in oncology.
In conclusion, HOXB-AS3 plays a multifaceted role in tumorigenesis and tumor progression, potentially serving as a cancer-specific biomarker for diagnosis, prognosis, and treatment. Its involvement in tumorigenesis is complex, encompassing both RNA and protein levels. The recent discovery of its translation into a micro-peptide adds a new dimension to its functional characterization. Future research should aim to resolve the ongoing debate regarding HOXB-AS3's RNA and protein functions and their respective contributions to tumorigenesis. Comprehensive studies investigating the interplay between HOXB-AS3 RNA and protein levels will provide valuable insights into its therapeutic potential as a cancer treatment target.
Abbreviations
AML: Acute myeloid leukemia; AUC: Area under the curve; CRC: Colorectal cancer; DFI: Disease-free interval; DSS: Disease-specific survival; EBP1: ErbB3-binding protein 1; EC: Endometrial carcinoma; EMT: Epithelial-mesenchymal transition; HOXB-AS3: HOXB Cluster Antisense RNA 3; LncRNAs: Long non-coding RNAs; LSCs: Leukemic stem cells; OS: Overall survival; ROC: Receiver operating characteristic; TCGA: The Cancer Genome Atlas.
Supplementary Material
Supplementary table.
Acknowledgements
Author Contributions
HLL designed and supervised the study and revised the manuscript. HZW and JRY wrote the manuscript draft. HZW, JRY, and MQZ collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Availability of Data and Materials
The datasets analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/), Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/), and UCSC Xena (https://xenabrowser.net/datapages/). And the datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chhikara BS, Parang K. Global Cancer Statistics 2022: the trends projection analysis. Chemical Biology Letters. 2023;10:451
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
3. Xia C, Dong X, Li H, Cao M, Sun D, He S. et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chinese medical journal. 2022;135:584-90
4. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72:7-33
5. Sarhadi VK, Armengol G. Molecular Biomarkers in Cancer. Biomolecules. 2022;12:1021
6. Kato S, Alsafar A, Walavalkar V, Hainsworth J, Kurzrock R. Cancer of Unknown Primary in the Molecular Era. Trends in cancer. 2021;7:465-77
7. Hackl H, Stocker G, Charoentong P, Mlecnik B, Bindea G, Galon J. et al. Information technology solutions for integration of biomolecular and clinical data in the identification of new cancer biomarkers and targets for therapy. Pharmacology & therapeutics. 2010;128:488-98
8. Dai M, Chen S, Teng X, Chen K, Cheng W. KRAS as a Key Oncogene in the Clinical Precision Diagnosis and Treatment of Pancreatic Cancer. Journal of Cancer. 2022;13:3209-20
9. Bou-Dargham MJ, Draughon S, Cantrell V, Khamis ZI, Sang QA. Advancements in Human Breast Cancer Targeted Therapy and Immunotherapy. Journal of Cancer. 2021;12:6949-63
10. McCabe EM, Rasmussen TP. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Seminars in cancer biology. 2021;75:38-48
11. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer research. 2017;77:3965-81
12. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nature reviews Molecular cell biology. 2023;24:430-47
13. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature reviews Molecular cell biology. 2021;22:96-118
14. Li X, Huang H, Liu M, Luo H. Tumor Suppressor LncRNA on Chromosome 8p12 (TSLNC8): A Concise Review in Human Malignancies. Journal of Cancer. 2023;14:2867-77
15. Yan H, Bu P. Non-coding RNA in cancer. Essays in biochemistry. 2021;65:625-39
16. Liu Y, Liu X, Lin C, Jia X, Zhu H, Song J. et al. Noncoding RNAs regulate alternative splicing in Cancer. Journal of experimental & clinical cancer research: CR. 2021;40:11
17. Lei L, Peng G, Luo H, Li W. SRY-box transcription factor 21 antisense divergent transcript 1: Regulatory roles and clinical significance in neoplastic conditions and Alzheimer's Disease. Journal of Cancer. 2023;14:3258-74
18. Cao Z, Oyang L, Luo X, Xia L, Hu J, Lin J. et al. The roles of long non-coding RNAs in lung cancer. Journal of Cancer. 2022;13:174-83
19. Zheng QX, Wang J, Gu XY, Huang CH, Chen C, Hong M. et al. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;135:111169
20. Li YH, Hu YQ, Wang SC, Li Y, Chen DM. LncRNA SNHG5: A new budding star in human cancers. Gene. 2020;749:144724
21. Lin H, Li J, Wang M, Zhang X, Zhu T. Exosomal Long Noncoding RNAs in NSCLC: Dysfunctions and Clinical Potential. Journal of Cancer. 2023;14:1736-50
22. Kang J, Abudurufu M, Zhang S, Jiang W, Luo H. lncRNA VIM-AS1 acts as a prognostic biomarker and promotes apoptosis in lung adenocarcinoma. Journal of Cancer. 2023;14:1417-26
23. Shenoy US, Adiga D, Kabekkodu SP, Hunter KD, Radhakrishnan R. Molecular implications of HOX genes targeting multiple signaling pathways in cancer. Cell biology and toxicology. 2022;38:1-30
24. Morgan R, Hunter K, Pandha HS. Downstream of the HOX genes: Explaining conflicting tumour suppressor and oncogenic functions in cancer. International journal of cancer. 2022;150:1919-32
25. Fan F, Mo H, Zhang H, Dai Z, Wang Z, Qu C. et al. HOXA5: A crucial transcriptional factor in cancer and a potential therapeutic target. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2022;155:113800
26. Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo X. et al. Homeobox Genes in Cancers: From Carcinogenesis to Recent Therapeutic Intervention. Frontiers in oncology. 2021;11:770428
27. Yao Q, Wang C, Wang Y, Zhang X, Jiang H, Chen D. The integrated comprehension of lncRNA HOXA-AS3 implication on human diseases. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2022;24:2342-50
28. Zeng C, Ye S, Chen Y, Zhang Q, Luo Y, Gai L. et al. HOXA-AS3 Promotes Proliferation and Migration of Hepatocellular Carcinoma Cells via the miR-455-5p/PD-L1 Axis. Journal of immunology research. 2021;2021:9289719
29. Qu F, Zhu B, Hu YL, Mao QS, Feng Y. LncRNA HOXA-AS3 promotes gastric cancer progression by regulating miR-29a-3p/LTβR and activating NF-κB signaling. Cancer cell international. 2021;21:118
30. Chen W, Li Q, Zhang G, Wang H, Zhu Z, Chen L. LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. Journal of cellular and molecular medicine. 2020;24:11755-67
31. Lin S, Zhang R, An X, Li Z, Fang C, Pan B. et al. LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung carcinoma cells. Oncogenesis. 2019;8:60
32. Liu T, Wang H, Yu H, Bi M, Yan Z, Hong S. et al. The Long Non-coding RNA HOTTIP Is Highly Expressed in Colorectal Cancer and Enhances Cell Proliferation and Invasion. Molecular therapy Nucleic acids. 2020;19:612-8
33. Shang A, Wang W, Gu C, Chen C, Zeng B, Yang Y. et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. Journal of experimental & clinical cancer research: CR. 2019;38:411
34. Sun Y, Zeng C, Gan S, Li H, Cheng Y, Chen D. et al. LncRNA HOTTIP-Mediated HOXA11 Expression Promotes Cell Growth, Migration and Inhibits Cell Apoptosis in Breast Cancer. International journal of molecular sciences. 2018;19:472
35. Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y. et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell death & disease. 2018;9:85
36. Raju GSR, Pavitra E, Bandaru SS, Varaprasad GL, Nagaraju GP, Malla RR. et al. HOTAIR: a potential metastatic, drug-resistant and prognostic regulator of breast cancer. Molecular cancer. 2023;22:65
37. Wong CH, Li CH, Man Tong JH, Zheng D, He Q, Luo Z. et al. The establishment of CDK9/RNA PolII/H3K4me3/DNA methylation feedback promotes HOTAIR expression by RNA elongation enhancement in cancer. Molecular therapy: the journal of the American Society of Gene Therapy. 2022;30:1597-609
38. Hu CY, Su BH, Lee YC, Wang CT, Yang ML, Shen WT. et al. Interruption of the long non-coding RNA HOTAIR signaling axis ameliorates chemotherapy-induced cachexia in bladder cancer. Journal of biomedical science. 2022;29:104
39. Chen J, Hou SF, Tang FJ, Liu DS, Chen ZZ, Zhang HL. et al. HOTAIR/Sp1/miR-199a critically regulates cancer stemness and malignant progression of cutaneous squamous cell carcinoma. Oncogene. 2022;41:99-111
40. Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H. et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Molecular cell. 2017;68:171-84.e6
41. Xu S, Jia G, Zhang H, Wang L, Cong Y, Lv M. et al. LncRNA HOXB-AS3 promotes growth, invasion and migration of epithelial ovarian cancer by altering glycolysis. Life sciences. 2021;264:118636
42. Xing Y, Sun X, Li F, Jiang X, Jiang A, Li X. et al. Long non-coding RNA (lncRNA) HOXB-AS3 promotes cell proliferation and inhibits apoptosis by regulating ADAM9 expression through targeting miR-498-5p in endometrial carcinoma. The Journal of international medical research. 2021;49:3000605211013548
43. Jiang W, Kai J, Li D, Wei Z, Wang Y, Wang W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. Journal of cellular physiology. 2020;235:7194-203
44. Zhuang XH, Liu Y, Li JL. Overexpression of long noncoding RNA HOXB-AS3 indicates an unfavorable prognosis and promotes tumorigenesis in epithelial ovarian cancer via Wnt/β-catenin signaling pathway. Bioscience reports. 2019;39:BSR20190906
45. Papaioannou D, Petri A, Dovey OM, Terreri S, Wang E, Collins FA. et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nature communications. 2019;10:5351
46. Huang HH, Chen FY, Chou WC, Hou HA, Ko BS, Lin CT. et al. Long non-coding RNA HOXB-AS3 promotes myeloid cell proliferation and its higher expression is an adverse prognostic marker in patients with acute myeloid leukemia and myelodysplastic syndrome. BMC cancer. 2019;19:617
47. Zhang XM, Chen H, Zhou B, Zhang QY, Liao Y, Wang JS. et al. lncRNA HOXB-AS3 promotes hepatoma by inhibiting p53 expression. European review for medical and pharmacological sciences. 2018;22:6784-92
48. Zhou Q, Kong D, Li W, Shi Z, Liu Y, Sun R. et al. LncRNA HOXB-AS3 binding to PTBP1 protein regulates lipid metabolism by targeting SREBP1 in endometrioid carcinoma. Life sciences. 2023;320:121512
49. Tseng CF, Chen LT, Wang HD, Liu YH, Shiah SG. Transcriptional suppression of Dicer by HOXB-AS3/EZH2 complex dictates sorafenib resistance and cancer stemness. Cancer science. 2022;113:1601-12
50. Wu C, Cui J, Huo Y, Shi L, Wang C. Alternative splicing of HOXB-AS3 underlie the promoting effect of nuclear m6A reader YTHDC1 on the self-renewal of leukemic stem cells in acute myeloid leukemia. International journal of biological macromolecules. 2023;237:123990
51. Niu L, Wu Z. Identification and validation of oxeiptosis-associated lncRNAs and prognosis-related signature genes to predict the immune status in uterine corpus endometrial carcinoma. Aging. 2023;15:4236-52
52. Leng F, Miu YY, Zhang Y, Luo H, Lu XL, Cheng H. et al. A micro-peptide encoded by HOXB-AS3 promotes the proliferation and viability of oral squamous cell carcinoma cell lines by directly binding with IGF2BP2 to stabilize c-Myc. Oncology letters. 2021;22:697
53. Pelcovits A, Niroula R. Acute Myeloid Leukemia: A Review. Rhode Island medical journal (2013). 2020;103:38-40
54. Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood reviews. 2017;31:63-76
55. Chopra M, Bohlander SK. The cell of origin and the leukemia stem cell in acute myeloid leukemia. Genes, chromosomes & cancer. 2019;58:850-8
56. Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 2014;28:2276-82
57. Passegué E, Weisman IL. Leukemic stem cells: where do they come from? Stem cell reviews. 2005;1:181-8
58. Wang T, Cui Y, Jin J, Guo J, Wang G, Yin X. et al. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic acids research. 2013;41:4743-54
59. Badwelan M, Muaddi H, Ahmed A, Lee KT, Tran SD. Oral Squamous Cell Carcinoma and Concomitant Primary Tumors, What Do We Know? A Review of the Literature. Current oncology (Toronto, Ont). 2023;30:3721-34
60. Misra S, Chaturvedi A, Misra NC. Management of gingivobuccal complex cancer. Annals of the Royal College of Surgeons of England. 2008;90:546-53
61. Xu L, Li X, Cai M, Chen J, Li X, Wu WK. et al. Increased expression of Solute carrier family 12 member 5 via gene amplification contributes to tumour progression and metastasis and associates with poor survival in colorectal cancer. Gut. 2016;65:635-46
62. Wang J, Li S, Liu Y, Zhang C, Li H, Lai B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer medicine. 2020;9:361-73
63. Li P, Fang Q, Yang Y, Chen D, Du W, Liu F. et al. Survival Significance of Number of Positive Lymph Nodes in Oral Squamous Cell Carcinoma Stratified by p16. Frontiers in oncology. 2021;11:545433
64. Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor perspectives in medicine. 2014;4:a014241
65. Aoki Y, Han Q, Kubota Y, Masaki N, Obara K, Tome Y. et al. Oncogenes and Methionine Addiction of Cancer: Role of c-MYC. Cancer genomics & proteomics. 2023;20:165-70
66. Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013;3:a014217
67. Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Advances in cancer research. 2010;107:163-224
68. Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5546-53
69. Marconi GD, Della Rocca Y, Fonticoli L, Melfi F, Rajan TS, Carradori S. et al. C-Myc Expression in Oral Squamous Cell Carcinoma: Molecular Mechanisms in Cell Survival and Cancer Progression. Pharmaceuticals (Basel, Switzerland). 2022;15:890
70. Li S, Zhang S, Chen J. c-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer gene therapy. 2019;26:400-10
71. Saleembhasha A, Mishra S. Novel molecules lncRNAs, tRFs and circRNAs deciphered from next-generation sequencing/RNA sequencing: computational databases and tools. Briefings in functional genomics. 2018;17:15-25
72. Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Advances in experimental medicine and biology. 2017;1008:1-46
73. Ren S, Li G, Liu C, Cai T, Su Z, Wei M. et al. Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma. Oncology reports. 2016;36:1861-7
74. Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R. et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Molecular cancer. 2015;14:94
75. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome biology. 2017;18:206
76. Li R, Wang X, Zhu C, Wang K. lncRNA PVT1: a novel oncogene in multiple cancers. Cellular & molecular biology letters. 2022;27:84
77. Guzel E, Okyay TM, Yalcinkaya B, Karacaoglu S, Gocmen M, Akcakuyu MH. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. Northern clinics of Istanbul. 2020;7:81-6
78. Zhang H, Yao B, Tang S, Chen Y. LINK-A Long Non-Coding RNA (lncRNA) Participates in Metastasis of Ovarian Carcinoma and Upregulates Hypoxia-Inducible Factor 1 (HIF1α). Medical science monitor: international medical journal of experimental and clinical research. 2019;25:2221-7
79. Liu J, Song W, Li J, Li X, Zhao R, Gong T. LINK-A lncRNA is upregulated in metastatic non-small cell lung cancer and is associated with poor prognosis. Oncology letters. 2019;18:3049-57
80. Zhang Y, Lu P, Du H, Zhang L. LINK-A lncRNA Promotes Proliferation and Inhibits Apoptosis of Mantle Cell Lymphoma Cell by Upregulating Survivin. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:365-70
81. Zhao B, Liu K, Cai L. LINK-A lncRNA functions in the metastasis of osteosarcoma by upregulating HIF1α. Oncology letters. 2019;17:5005-11
82. Yao X, Wang T, Sun MY, Yuming Y, Guixin D, Liu J. Diagnostic value of lncRNA HOTAIR as a biomarker for detecting and staging of non-small cell lung cancer. Biomarkers. 2022;27:526-33
83. Lou ZH, Xu KY, Qiao L, Su XQ, Ou-Yang Y, Miao LB. et al. Diagnostic Potential of the Serum lncRNAs HOTAIR, BRM and ICR for Hepatocellular Carcinoma. Front Biosci (Landmark Ed). 2022;27:264
84. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C. et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Molecular cancer. 2018;17:74
Author contact
![]() Corresponding author: Hongliang Luo, Department of Gastrointestinal Surgery, the Second Affiliated Hospital of Nanchang University, No. 1 Minde Road, Nanchang 330008, Jiangxi Province, China. E-mail: ndefy13028edu.cn. Tel: +86 13097280001.
Corresponding author: Hongliang Luo, Department of Gastrointestinal Surgery, the Second Affiliated Hospital of Nanchang University, No. 1 Minde Road, Nanchang 330008, Jiangxi Province, China. E-mail: ndefy13028edu.cn. Tel: +86 13097280001.

 Global reach, higher impact
Global reach, higher impact