Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(4):990-998. doi:10.7150/jca.90321 This issue Cite
Research Paper
MiRNA-766-3p inhibits gastric cancer via targeting COL1A1 and regulating PI3K/AKT signaling pathway
1. School of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China.
2. Department of Gastroenterology, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200437, China.
3. Central Laboratory, Shanghai Skin Disease Hospital, School of Medicine, Tongji University, Shanghai 200443, China.
4. Shanghai Institute of Stem Cell Research and Clinical Translation, Shanghai 200120, China.
* These authors contributed equally to this work and should be considered co-first authors.
Received 2023-9-19; Accepted 2023-11-17; Published 2024-1-1
Abstract
Objective MiRNA-766-3p has been shown to be associated with a variety of cancers. However, few studies have been done in gastric cancer (GC). This study explores the mechanism of miR-766-3p in GC.
Methods The potential targets of microRNA (miRNA) were predicted using Tarbase and Targetscan databases. The results are intersected with differential genes (DEGs) (fold change > 1.5, P < 0.05) in gastric cancer to obtain potential core targets. The hub targets screened by constructing PPI networks (degree > 5, expression > 0.5). Validating the differential expression and expression in immunohistochemistry of these targets through the database. And the binding sites between miRNAs and mRNAs were verified using dual-luciferase Assay. Finally, qRT-PCR and Western Blot experiments were conducted to validate the hub targets and signal pathways.
Results The potential hub targets from the PPI network were THBS2, COL1A1, FGG, FGB, and PLAU. Combining database, luciferase Assay and experimental validation, miR-766-3p can sponge COL1A1 and it plays the most important role in gastric cancer progression. In GC, COL1A1 was upregulated and the enrichment analysis revealed that COL1A1 regulates PI3K/AKT signal pathway, and AKT is also highly expressed in gastric cancer.
Conclusion The miR-766-3p can inhibit the progression of gastric cancer by targeting COL1A1 and regulating the PI3K/AKT signal pathway. It could be a potential therapy option for the GC.
Keywords: MiRNA-766-3p, COL1A1, PI3K/AKT signal pathway, Gastric cancer.
Introduction
Gastric cancer (GC) remains a major cancer worldwide, accounting for over one million new cases in 2020 and an anticipated 769,000 deaths, ranking fourth for mortality and fifth for incidence globally [1]. Surgery and chemotherapy remain the most effective treatment modality and gene therapy have seen significant advancements in the past decade [2]. However, due to the low rate of early detection and diagnosis, many patients with gastric cancer have very poor survival, and the overall 5-year survival rate of GC patients is less than 50% in China [3,4]. Gastric cancer development is a complicated process that involves many genetic and epigenetic changes in oncogenes, tumor suppressor genes, DNA repair genes, cell cycle regulators, and signaling molecules [5]. Therefore, it is crucial to elucidate the pathogenesis of gastric cancer by identifying novel targets for treatment.
MicroRNA (miRNA), 18-22 nucleotides in length, is a class of small noncoding RNA molecules with highly conserved. It is generally considered that miRNAs influence gene expression at the transcriptional level via binding to mRNA [6]. Much evidence demonstrates that miRNAs correlate with various tumors, such as breast cancer, lung cancer and ovarian cancer [7-9]. It contributes to cancer through a nuclear function that affects gene transcription and epigenetic states [10]. Importantly, miRNA is also associated with gastric cancer progression. MiR-34a inhibits the proliferation and invasion of gastric cancer by regulating PDL1[11]. MiR-1262 inhibits gastric cardia cancer by targeting the oncogene ULK1[12]. MiR-542-3p suppresses cell proliferation by targeting the oncogene astrocyte-elevated gene-1[13]. This evidence shows that miRNAs involved in gastric cancer progression.
Our previous work demonstrated that miR-766-3p plays a kernel role in GC by identifying the differentially expressed (DE) miRNAs of GC [14]. The study shows that the miR-766-3p contributes to anti-inflammatory responses and stops the inflammatory carcinoma transformation by inhibiting NF-κB signaling indirectly [15]. It has also been demonstrated to target a variety of oncogenes, such as hepatocellular carcinoma, colorectal cancer, renal cell carcinoma and thyroid carcinoma. The miR-766-3p/FOSL2 axis plays an oncogenic role in hepatocellular carcinoma [16]. It inhibits proliferation in colorectal cancer cells via the PI3K/AKT pathway when HNF4G is down-regulated [17]. MiR-766-3p also targets and inhibits SF2 expression and promotes the proliferation of renal cell carcinoma cells [18]. Circ_0059354 accelerates the growth of papillary thyroid cancer by increasing ARFGEF1 levels via miR-766-3p sponging [19]. These studies have shown that miR-766-3p plays an important role in cancer. However, few studies have been conducted to explore the mechanisms of miR-766-3p in gastric cancer. In this paper, the core targets and signal pathway of miR-766-3p in gastric cancer was further analyzed. We identified COL1A1 as the core target of miR-766-3p through database prediction and screening. The binding sites between miR-766-3p and COL1A1 were verified using dual luciferase Assay. Biological Functional Analysis was used and found that COL1A1 correlates with PI3K/AKT signal pathway. Experiments were used to validate the conclusions of the data analyses.
Materials and methods
Patients and samples
From July 2022 to May 2023, a total of 60 clinical tissue samples (30 tumor samples and 30 adjacent normal samples) were collected from patients at Yueyang Hospital of Integrated Traditional Chinese and Western (Table 1).
Patient characteristics (n=30)
| n (%) | |
|---|---|
| Sex | |
| Male | 18 (60) |
| Female | 12 (40) |
| Age | |
| >70 | 4 (13) |
| 50-70 | 21 (70) |
| <50 | 5 (17) |
| Location | |
| Lower third | 19 (63) |
| Middle third | 6 (20) |
| Upper third | 5 (17) |
| Stage | |
| Ⅰ | 10 (33) |
| Ⅱ | 8 (27) |
| Ⅲ | 8 (27) |
| Ⅳ | 4 (13) |
| T | |
| T1 | 10 (33) |
| T2 | 4 (13) |
| T3 | 7 (24) |
| T4 | 9 (30) |
| N | |
| N0 | 11 (37) |
| N1 | 11 (37) |
| N2 | 2 (6) |
| N3 | 3 (10) |
| Nx | 3 (10) |
| M | |
| M0 | 25 (83) |
| M1 | 2 (6) |
| Mx | 3 (10) |
Inclusion criteria: (1) Clinical specimens were confirmed to be gastric cancer by histopathological examination and there was at least one solid or measurable extra-gastric lesion; (2) 18 years old ≤ 85 years old; (3) Eastern Cooperative Oncology Group (ECOG) score of 0-2; (4) Patients or authorized relatives of the patients signed the informed consent before enrolment. Exclusion criteria: (1) patients who have received radiotherapy or chemotherapy; (2) history of other tumors within 5 years; (3) patients in pregnancy or lactation; (4) combined organ failure or other serious diseases; (5) combined neurological or psychiatric history. And paraneoplastic tissue is taken from at least 4 cm away from the tumor lesion. All tissue samples were immediately frozen and preserved in liquid nitrogen until further use. Samples were then kept at -80°C for RNA protein extraction.
MiR766-3p targets prediction and enrichment analysis
The miRNA target genes were predicted using two databases involving TarBase (http://www.diana.pcbi.upenn.edu/tarbase) (v8.0) and TargetScan (http://www.targetscan.org/vert_72/), which contained the largest collection of manually curated experimental data. The signal pathway of the hub target was enriched using DAVID (https://david.ncifcrf.gov) online.
Differentially expressed genes (DEGs) identification
The mRNA dataset (GSE118916) of gastric cancer was obtained from the GEO database (http://www.diana.pcbi.upenn.edu/tarbase), which includes 30 GC tissue samples and 30 normal samples. The mRNA levels of all samples were standardized using DESeq software, and the difference significance test of reads was performed using NB (Negative binomial distribution). R package was used to identify the differentially expressed genes (DEGs) between the Tumor Group and Normal Group (fold change > 1.5, P < 0.05).
Protein-protein interaction (PPI) network construction
The DEGs were used to construct a PPI network by the STRING (v11.5), and CytoHubba clusters were used to gain hub genes. The determinate nodes were considered as potential hub mRNAs, and they will be further validated in database and experiments.
Core genes validated by databases
Data from the TCGA (The Cancer Genome Atlas) and the GTEx (Genotype-Tissue Expression) projects were used to conduct expression and survival analyses for possible indicators. The Human Protein Atlas (HPA) database (https://www.proteinatlas.org) was then used to obtain core proteins immunohistochemistry results between stomach glandular cells and tumor cells.
Dual-luciferase reporter assay
The binding sites of miR-766-3p and COL1A1 were calculated and predicted using miRanda (v3.3). The wild-type and mutant plasmids of COL1A1 (psiCHECK-2-WT-COL1A1 3′UTR, psiCHECK-2-MUT-COL1A1 3′UTR) were constructed and provided by Yilaibo (http://www.shyilaibo.com). Luciferase activity was assessed with a dual luciferase kit (E1901; Promega, USA) 48 h after co-transfection of each plasmid with miR-NC or miR-766-3p.
qRT-PCR validation
Total RNA was isolated using FreeZol Reagent 200rxns (R711-01, Vazyme). Reverse transcription was operated using a miRNA 1st Strand cDNA Synthesis Kit (MR101-01, Vazyme), and HiScript II Q RT Super Mix for qPCR (R223-01, Vazyme). Then, using the miRNA Universal SYBR PCR MasterMix (MQ101-01, Vazyme) and ChamQ SYBR qPCR Master Mix (Q321-02, Vazyme), the levels of miRNA and mRNA expression were determined. GAPDH and U6 were employed as endogenous controls for mRNA and miRNA expression levels, respectively. Finally, the relative RNA expression levels were determined using the 2-ΔΔCt technique. Table 2 displays the primer sequences.
Western blot
RAPI reagents (epizyme) were used to extract total proteins. Protein samples were loaded and separated by 7.5% SDS-PAGE before being transferred to NC membranes and blocked at room temperature for 1 hour. The membranes were washed five times with TBST before being incubated overnight at 4°C with anti-COL1A1 (1:1000, 72026T, CST) and anti-GAPDH (1:1000, 5174S, CST). The membranes were then treated for 1 hour at room temperature with horseradish peroxidase (HRP) conjugated second antibodies (Proteintech). In a dark chamber, the films were developed and fixed with an ECL solution. Image J 1.2.4 (NIH, USA) was used to semi-quantify protein expression.
Statistical analyses
All variables are provided as mean SD and statistical analysis was conducted using the SPSS 23.0 program (IBM Analytics). The groups were compared using two-tailed Student's t-tests, and a P value of 0.05 was regarded as statistically significant. Plotting was done using GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA, USA).
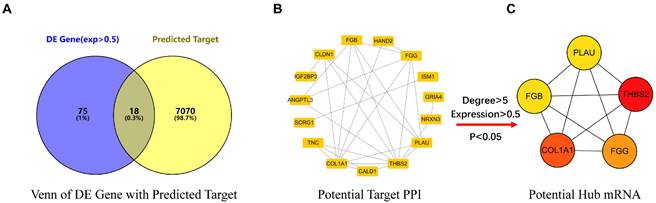
Potential mRNA of miR-766-3p related to gastric cancer screening. (A) The Venn diagram of DE Gene and predicted target taking the intersection; (B) The 18 DE mRNAs from the Venn diagram were used to build the PPI network; (C) The 5 core DE mRNAs were screened from PPI network.
The primer sequences obtained from the NCBI database
| Gene name | Forward primer (5′- 3′) | Reverse primer (5′- 3′) |
|---|---|---|
| miR-766-3p | CGACTCCAGCCCCACAGC | AGTGCAGGGTCCGAGGTATT |
| COL1A1 | TCGGAGGAGAGTCAGGAA | ACACAAGGAACAGAACAGTC |
| GAPDH | CCTGCCTCTACTGGCGCTGC | GCAGTGGGGACACGGAAGGC |
| U6 | CTCGCTTCGGCAGAACA | ACGCTTCACGAATTTGCGT |
| PIK3CA | CCTGCTTTTGGAGTCCTATTGT | ATCTGGTCGCCTCATTTGC |
| AKT1 | CTCTTTCCAGACCCACGACC | ACAGGTGGAAGAACAGCTCG |
Results
Potential targets of miR-766-3p related to gastric cancer
Using the prediction programs, the miR-766-3p target genes were predicted. As a result, a total of 7088 targets were collected from Tarbase and TargetScan. Using the R package, 93 DE mRNAs were accessed by analyzing 30 gastric cancer clinical microarray data from the GEO database. In this work, we intersected the 93 DE mRNAs with 7088 predicted targets and required target expression abundance greater than 0.5. Following the preceding stages, 18 candidate targets were obtained eventually (Fig. 1A).
PPI network construction and hub mRNAs screening
Based on the STRING database, the PPI network was constructed by 18 DE mRNAs (Fig. 1B). We employed the CytoHubba cluster (Top5) to screen the key mRNA of the PPI, and five core targets were eventually selected (THBS2, COL1A, FGG, FGB, PLAU) (Fig. 1C, Table 3).
The distributions of potential hub mRNA from the network
| Cluster | Hub miRNA | Degree | Expression | P-value | Status |
|---|---|---|---|---|---|
| 1 | THBS2 | 9 | 0.5 | 8.06E-06 | Up |
| 2 | COL1A1 | 8 | 0.6 | 5.56E-05 | Up |
| 3 | FGG | 6 | 0.9 | 0.003 | Down |
| 4 | FGB | 5 | 0.6 | 0.014 | Down |
| 5 | PLAU | 5 | 0.5 | 0.001 | Up |
Validation core markers based on TCGA and HPA database
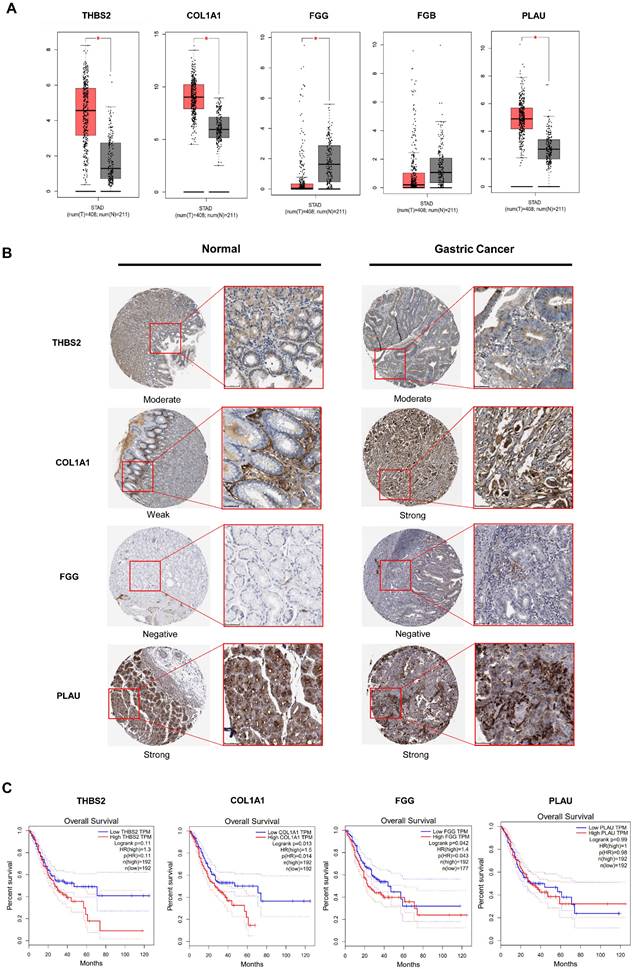
Gastric cancer (n = 408) and normal (n = 211) expression of five key genes was assessed using GEPIA, respectively. The result revealed that the expression of THBS2, COL1A, FGG, and PLAU was considerably different (Fig. 2A). To further analyze the levels of 4 targets, we used the HPA database to validate the tissue expression in immuno-histochemistry. Only one target showed variations in expression between tumor and normal tissues (Fig. 2B). The COL1A1 was negative in normal tissues but strongly expressed in tumors, and immuno-histochemistry revealed no differences in the expression of other targets. In addition, we investigated the prognostic values of COL1A1 in gastric cancer patients based on the overall survival (OS) calculation. According to the findings, the high mRNA levels of COL1A1 have statically significant (P = 0.014) of OS in gastric cancer patients (Fig. 2C). Other targets had no impact on overall survival. Based on the confirmation of the data presented above, it is possible to conclude that COL1A1 is very significant to gastric cancer and is an essential target for miR-766-3p.
Dual-luciferase reporter assay
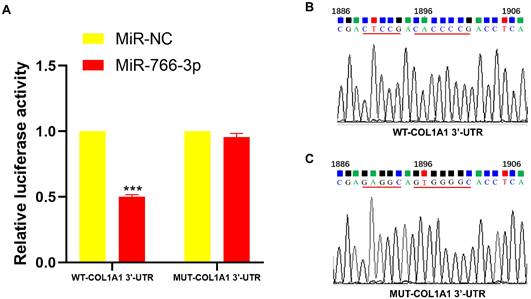
To verify the binding ability of miR-766-3p to COL1A1, a dual luciferase reporter assay was applied. The results showed that miR-766-3p significantly inhibited the luciferase activity of psiCHECK-2-WT-COL1A1, whereas miR-NC did not inhibit psiCHECK-2- MUT-COL1A1 (Fig. 3A-C).
Validation core target and signal pathway by qRT-PCR and Western Blot
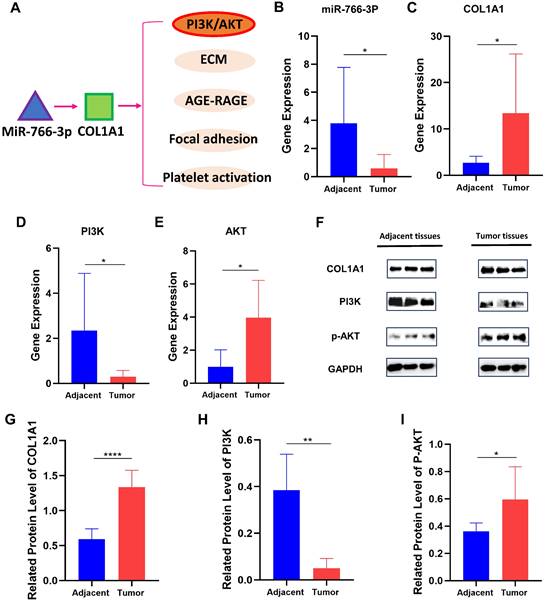
We analyze the downstream pathway of COL1A1 by the Davide database and get many signal pathways (Fig. 4A). Of them, PI3K/AKT signal pathway is a fundamental one, with frequent oncogenic alterations in GC [20]. Thus, we detected expression levels of miR-766-3p, COL1A1and PI3K/AKT of 30 patients' stomach carcinoma tissues and adjacent normal tissues via RT-qPCR and Western Blot. As a result, we found COL1A1 had a significantly higher expression level in tumor tissues than in normal ones, while miR-766-3p, conversely, had a significantly lower expression level in tumor tissues (Fig. 4B, C). Regarding the PI3K/AKT signal pathway, PI3K is low-expressed and AKT is over-expressed in gastric cancers, and there is negative regulation between them (Fig. 4D, E). At the level of protein expression as shown in Fig. 3F. COL1A1 also differs significantly between tumor and normal tissues (Fig. 4G). Relative quantitative protein expression levels of PI3K/AKT are consistent with the mRNA level (Fig. 4H, I).
GEPIA and TCGA datasets were used for expression and survival analysis for prospective targets. (A) Based on the TCGA database, the expression of 5 potential markers was estimated (Tumor group, n = 408; Normal group, n = 211); (B) The immunohistochemistry results of the THBS2, COL1A1, FGG, PLAU between human gastric tumor and normal tissues; (C) Survival analysis was used to determine the prognostic significance of targets in gastric cancer patients.
The dual-luciferase reporter assay confirmed the connection of miR-766-3p and COL1A. (A) The link between miR-766-3p and COL1A1. (B) Construction of COL1A1 wild-type plasmid. (C) Construction of COL1A1 mutant plasmids. Statistical significance is expressed as ****P < 0.0001, ***P<0.001, **P<0.01 and *P<0.05.
The expression levels of miR-766-3p, COL1A1 and PI3K/AKT in different stomach tumor tissues and normal gastric tissues. (A) The downstream pathway of COL1A1; (B) MiR-766-3p mRNA expression; (C) COL1A1 mRNA expression; (D) PI3K mRNA expression; (E) AKT mRNA expression; (F) COL1A1, PI3K and p-AKT protein expression in normal tissue and GC tissue; (G) The relative quantitative expression of COL1A1. (H) The relative quantitative expression of PI3K. (I) The relative quantitative expression of P-AKT. Statistical significance is expressed as ****P < 0.0001, ***P<0.001, **P<0.01 and *P<0.05.
Discussion
MiRNAs as a class of regulatory factors involved in tumor regulation, and many studies show that miRNAs influence tumor progression by performing many functions, including cell division, cell differentiation, angiogenesis, migration, apoptosis and oncogenesis [21,22]. For example, by directly binding to the PTEN, miR-21 may increase the proliferation, invasion, and migration of GC cells [23]. And miR-21 could promote GC via activating the PI3K/AKT pathway [24]. In GC cells, miR-375 is dysregulated, which promotes the growth of the PI3K/Akt pathway and cell survival [25]. In our previous study, by creating the circRNA-miRNA-mRNA (CMM) network and the protein-protein interaction (PPI) network, we discovered that miR-766-3p plays a crucial role [14]. However, the mechanism of miR-766-3p in GC is unclear.
In this study, the potential targets of miR-766-3p were predicted and screened by using Tarbase and Targetscan databases, and COL1A1 was identified as the core target of miR-766-3p by combining TCGA and HPA databases. The qRT-PCR and Western Blot also demonstrated that the miR-766-3p and COL1A1 affect the progression of GC. According to bioinformatic analysis, COL1A1 is a hydrophilic, negatively charged secreted protein that is crucial for the formation of collagen structures and cell adhesion [26]. As we all know, COL1A1 has been found to be elevated in many cancers and affects various signal pathways, such as gastric, colorectal, breast and thyroid tumors [27-31]. And miRNA-98 regulation has been demonstrated to cause lower COL1A1 mRNA levels [32]. Interestingly, our study also found that miR-766-3p can downregulate the high level of COL1A1 in GC. The enrichment analysis revealed that the COL1A1 was highly correlated with PI3K/AKT signal pathway, and it promoted the activation of PI3K/AKT [33]. In GC, the PI3K/AKT signal pathway plays an important role. For example, SLC1A3 through the PI3K/AKT pathway to hasten the growth of gastric cancer [34]. LGR6 may accelerate the development of GC via the PI3K/AKT/mTOR pathway [35]. The activation of NF-B and the PI3K/AKT/SP1 axis is necessary for the UBAP2L-induced EMT process in GC cells [36]. Gastric cancer is prevented from developing and metastasizing by BFAR via the PI3K/AKT/mTOR signal pathway [37]. These studies show that the PI3K/AKT signaling pathway influences the progression of GC. Furthermore, multiple pieces of evidence have demonstrated that miRNAs can regulate PI3K/Akt signal directly and partially identified the processes behind their oncogene or tumor-suppressor functions in GC. MiR-196b was found to accelerate GC tumor growth by promoting cell cycle and cell proliferation, possibly by activating the PI3K/Akt/mTOR pathway [38]. On the other hand, miR-181d and miR-203 were found to reduce GC cell growth by targeting PIK3CA and, as a result, attenuated Akt activation [39]. It indicates that the PI3K/AKT signal pathway is involved in the occurrence and progression of gastric cancer.
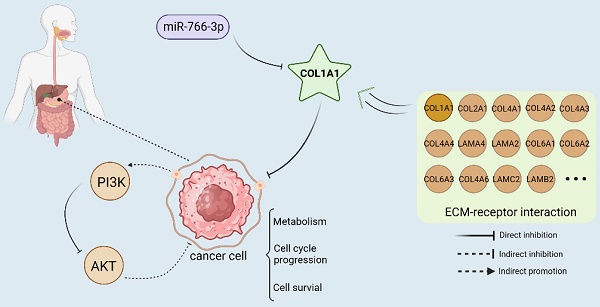
MiRNA-766-3p inhibited gastric cancer via targeting COL1A1 and regulating the PI3K/AKT signaling pathway. (Created with BioRender.com)
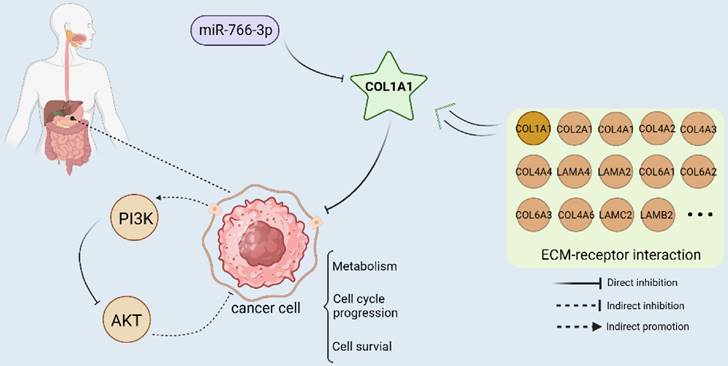
In summary, we demonstrate that miR-766-3p is under-expressed in the tumor tissues of gastric cancer patients. The low expression of 766 was significantly associated with advanced TNM stage, primary tumor and lymph nodes (Fig. S1, Table S2). COL1A1 is an important target of miR-766-3p and is inversely correlated with the expression of miR-766-3p. The luciferase Assay indicated that miR-766-3p suppresses gastric cancer through directly targeting the 3'-UTR of COL1A1 mRNA and down-regulating the levels of COL1A1. It can also participate in the progression of gastric cancer by influencing the PI3K/AKT signal pathway. Meanwhile, COL1A1 is one of the main components of the extracellular matrix (ECM). ECM is also the key component of the tumor micro-environment (TME), which has complementary effects on the development and metastasis of tumors in diverse ways [40]. MiR-766-3p/COL1A1/PI3K/AKT may inhibit gastric cancer progression through suppressing ECM, metabolism, cell cycle progression and cell survival (Fig. 5). Our findings suggest a potential molecular basis for the genesis and progression of gastric cancer, which may lead to new approaches to diagnostics and treatment. In the future, it may represent a potential treatment strategy against the GC.
Abbreviations
GC: gastric cancer; miRNA: microRNA; DEGs: differential genes; TCGA: The Cancer Genome Atlas; GTEx: Genotype-Tissue Expression; HPA: Human Protein Atlas; CMM: circRNA-miRNA-mRNA; ECM: extracellular matrix; TME: tumor micro environment.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
Funding
This study was supported by the Natural Science Foundation of Shanghai Science and Technology Commission (No. 20ZR1459300), the Key Program of Yueyang Hospital of Shanghai University of Traditional Chinese Medicine (No.2019YYZ01), the Special Program of Yueyang Hospital of Shanghai University of Traditional Chinese Medicine (No. 2021yygm06), the Peak Disciplines (Type IV) of Institutions of Higher Learning in Shanghai, the Zhu Shengliang National Famous Elderly Chinese Medicine Experts Inheritance Workshop Construction Project, National Chinese Medicine Human Education Letter [2022] No. 75, Shanghai Municipal Hospital Gastroenterology clinical competence improvement and advancement specialist alliance(SHDC22021311) and Shanghai Yueyang Hospital TCM Speciality Construction Project (YW (2023-2024)-01-08).
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics approval
All procedures performed in studies involving human and animal participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Bioethics Committee of the Medical University of Yueyang Hospital of Integrated Traditional Chinese and Western.
Author contributions
Hongmei Ni, Shengquan Fang, and Qilong Chen designed, conceptualized and supervised the research; ShengHu and Caiyun Zhang analyzed the data; Yue Zhou, Ming Han, Jingjing Li and Fulong Li provided clinical data; Yujie Ding and Mengyuan Zhang performed the experiments, analyzed the data, made the figures and wrote the first draft. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Deo SVS, Sharma J, Kumar S. GLOBOCAN 2020 Report on Global Cancer Burden: Challenges and Opportunities for Surgical Oncologists. Ann Surg Oncol. 2022;29:6497-500
2. Al-Batran S-E. Newest Knowlege in gastric cancer: multimodal therapy of gastric cancer. Annals of Oncology. 2017;28:ix35
3. Coit DG, Thompson JA, Algazi A. et al. Melanoma, Version 2.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016;14:450-73
4. Zhou J, Sun H, Wang Z. et al. Guidelines for the Diagnosis and Treatment of Hepatocellular Carcinoma (2019 Edition). Liver Cancer. 2020;9:682-720
5. Molaei F. et al. Molecular Signaling in Tumorigenesis of Gastric Cancer. IBJ. 2018;22:217-30
6. Smolarz B, Durczyński A, Romanowicz H, Szyłło K, Hogendorf P. miRNAs in Cancer (Review of Literature). IJMS. 2022;23:2805
7. Nurzadeh M, Naemi M, Sheikh Hasani S. A comprehensive review on oncogenic miRNAs in breast cancer. J Genet. 2021;100:15
8. Powrózek T, Małecka-Massalska T. MiRNA and lung cancer radiosensitivity: a mini-review. European Review for Medical and Pharmacological Sciences. 2019;23:8422-8
9. Ghafouri-Fard S, Shoorei H, Taheri M. miRNA profile in ovarian cancer. Experimental and Molecular Pathology. 2020;113:104381
10. Liu J, Yang T, Huang Z, Chen H, Bai Y. Transcriptional regulation of nuclear miRNAs in tumorigenesis (Review). Int J Mol Med. 2022;50:92
11. Yong H, Fu J, Gao G, Shi H, Zheng D, Zhou X. MiR-34a suppresses the proliferation and invasion of gastric cancer by modulating PDL1 in the immune microenvironment. Molecular and Cellular Probes. 2020;53:101601
12. Zheng Y, Xie M, Zhang N. et al. miR-1262 suppresses gastric cardia adenocarcinoma via targeting oncogene ULK1. J Cancer. 2021;12:1231-9
13. Shen X, Si Y, Yang Z, Wang Q, Yuan J, Zhang X. MicroRNA-542-3p suppresses cell growth of gastric cancer cells via targeting oncogene astrocyte-elevated gene-1. Med Oncol. 2015;32:361
14. Zhang M, Ding Y, Hu S. et al. Transcriptomics and systems network-based molecular mechanism of herbal formula Huosu-Yangwei inhibited gastric cancer in vivo. J Ethnopharmacol. 2023;316:116674
15. Hayakawa K, Kawasaki M, Hirai T. et al. MicroRNA-766-3p Contributes to Anti-Inflammatory Responses through the Indirect Inhibition of NF-κB Signaling. IJMS. 2019;20:809
16. Li Z, Liu Y, Yan J. et al. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging. 2020;12:2485-97
17. He X-X, Luo S-S, Qin H-Q, Mo X-W. MicroRNA-766-3p-mediated downregulation of HNF4G inhibits proliferation in colorectal cancer cells through the PI3K/AKT pathway. Cancer Gene Ther. 2022;29:803-13
18. Chen C, Xue S, Zhang J. et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma: miR-766-3p represses cell growth in RCC. Int J Cancer. 2017;141:1867-78
19. Li Z, Xu J, Guan H, Lai J, Yang X, Ma J. Circ_0059354 aggravates the progression of papillary thyroid carcinoma by elevating ARFGEF1 through sponging miR-766-3p. J Endocrinol Invest. 2022;45:825-36
20. Hu M, Zhu S, Xiong S, Xue X, Zhou X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol Rep. 2019.
21. Mardani R, Jafari Najaf Abadi MH, Motieian M. et al. MicroRNA in leukemia: Tumor suppressors and oncogenes with prognostic potential. Journal Cellular Physiology. 2019;234:8465-86
22. Miao J, Regenstein JM, Xu D. et al. The roles of microRNA in human cervical cancer. Archives of Biochemistry and Biophysics. 2020;690:108480
23. Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB, Cheng XC. MicroRNA-21 gene and cancer. Med Oncol. 2013;30:376
24. Dejana E, Tournier-Lasserve E, Weinstein BM. The Control of Vascular Integrity by Endothelial Cell Junctions: Molecular Basis and Pathological Implications. Developmental Cell. 2009;16:209-21
25. Xu Y, Jin J, Liu Y. et al. Snail-Regulated MiR-375 Inhibits Migration and Invasion of Gastric Cancer Cells by Targeting JAK2. Tan M, Ed. PLoS ONE. 2014;9:e99516
26. Xiang G, Huang L, Zhang X. et al. Molecular Characteristics and Promoter Analysis of Porcine COL1A1. Genes. 2022;13:1971
27. Li Y, Sun R, Zhao X, Sun B. RUNX2 promotes malignant progression in gastric cancer by regulating COL1A1. CBM. 2021;31:227-38
28. Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Report. 2018.
29. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target, Discovery medicine - X-MOL [Internet]
30. Huang C, Yang X, Han L. et al. The prognostic potential of alpha-1 type I collagen expression in papillary thyroid cancer. Biochemical and Biophysical Research Communications. 2019;515:125-32
31. Li X, Sun X, Kan C. et al. COL1A1: A novel oncogenic gene and therapeutic target in malignancies. Pathology - Research and Practice. 2022;236:154013
32. McDaniel K, Huang L, Sato K. et al. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. Journal of Biological Chemistry. 2017;292:11336-47
33. Park JH, Kim SR, An HJ, Kim WJ, Choe M, Han JA. Esculetin promotes type I procollagen expression in human dermal fibroblasts through MAPK and PI3K/Akt pathways. Mol Cell Biochem. 2012;368:61-7
34. Xu L, Chen J, Jia L, Chen X, Awaleh Moumin F, Cai J. SLC1A3 promotes gastric cancer progression via the PI3K/AKT signalling pathway. J Cell Mol Med. 2020;24:14392-404
35. Ke J, Ma P, Chen J, Qin J, Qian H. LGR6 promotes the progression of gastric cancer through PI3K/AKT/mTOR pathway. OTT. 2018 Volume 11: 3025-33
36. Li O, Zhao C, Zhang J. et al. UBAP2L promotes gastric cancer metastasis by activating NF-κB through PI3K/AKT pathway. Cell Death Discov. 2022;8:123
37. Chen G, Zhang H, Sun H. et al. Bufalin targeting BFAR inhibits the occurrence and metastasis of gastric cancer through PI3K/AKT/mTOR signal pathway. Apoptosis. 2023.
38. Li N, Wang W, Xu B, Gong H. miR-196b regulates gastric cancer cell proliferation and invasion via PI3K/AKT/mTOR signaling pathway. Oncology Letters. 2016;11:1745-9
39. Zhou X-K, Liang M, Shi B. et al. Downregulation of miR203 induces overexpression of PIK3CA and predicts poor prognosis of gastric cancer patients. DDDT. 2015 3607
40. Wang M, Zhao J, Zhang L. et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8:761-73
Author contact
![]() Corresponding author: Hongmei Ni, email: nihm20032003com; Shengquan Fang, email: fsq20032003com; Qilong Chen, email: cqlw1975com.
Corresponding author: Hongmei Ni, email: nihm20032003com; Shengquan Fang, email: fsq20032003com; Qilong Chen, email: cqlw1975com.

 Global reach, higher impact
Global reach, higher impact