Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(5):1191-1202. doi:10.7150/jca.90439 This issue Cite
Research Paper
Higher concentration of P7C3 than required for neuroprotection suppresses renal cell carcinoma growth and metastasis
1. Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics, Institute of Urology, Peking University Shenzhen Hospital, Shenzhen, Guangdong, China.
2. Department of Urology, Beijing Friendship Hospital, Capital Medical University, Beijing, China.
3. Department of Neurosurgery, Peking University Shenzhen Hospital, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen, Guangdong Province, China.
4. Department of Urology, Peking University First Hospital, Beijing, China.
5. Institute of Precision Medicine, Peking University Shenzhen Hospital, Shenzhen, China.
6. Department of Urology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
* These authors contributed equally to this work.
Received 2023-9-23; Accepted 2023-11-26; Published 2024-1-1
Abstract
Background: P7C3 is a novel compound that has been widely applied in neurodegenerative diseases and nerve injury repair. Here, we show that higher concentrations of P7C3 than are required for in vivo neuroprotection have the novel function of suppressing renal cell carcinoma (RCC) proliferation and metastasis.
Methods: Colony formation, CCK-8 and EdU assay were applied to evaluate RCC cell proliferation. Wound healing and transwell assay were used to measure RCC cell migration and invasion. Flow cytometry assay was employed to detect RCC cell apoptosis and cell cycle. qRT-PCR assay was carried out to measure ribonucleotide reductase subunit M2 (RRM2) mRNA expression level, while western blot assay was utilized to detect the expression level of target proteins. RCC cell growth in vivo was determined by xenografts in mice.
Results: We observed that high concentrations of P7C3 could restrain the proliferation and metastasis of RCC cells and promote cell apoptosis. Mechanistically, this new effect of higher dose of P7C3 was associated with reduced expression of RRM2, and the beneficial efficacy of P7C3 in RCC was blocked when suppression of RRM2 was prevented. When RRM2 suppression was permitted, the cGAS-STING pathway was activated by virtue of RRM2/Bcl-2/Bax signaling. Lastly, intraperitoneal injection of this high level of P7C3 in mice potently inhibited tumor growth.
Conclusion: In conclusion, we show here that P7C3 that exerts an anti-cancer effect in RCC. Our study indicated that P7C3 might act as a novel drug for RCC in the future. The regulatory signal pathway RRM2/Bcl-2/BAX/cGAS-STING might present novel insight to the potential mechanism of RCC development.
Keywords: P7C3, renal cell carcinoma, RRM2, cGAS-STING, BAX
1. Introduction
Renal cell carcinoma (RCC) is the second most lethal urologic tumor. It was estimated that there were 400,000 new cases of renal cancer worldwide and 175,000 people died from this disease [1]. Partial nephrectomy is suitable for those patients with earlier stage renal cancer and their 5-year survival rate reached to 90% [2]. However, approximately 30% of renal cancer patient were diagnosed as an advanced stage at their first exanimation [3]. Despite aggressive therapy were given, it is inevitable that high rates of recurrence and metastasis, causing extremely low 5-year survival rate [4]. Hence, it is imperative to investigate novel medicine to fight against renal cancer.
P7C3, a small molecule, was first discovered in 2010 that could efficiently cross the blood-brain barrier [5]. As a neuroprotective compound, P7C3 has a good effect on neurodegenerative diseases and nerve injury repair [6]. It was shown to protect newborn neurons and promote neurogenesis in adult mice. P7C3 can maintain the survival of nerve cell via activating nicotinamide phosphoribosyltransferase (NAMPT) activity and recovering the level of cellular nicotinamide adenine dinucleotide (NAD+) [7, 8]. Several studies suggested that the derivatives of P7C3 (P7C3-A20 and P7C3-S243) could protect neuroprotective activity in animal models of Parkinson's disease and amyotrophic lateral sclerosis [9-13]. Recent study has showed that P7C3 suppressed the growth and metastasis of glioma via inhibiting PGK1 expression and blocking the process of aerobic glycolysis [14]. However, the role of P7C3 in renal cancer remains unclear.
In our study, we discovered that P7C3 could restrained the proliferation and metastasis of RCC cells and promoted cell apoptosis. Mechanically, P7C3 directly targeted on ribonucleotide reductase subunit M2 (RRM2) and suppressed the mRNA and protein expression of RRM2. RRM2 is the subunit of ribonucleotide reductase, getting involved in the production of deoxynucleic acid and nucleotide metabolism. Numerous studies suggest that RRM2 was dysregulated in various cancers, promoting the progression of cancers, such as renal cancer, liver cancer, lung cancer, breast cancer, colon cancer, ovarian cancer and bladder cancer [15-21]. In renal cancer, RRM2 was dramatically overexpressed in tumor tissue, causing poor prognosis of cancer patients [22]. OSAKO et al. discovered that RRM2 promoted RCC cell proliferation and impaired cell apoptosis in vitro [23]. Xiong et al. found that RRM2 expression was dramatically increased in sunitinib-resistant RCC patient tissues and cells. RRM2 could enhance the sunitinib-resistant of RCC cells via stabilizing ANXA1 protein to activate PI3K/AKT signal pathway [24]. However, we observed that RRM2 could accelerate the progression of RCC via suppressing the activity of Bcl-2/BAX/cGAS-STING signal pathway. In general, our study first reports the role of P7C3 in RCC progression in vitro and in vivo, expounding the molecular mechanism of P7C3 on the anti-cancer effects. These findings may provide a novel method to fight against renal cancer.
2. Material and methods
2.1 Cell lines
Human RCC lines Caki-1 and 786-O were provided by American Type Culture Collection (ATCC), USA. All cells included in this study were grown in DMEM medium (Gibco, Carlsbad, CA, USA) mixed with 10% fetal bovine serum (FBS) (Gibco, South America) and 1% antibiotics. All cells were cultured in a humidified incubator containing 5% CO2 at 37 °C.
2.2 Quantitative real-time PCR assay
Total RNA from RCC cell lines was extracted by utilizing TRIzol reagent. Then, a reverse transcription kit with a gDNA remover (TAKARA, JAPAN) was applied to reverse total RNAs. Quantification of mRNA was using by PrimeScript One Step RT-PCR Kit (TAKARA, JAPAN). The reaction of quantitative real-time PCR assay was carried out by a Roche LightCycler® 480II PCR instrument (Basel, Switzerland). GAPDH was used as an internal control. 2-ΔΔCT method was applied to calculate the relative RNA expression levels.
2.3 Cell transfection
The coding sequence of RRM2 was cloned into pcDNA3.1 vector. pcDNA3.1-negative control (NC) and pcDNA3.1-RRM2 were purchased from GenePharma (Suzhou, China). The plasmid of pcDNA3.1-RRM2 was extracted by using the Plasmid Maxi Kit (OMEGA, USA). The plasmid of pcDNA3.1-RRM2 or pcDNA3.1-NC were transfected into Caki-1 and 786-O cells by utilizing Lipofectamine 3000 (Invitrogen, USA) according to the instructions.
2.4 Cell proliferation assay
A colony-formation assay and an EdU assay were employed to detect the proliferation of RCC cells. For the colony-formation assay, the treated cells were seeded into a 6-well plate at a density of 1000 cells per well and cultured in the incubator for 14 days. Then, the treated cells were washed by PBS solution, fixed by 4% paraformaldehyde and stained with 0.1% crystal violet. The stained cells were imaged and washed with 33% glacial acetic acid. The absorbance of scrubbing solution was detected by using a microplate reader at 550 nm. For EdU assay, treated cells were seeded in a 12-well plate covered with carry with glass. An EdU assay kit (Ribobio, Guangzhou, China) was applied to performed the EdU assay. Cells adhered to carry with glass were photographed by using a microscope (Leica, Germany) under a fluorescence mode. The proportion of EdU positive cells to DAPI-stained cells was equal to the proliferation rate of treated cells.
2.5 Cell migration and invasion assays
A scratch test assay and a transwell assay was carried out to measure the migratory ability of RCC cells. For scratch test assay, RCC cells cultured in a 6-well plate and grown to 100% confluence were exposed to 30 μM of P7C3. A 200 ul pipette tip was applied to generate a clear scar in the cell layer. Images of cells were taken by a microscope (Leica, Germany) at 0 h and 24 h. For the transwell migration assay, RCC cells with 100 ul of serum-free medium were added in the upper chamber, and the lower chamber was stuffed with 500 ul of DMEM medium with 10% FBS. After incubation for 48 h, RCC cells crossed over the chamber were 4% paraformaldehyde, stained with 0.1% crystal violet and imaged. The stained cells were washed by 33% glacial acetic acid, and the absorbance of scrubbing solution were measured by a microplate reader at 550 nm. The experimental method of the transwell invasion assay was identified with that of the transwell migration assay. However, the upper chambers were covered with diluted Matrigel (BD Biosciences, USA).
2.6 Cell cycle assay
RCC cells seeded in to a 6-well plate were exposed to 30 μM of P7C3. Cells were washed with PBS solution twice, fixed with 70% cold ethyl alcohol and incubated at 4℃ overnight. Each sample was mixed with RNase enzyme and incubated at room temperature for half an hour. Then, each sample was incubated with PI kit. Finally, cell cycle detection was determined by a flow cytometry (Beckman, CA, USA).
2.7 Cell apoptosis assay
RCC cells seeded in to a 6-well plate were exposed 30 μM of P7C3. Cells were harvested after digestion of pancreatin without EDTA. Then, cells were resuspended after washing with PBS solution. Finally, all samples were added with Annexin V and PI kit. Cell apoptosis was determined by a flow cytometry (Beckman, CA, USA).
2.8 RNA-sequence assay
The data set included Caki1 and 786-O cell lines treated with 30 μM of P7C3 or PBS solution in thrice replicates. 1ug total RNA was applied for cDNA libraries using cDNA-PCR Sequencing Kit (SQK-LSK110+EXP-PCB096). Transcripts were validated against known reference transcript annotations with gffcompare. Gene function was annotated based on NCBI, Protein family, KOG/COG/eggNOG, KEGG and GO databases. Differential expression analysis of two conditions/groups was performed using the DESeq2 R package. Genes with a FDR < 0.01 and foldchange ≥ 2 found by DESeq2 were regarded as differentially expressed.
2.9 Tandem mass tag proteomics analysis
Total protein from Caki-1 and 786-O cell lines were treated with 30 μM of P7C3 or PBS solution were extracted and determined by using a BCA kit (Thermo Fisher Scientific, USA). Total protein was separated by 12.5% SDS-PAGE gel. Filter-aided sample preparation was performed by using SDT buffer and UA buffer. The produced peptides were collected as a filtrate. Then, the peptide mixture was labelled by utilizing a TMT kit (Thermo Fisher Scientific, USA). Agilent 1260 Infinity II HPLC system was applied to segregate the labeled peptides. Each component of labeled peptides was analyzed by using nano LC-MS/MS system. LC-MS/MS analysis was performed by using a Q Exactive plus mass Spectrometer (Thermo Fisher Scientific, USA) and Easy nLC (Thermo Fisher Scientific, USA) for one hour and a half. Mascot engine embedded into Proteome Discoverer 2.2 (Thermo Fisher Scientific, USA) was applied to analyze the MS/MS raw data.
2.10 Western blotting assay
Total protein from RCC cells was extracted using RIPA lysis buffer (Beyotime, China). Total protein was boiled and quantified by using BCA protein assay kit (Abcam, UK). Equal amounts of protein were separated by 10% SDS-PAGE gels and transferred onto 0.45 μm PVDF membranes (Millipore, USA). The membranes were blocked by 5% skim milk in TBST solution and washed with TBST solution three times. Then, the membranes were incubated with a primary antibody at 4 °C for 12 h, followed by a secondary antibody at room temperature for 2h. At last, the membranes were soaked in DAB Horseradish Peroxidase Color Development Kit (Beyotime, China). All membranes were exposed to a BioSpectrum 600 Imaging System (UVP, USA).
2.11 Immunofluorescence assay
The transfected cells were seeded into 24-well plates covered with glass coverslips for a day. The cells were washed by PBS solution and fixed with 4% of paraformaldehyde. After incubation of skim milk, the glass coverslips were incubated with primary antibodies against RRM2 overnight. Finally, images were captured by using a fluorescence microscope after incubation of fluorescent secondary antibody.
2.12 Xenografts in mice
The growth of RCC cells in vivo was determined by Xenografts in mice. The tumor transplantation experiment was examined and approved by the Ethics Committee of Peking University Shenzhen Hospital. All animal care and experimental procedures were in line with the requirement of animal ethics. 12 4-week-old male BALB/c nude mice were randomized to NC group and P7C3-treated group. Approximately 5×106 786-O cells were injected into the backs of nude mice. Until the size of xenograft tumors reached to 100mm3, mice in P7C3-treated group was peritoneal injection with 10 mg/kg/body P7C3 daily, while negative control group was given intraperitoneal injections of normal saline daily. The size of all xenograft tumors was measured by dividing rule per week on day 14 after beginning P7C3 treatment. All nude mice were sacrificed by carbon dioxide after the injection for 4 weeks, and the xenograft tumors were weighed.
2.13 Statistical analysis
All date from three times independent replication experiment were analyzed by using SPSS 20.0 and GraphPad Prism. All date was presented as Mean ± S.E. Student's t-test was applied to analyze the difference in two groups. ANOVA test was employed to analyze the results of CCK-8 assay. P < 0.05 was considered statistically significant in all results.
3. Results
3.1 P7C3 suppressed the proliferation of RCC cells
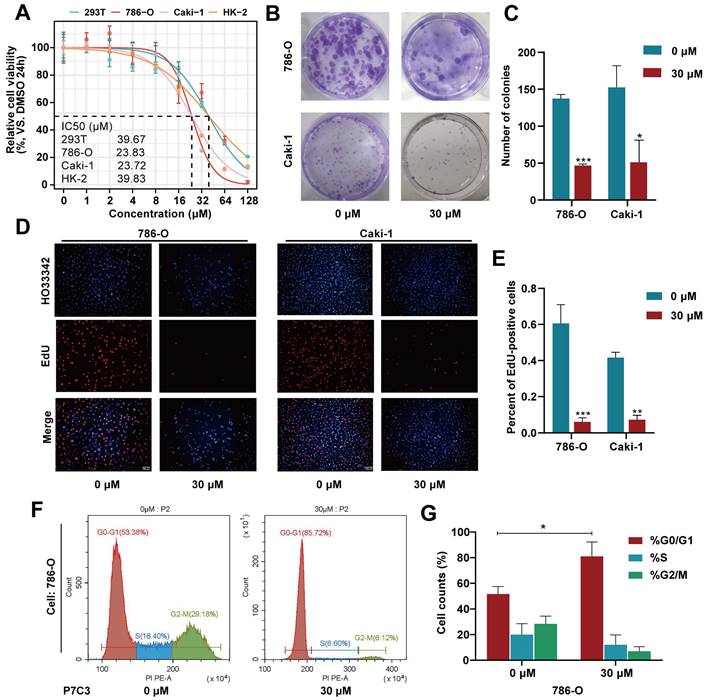
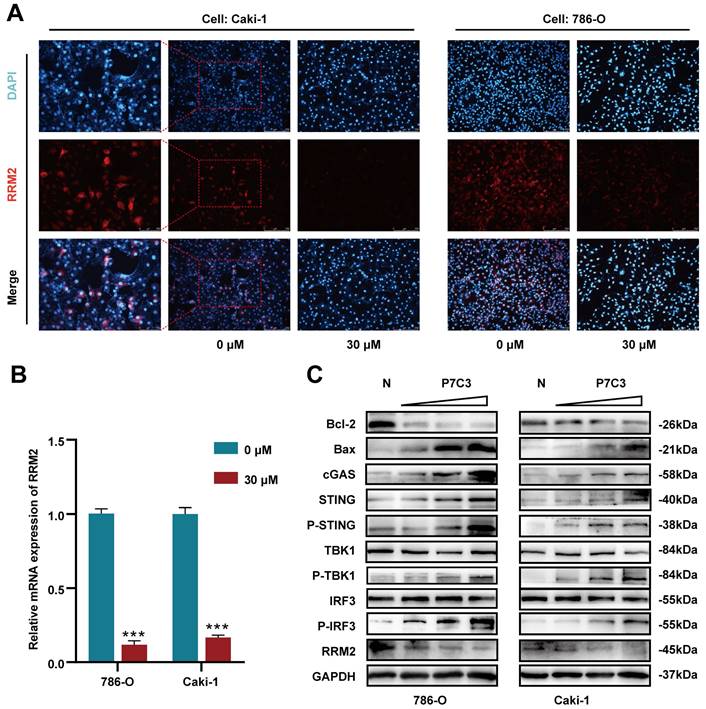
As shown in Fig. 1A, the IC50 value of P7C3 in 293-T, 786-O, Caki-1 and HK-2 was 39.67μM, 23.83μM, 23.72μM and 39.83μM, respectively (Fig. 1A). Colony formation assay presented that the colony numbers of 786-O and Caki-1 cells decreased dramatically while cells were exposed to 30 μM of P7C3 (Fig. 1B and C). In addition, the results of EdU assay were similar to that of Colony formation assay. EDU-positive cells were significantly impaired while 786-O and Caki-1 cells were exposed to 30 μM of P7C3 (Fig. 1D and E). Cell cycle assay showed that P7C3 caused a reduction of S phase (from 16.4% to 6.6% in 786-O cell) and an decrease of G2/M phase (from 29.18% to 6.12% in 786-O cell) (Fig. 1F and G). These results indicating that P7C3 could suppressed the proliferation of RCC cells.
3.2 P7C3 suppressed the metastasis and promoted the apoptosis of RCC cells
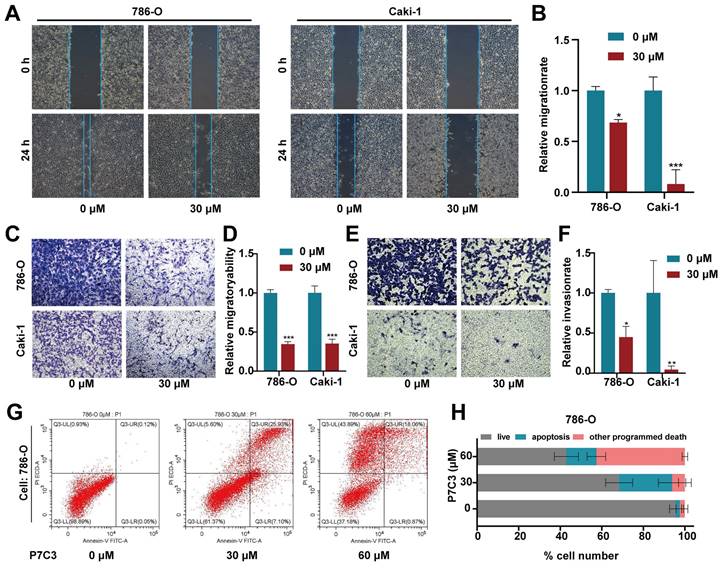
The scratch assay and transwell assay were conducted to detect the migration and invasion of RCC cell. The results of scratch assay showed that P7C3 significantly impaired the migration of Caki-1 and 786-O cells (Fig. 2A and B). Transwell migration assay demonstrated that the number of cells crossed the chamber in P7C3-treated group were dramatically less than that in negative control group (Fig. 2C and D). Transwell invasion assay presented that P7C3 significantly impaired the invasion of RCC cells (Fig. 2E and F). Flow Cytometry analysis was applied to measure cell apoptosis. As shown in Fig. 2G and H, 30 μM of P7C3 obviously facilitated the apoptosis of RCC cells, whereas 60 μM of P7C3 dramatically promoted the death and apoptosis of RCC cells. These results demonstrated that P7C3 restrained the migration and invasion of RCC cells and accelerated the apoptosis of RCC cells.
3.3 P7C3-mediated suppression of RCC is associated with reduced expression of RRM2
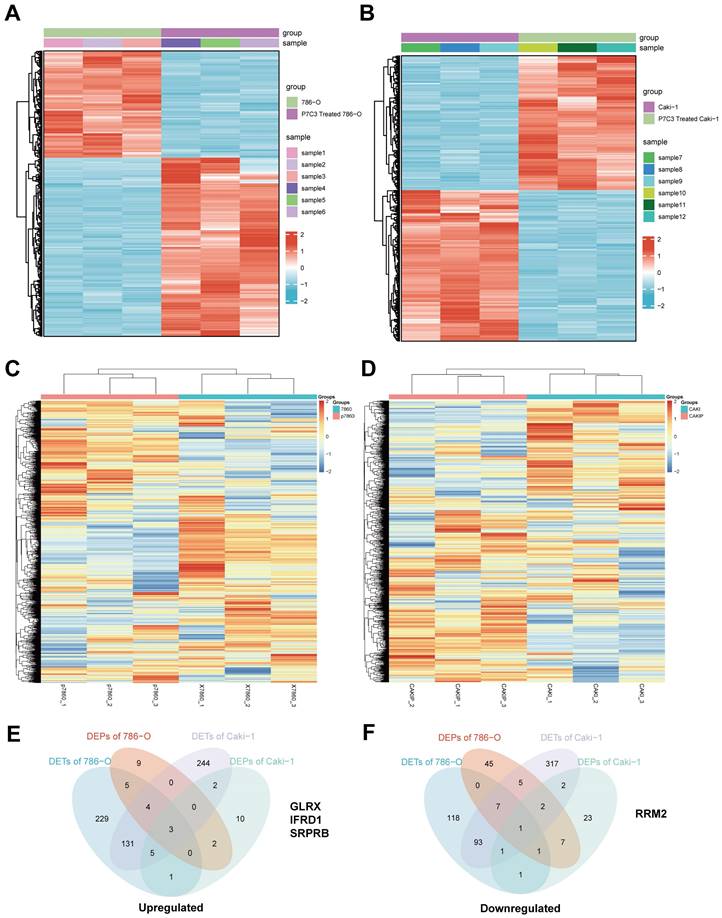
To elucidate the molecular mechanism of anticancer effects mediated by PC73, we applied high throughput sequencing and observed that 507 transcripts are overexpressed in 786-O cell treated with P7C3, while 303 transcripts are down-regulated (Fig. 3A). Meanwhile, 530 transcripts are overexpressed in Caki-1 cell treated with P7C3, while 303 transcripts are down-regulated (Fig. 3B). 23 proteins are overexpressed in 786-O cell treated with P7C3, while 74 proteins are down-regulated (Fig. 3C). 23 proteins are overexpressed in Caki-1 cell treated with P7C3, while 38 proteins are down-regulated (Fig. 3D). 4 genes are the complication of the results of RNA-seq and proteomics data, including GLRX, IFRD1, SRPRB, and RRM2 (Fig. 3E and F). RRM2 expression was significantly decreased when cells were treated with P7C3, while the expression level of GLRX, IFRD1, SRPRB and were augmented obviously upon P7C3 treatment. Further experiments demonstrated that P7C3 not only suppressed RRM2 mRNA expression, but also impaired RRM2 protein expression (Fig. 4A and B). Previous study suggested that knockdown of RRM2 cause cell apoptosis via accelerating the degradation of Bcl-2 protein [25]. Another study showed that Bcl-2 associated X protein (BAX) could activate the cGAS-STING pathway via inducing mitochondrial damage and mtDNA leakage to the cytosol [26]. We suspected that P7C3 could activate the cGAS-STING pathway via regulating RRM2/Bcl-2/BAX signal pathway. As shown in Fig 4C, P7C3 suppressed RRM2 protein expression to inhibited Bcl-2 protein expression and increase BAX protein expression, which activating cGAS-STING pathway. In addition, P7C3 enhanced the phosphorylation of STING, TBK1 and IRF3. These results suggested that P7C3 could inhibited RRM2 expression to activate the cGAS-STING pathway.
P7C3 suppress the proliferation of RCC cells. (A) The IC50 value of 293T, 786-O, Caki-1 and HK-2 cells treated with P7C3. (B and C) The colony numbers of 786-O and Caki-1 cells treated with 30 μM of P7C3 or PBS solution. (D and E) The proliferative rate of 786-O and Caki-1 cells treated with 30 μM of P7C3 or PBS solution. (F and G) Cell cycle of 786-O and Caki-1 cells treated with 30 μM of P7C3 or PBS solution. *p < 0.05, **p < 0.01, ***p < 0.001.
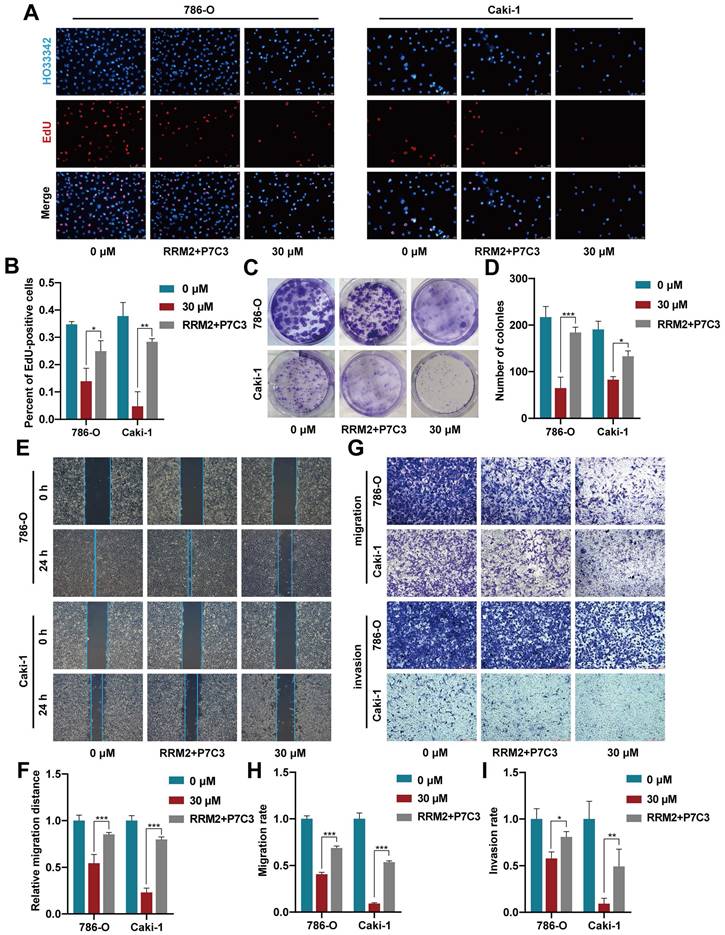
3.4 Increased RRM2 expression reverses the restraining influence mediated by P7C3 inhibition on cell growth and metastasis
To detect whether P7C3 exerts its anticancer effect by modulating RRM2 expression, rescue assay between P7C3 and RRM2 was carried out. EdU assay demonstrated that increased RRM2 expression obviously resisted the P7C3-induced proliferation inhibition in RCC cells (Fig. 5A and B). Colony formation assay presented that forced RRM2 expression significantly retrieved the colony numbers reduction mediated by P7C3 treatment in RCC cells (Fig. 5C and D). Wound-healing assay demonstrated that augmented RRM2 expression reversed RCC cell migration suppression induced by P7C3 (Fig. 5E and F). Finally, transwell migration and invasion assay showed that the inhibitory effect of P7C3 treatment on cell migration and invasion could be reversed while RRM2 expression was enhanced (Fig. 5G-I). These results indicated that overexpression of RRM2 reversed inhibitory effect of P7C3 treatment on cell growth and metastasis.
P7C3 suppress RCC metastasis and promote cell apoptosis. (A and B) The migration distance of 786-O and Caki-1 cells treated with 30 μM of P7C3 or PBS solution. (C and D) P7C3 restrained the migration of 786-O and Caki-1 cells. (E and F) P7C3 restrained the invasion of 786-O and Caki-1 cells. (G and H) The apoptosis rate of 786-O and Caki-1 cells treated with PBS solution, 30 μM of P7C3 and 60 μM of P7C3. *p < 0.05, **p < 0.01, ***p < 0.001.
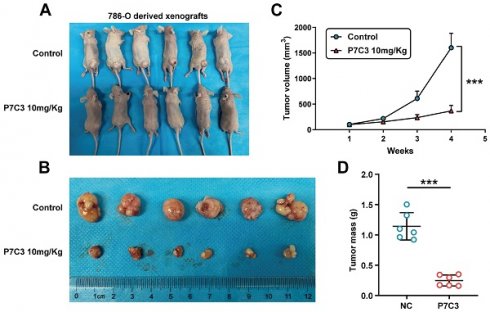
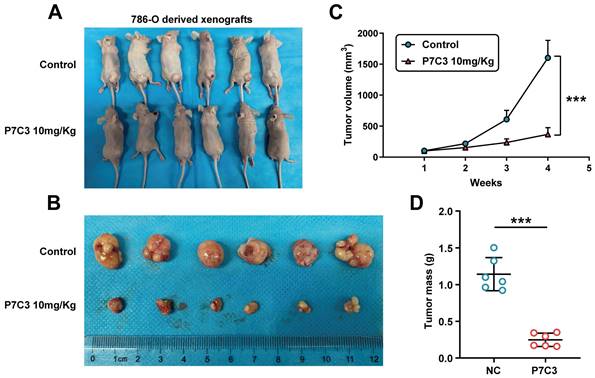
3.5 P7C3 restrained RCC growth in vivo
To investigate the role of P7C3 in RCC cell growth in vivo, 786-O cells were injected into the back of nude mice to produce a xenograft tumor model. Mice were peritoneal injection with P7C3 plus physiological saline was while tumors grew to 100 mm3 (Fig. 6A). In the xenograft tumor model, the transplanted tumors treated with P7C3 were much smaller than those treated with physiological saline (Fig. 6B). Apparently, PC73 inhibited the growth rate and weight of tumors (Fig. 6C and D). These results demonstrated that P7C3 inhibited the growth of RCC cells in vivo.
4. Discussion
Renal cell carcinoma is a common malignant tumor in the urinary system, accounting for 2%-3% of adult malignant tumors [1]. RCC is occult, and there are no obvious symptoms in the early stage. However, approximately 25-30% of RCC patients have distant metastasis at the first time of diagnosis [27]. Palliative treatment is applied for metastatic RCC patients which is not sensitive to radiotherapy and chemotherapy, with only 10% 5-year survival rate [28]. In addition, about 18-23 % limited RCC patients developed to distant metastasis after surgery, causing poor prognosis [29]. Hence, it is urgent to explore novel method to fight against RCC.
P7C3, a novel compound, containing screening Pool #7 (P7) and aminopropyl carbazole Compound #3 (C3) was discovered to enhance the survival of newborn neurons, instead of accelerating the proliferation rate [11]. P7C3 compounds have been shown to be safe and neuroprotective in nonhuman primates [30]. Numerous studies showed that P7C3 compounds contributes to the treatment of traumatic brain injury [6, 9, 31-35], Alzheimer's disease [36], amyotrophic lateral sclerosis [37], Parkinson's disease [10, 38] and stroke [39, 40].
Differentially expressed genes and proteins in RCC cells and RCC cells treated with 30 μM of P7C3. (A) The cluster heat maps show differentially expressed genes in 786-O cell and 786-O cell treated with 30 μM of P7C3. (B) The cluster heat maps show differentially expressed genes in Caki-1 cell and Caki-1 cell treated with 30 μM of P7C3. (C) The cluster heat maps show differentially expressed proteins in 786-O cell and 786-O cell treated with 30 μM of P7C3. (D) The cluster heat maps show differentially expressed proteins in Caki-1 cell and Caki-1 cell treated with 30 μM of P7C3. (E) The Venn diagram exhibiting the overlap between up-regulated gene cluster and protein cluster heat maps. (F) The Venn diagram exhibiting the overlap between down-regulated gene cluster and protein cluster heat maps.
P7C3 activates cGAS-STING signal pathway. (A) The protein expression of RRM2 in 786-O and Caki-1 treated with 30 μM of P7C3 or PBS solution. (B) The Mrna expression of RRM2 in 786-O and Caki-1 treated with 30 μM of P7C3 or PBS solution. (C) Western blot analysis of RRM2, Bcl-2, BAX, cGAS, STING, p-STING, TBK1, p-TBK1, IRF3, p-IRF3 in 786-O and Caki-1 cells treated with different concentration of P7C3 or PBS solution. *p < 0.05, **p < 0.01, ***p < 0.001.
In a zebrafish model of retinal degeneration, P7C3 effectually restrained the apoptosis of retinal [41]. In preclinical models of depression and anxiety, P7C3 could suppress the progression of hippocampal cell death [37, 42]. Wallker et al. discovered that the P7C3 compounds play a role in antidepressant efficacy in mice via enhancing hippocampal neurogenesis [43]. In addition, they also discovered that P7C3-A20 contributed to the treatment of post-traumatic stress associated depression and anxiety, as well as prenatal maternal stress [44-46]. In preclinical models of peripheral neuropathy caused by chemotherapy and Alzheimer's disease, P7C3 could reverse the progress of neurological dysfunction and nerve cell death [36, 47, 48]. Most important of all, the potency, stability and safety of P7C3 were proved by several studies [32]. Recently, the anti-cancer effects of P7C3 were proved in glioma, opening up a new vision of P7C3 application.
RRM2 overexpression reverses P7C3-induced reduce in cell growth and metastasis in RCC cells. (A-D) Augment of RRM2 reversed P7C3-induced proliferation suppression. (E-I) Augment of RRM2 reversed P7C3-induced migration and invasion suppression. *p < 0.05, **p < 0.01, ***p < 0.001.
RRM2 suppress RCC cell growth in vivo. (A and B) The size of transplanted tumors treated with P7C3 were much smaller than negative control group. (C) The growth rate of transplanted tumors in P7C3-treated group and negative control group. (D) The quality of transplanted tumors in P7C3-treated group and negative control group. *p < 0.05, **p < 0.01, ***p < 0.001.
In our study, we observed that high concentration of P7C3 could inhibit the growth of RCC cells in vitro and in vivo. Vazquez-Rosa et al. showed that low concentration of P7C3-A20 (0.3, 1, 3, and 5 μM) could recover the blood-brain barrier, suppress chronic neurodegeneration and restore cognition in mice with traumatic brain injury, while these concentrations of P7C3 have no effect in the RCC culture system. We suspected that higher doses of P7C3 than are needed to protect nerve cells are able to shut down cancer cell proliferation. Secondly, the metabolic rate of cancer cells is different from that in nerve cells. Thirdly, low concentrations of P7C3 may be easily cross the blood-brain barrier instead of high concentrations of P7C3. We also observed that P7C3 significantly restrained RCC metastasis and enhance cell apoptosis. However, 30 μM of P7C3 caused 25.93% of cell apoptosis and 5.6% of cell death, while 60 μM of P7C3 caused 18.06% of cell apoptosis and 43.89% of cell death. We suspected that high concentration of P7C3 might trigger other mechanisms of procedural death. Mechanistically, P7C3 could suppress the translation of RRM2. Numerous studies suggest that RRM2 expression was dramatically increased in various cancer tissues, causing unfavorable prognosis to these patients [15]. In vitro and in vivo assay demonstrated that RRM2 got involved in the progression of cancers, such as cell proliferation, cell apoptosis, cell metastasis, chemoresistance, energy metabolism and immune resistance [49, 50]. In melanoma, RRM2 could enhance melanoma grow and promote cellular senescence [51]. In breast cancer, RRM2 accelerated cell migration and invasion via regulating epithelial-mesenchymal transition (EMT) process [52]. In head and neck and lung cancers, knockdown of RRM2 caused cell apoptosis via accelerating the degradation of Bcl-2 [25]. In lung adenocarcinoma (LUAD), high expression of RRM2 is regarded as a potential independent risk factor and correlated with immune infiltration. Zhang et al. found that RRM2 could enhance the proliferation and metastasis of LUAD cells. In adddiotn, RRM2 could promote cell proliferation and suppress cell apoptosis via impairing the activity of cGAS/STING signaling pathway [53]. The cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway is regarded as an important innate immune pathway and activated by double-stranded DNA, promoting unique immune effector responses that can regulate different kinds of tumorigenesis, including cell growth, cell metastasis, chemotherapy resistance and immune escape [54-56]. cGAS can produce cyclic GMP-AMP (cGAMP), a distinct mixed-linkage second-messenger member molecule, binding and activing STING. STING activation launch the downstream transcriptional progression including nuclear factor (NF)-κB and interferon (IFN) regulatory factor 3 (IRF3) to augment the expression of chemokines, proinflammatory cytokines and type I IFNs [57, 58]. However, the regulatory pathway between RRM2 and cGAS/STING signaling pathway remains unclear. Recently, a study suggest that Bcl-2 associated X protein (BAX) could activate the cGAS-STING pathway via inducing mtDNA cytosolic leakage [59, 60]. Therefore, we suspected that P7C3 might activate cGAS-STING via regulating Bcl-2/BAX pathway. Western blot assay suggested that P7C3 obviously impaired Bcl-2 protein expression and increased BAX protein expression, indicating that P7C3 could activate cGAS-STING signal pathway via regulating BAX expression. Further experimental results confirmed that P7C3 could activate cGAS-STING signal pathway via regulating RRM2/Bcl-2/BAX pathway. Finally, rescue assays demonstrated that P7C3 exert its anti-cancer effects via regulating RRM2 expression.
In conclusion, we identified the novel compound P7C3 that exerts an anti-cancer effect in RCC. Further experiments demonstrated that P7C3 suppressed RRM2 expression to disequilibrate the balance between Bcl-2 and BAX and impair the activity of cGAS-STING signal pathway, inhibiting the development of RCC. Our study indicated that P7C3 might act as a novel drug for RCC in the future. The regulatory signal pathway RRM2/Bcl-2/BAX/ cGAS-STING might present novel insight to the potential mechanism of RCC development.
Acknowledgements
This work was sponsored by Guangdong High-level Hospital Construction Fund and sponsored by Shanghai Sailing Program (21YF1437200).
Data availability
The data used to support the findings of this study are available from the corresponding authors upon request.
Author contributions
Ge Shu, Chenchen Huang and Jianfa Li performed the experiments. Ge Shu, Wenjin Chen and Chenchen Huang prepared all the figures. Hui Shan did mass spectrometry analysis. Ge Shu and Jianfa Li wrote the manuscript. Jing Ye supervised this study. Yaoting Gui provided fund for the whole project.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Bhindi B, Lohse CM, Schulte PJ, Mason RJ, Cheville JC, Boorjian SA. et al. Predicting Renal Function Outcomes After Partial and Radical Nephrectomy. Eur Urol. 2019;75:766-72
3. Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894-906
4. Attalla K, Weng S, Voss MH, Hakimi AA. Epidemiology, Risk Assessment, and Biomarkers for Patients with Advanced Renal Cell Carcinoma. Urol Clin North Am. 2020;47:293-303
5. Pieper AA, Xie S, Capota E, Estill SJ, Zhong J, Long JM. et al. Discovery of a proneurogenic, neuroprotective chemical. Cell. 2010;142:39-51
6. Blaya MO, Wasserman JM, Pieper AA, Sick TJ, Bramlett HM, Dietrich WD. Neurotherapeutic capacity of P7C3 agents for the treatment of Traumatic Brain Injury. Neuropharmacology. 2019;145:268-82
7. Manickam R, Tur J, Badole SL, Chapalamadugu KC, Sinha P, Wang Z. et al. Nampt activator P7C3 ameliorates diabetes and improves skeletal muscle function modulating cell metabolism and lipid mediators. J Cachexia Sarcopenia Muscle. 2022;13:1177-96
8. Ryu SW, Kim YO, Kim HB, Oh SB, Choi JI, Yoon MH. Antinociceptive effect of intrathecal P7C3 via GABA in a rat model of inflammatory pain. Eur J Pharmacol. 2021;899:174029
9. Vazquez-Rosa E, Shin MK, Dhar M, Chaubey K, Cintron-Perez CJ, Tang X. et al. P7C3-A20 treatment one year after TBI in mice repairs the blood-brain barrier, arrests chronic neurodegeneration, and restores cognition. Proc Natl Acad Sci U S A. 2020;117:27667-75
10. De Jesus-Cortes H, Miller AD, Britt JK, DeMarco AJ, De Jesus-Cortes M, Stuebing E. et al. Protective efficacy of P7C3-S243 in the 6-hydroxydopamine model of Parkinson's disease. NPJ Parkinsons Dis. 2015;1:15010 -
11. Pieper AA, McKnight SL, Ready JM. P7C3 and an unbiased approach to drug discovery for neurodegenerative diseases. Chem Soc Rev. 2014;43:6716-26
12. MacMillan KS, Naidoo J, Liang J, Melito L, Williams NS, Morlock L. et al. Development of proneurogenic, neuroprotective small molecules. J Am Chem Soc. 2011;133:1428-37
13. Naidoo J, De Jesus-Cortes H, Huntington P, Estill S, Morlock LK, Starwalt R. et al. Discovery of a neuroprotective chemical, (S)-N-(3-(3,6-dibromo-9H-carbazol-9-yl)-2-fluoropropyl)-6-methoxypyridin-2-amine [(-)-P7C3-S243], with improved druglike properties. J Med Chem. 2014;57:3746-54
14. Chen W, Jia W, Wu C, Chen L, Sun K, Wang J. et al. The Neurogenic Compound P7C3 Regulates the Aerobic Glycolysis by Targeting Phosphoglycerate Kinase 1 in Glioma. Front Oncol. 2021;11:644492
15. Zuo Z, Zhou Z, Chang Y, Liu Y, Shen Y, Li Q. et al. Ribonucleotide reductase M2 (RRM2): Regulation, function and targeting strategy in human cancer. Genes Dis. 2024;11:218-33
16. Yang Y, Lin J, Guo S, Xue X, Wang Y, Qiu S. et al. RRM2 protects against ferroptosis and is a tumor biomarker for liver cancer. Cancer Cell Int. 2020;20:587
17. Khan P, Siddiqui JA, Kshirsagar PG, Venkata RC, Maurya SK, Mirzapoiazova T. et al. MicroRNA-1 attenuates the growth and metastasis of small cell lung cancer through CXCR4/FOXM1/RRM2 axis. Mol Cancer. 2023;22:1
18. Abdel-Rahman MA, Mahfouz M, Habashy HO. RRM2 expression in different molecular subtypes of breast cancer and its prognostic significance. Diagn Pathol. 2022;17:1
19. Lu AG, Feng H, Wang PX, Han DP, Chen XH, Zheng MH. Emerging roles of the ribonucleotide reductase M2 in colorectal cancer and ultraviolet-induced DNA damage repair. World J Gastroenterol. 2012;18:4704-13
20. Li H, Lei Y, Li S, Li F, Lei J. LncRNA PART1 Stimulates the Development of Ovarian Cancer by Up-regulating RACGAP1 and RRM2. Reprod Sci. 2022;29:2224-35
21. Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885-92
22. Guo X, Sun Z, Jiang S, Jin X, Wang H. Identification and validation of a two-gene metabolic signature for survival prediction in patients with kidney renal clear cell carcinoma. Aging (Albany NY). 2021;13:8276-89
23. Osako Y, Yoshino H, Sakaguchi T, Sugita S, Yonemori M, Nakagawa M. et al. Potential tumor-suppressive role of microRNA-99a-3p in sunitinib-resistant renal cell carcinoma cells through the regulation of RRM2. Int J Oncol. 2019;54:1759-70
24. Xiong W, Zhang B, Yu H, Zhu L, Yi L, Jin X. RRM2 Regulates Sensitivity to Sunitinib and PD-1 Blockade in Renal Cancer by Stabilizing ANXA1 and Activating the AKT Pathway. Adv Sci (Weinh). 2021;8:e2100881
25. Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z. et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19:3416-28
26. Cosentino K, Hertlein V, Jenner A, Dellmann T, Gojkovic M, Pena-Blanco A. et al. The interplay between BAX and BAK tunes apoptotic pore growth to control mitochondrial-DNA-mediated inflammation. Mol Cell. 2022;82:933-49 e9
27. Msaouel P, Genovese G, Tannir NM. Renal Cell Carcinoma of Variant Histology: Biology and Therapies. Hematol Oncol Clin North Am. 2023;37:977-92
28. Choueiri TK, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N Engl J Med. 2017;376:354-66
29. Dabestani S, Thorstenson A, Lindblad P, Harmenberg U, Ljungberg B, Lundstam S. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: a population-based study. World J Urol. 2016;34:1081-6
30. Bauman MD, Schumann CM, Carlson EL, Taylor SL, Vazquez-Rosa E, Cintron-Perez CJ. et al. Neuroprotective efficacy of P7C3 compounds in primate hippocampus. Transl Psychiatry. 2018;8:202
31. Shin MK, Vazquez-Rosa E, Koh Y, Dhar M, Chaubey K, Cintron-Perez CJ. et al. Reducing acetylated tau is neuroprotective in brain injury. Cell. 2021;184:2715-32 e23
32. Sridharan PS, Miller E, Pieper AA. Application of P7C3 Compounds to Investigating and Treating Acute and Chronic Traumatic Brain Injury. Neurotherapeutics. 2023
33. Yin TC, Britt JK, De Jesus-Cortes H, Lu Y, Genova RM, Khan MZ. et al. P7C3 neuroprotective chemicals block axonal degeneration and preserve function after traumatic brain injury. Cell Rep. 2014;8:1731-40
34. Dutca LM, Stasheff SF, Hedberg-Buenz A, Rudd DS, Batra N, Blodi FR. et al. Early detection of subclinical visual damage after blast-mediated TBI enables prevention of chronic visual deficit by treatment with P7C3-S243. Invest Ophthalmol Vis Sci. 2014;55:8330-41
35. Blaya MO, Bramlett HM, Naidoo J, Pieper AA, Dietrich WD. Neuroprotective efficacy of a proneurogenic compound after traumatic brain injury. J Neurotrauma. 2014;31:476-86
36. Voorhees JR, Remy MT, Cintron-Perez CJ, El Rassi E, Khan MZ, Dutca LM. et al. (-)-P7C3-S243 Protects a Rat Model of Alzheimer's Disease From Neuropsychiatric Deficits and Neurodegeneration Without Altering Amyloid Deposition or Reactive Glia. Biol Psychiatry. 2018;84:488-98
37. Tesla R, Wolf HP, Xu P, Drawbridge J, Estill SJ, Huntington P. et al. Neuroprotective efficacy of aminopropyl carbazoles in a mouse model of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2012;109:17016-21
38. De Jesus-Cortes H, Xu P, Drawbridge J, Estill SJ, Huntington P, Tran S. et al. Neuroprotective efficacy of aminopropyl carbazoles in a mouse model of Parkinson disease. Proc Natl Acad Sci U S A. 2012;109:17010-5
39. Loris ZB, Hynton JR, Pieper AA, Dietrich WD. Beneficial Effects of Delayed P7C3-A20 Treatment After Transient MCAO in Rats. Transl Stroke Res. 2018;9:146-56
40. Loris ZB, Pieper AA, Dietrich WD. The neuroprotective compound P7C3-A20 promotes neurogenesis and improves cognitive function after ischemic stroke. Exp Neurol. 2017;290:63-73
41. Asai-Coakwell M, March L, Dai XH, Duval M, Lopez I, French CR. et al. Contribution of growth differentiation factor 6-dependent cell survival to early-onset retinal dystrophies. Hum Mol Genet. 2013;22:1432-42
42. Choi SH, Bylykbashi E, Chatila ZK, Lee SW, Pulli B, Clemenson GD. et al. Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer's mouse model. Science. 2018 361
43. Walker AK, Rivera PD, Wang Q, Chuang JC, Tran S, Osborne-Lawrence S. et al. The P7C3 class of neuroprotective compounds exerts antidepressant efficacy in mice by increasing hippocampal neurogenesis. Mol Psychiatry. 2015;20:500-8
44. Bavley CC, Kabir ZD, Walsh AP, Kosovsky M, Hackett J, Sun H. et al. Dopamine D1R-neuron cacna1c deficiency: a new model of extinction therapy-resistant post-traumatic stress. Mol Psychiatry. 2021;26:2286-98
45. Schroeder R, Nguyen L, Pieper AA, Stevens HE. Maternal treatment with P7C3-A20 protects from impaired maternal care after chronic gestational stress. Behav Brain Res. 2022;416:113558
46. Schroeder R, Sridharan P, Nguyen L, Loren A, Williams NS, Kettimuthu KP. et al. Maternal P7C3-A20 Treatment Protects Offspring from Neuropsychiatric Sequelae of Prenatal Stress. Antioxid Redox Signal. 2021;35:511-30
47. Kemp SWP, Szynkaruk M, Stanoulis KN, Wood MD, Liu EH, Willand MP. et al. Pharmacologic rescue of motor and sensory function by the neuroprotective compound P7C3 following neonatal nerve injury. Neuroscience. 2015;284:202-16
48. Jiang B, Song L, Huang C, Zhang W. P7C3 Attenuates the Scopolamine-Induced Memory Impairments in C57BL/6J Mice. Neurochem Res. 2016;41:1010-9
49. Sharma A, Kumar P, Ambasta RK. Cancer Fighting SiRNA-RRM2 Loaded Nanorobots. Pharm Nanotechnol. 2020;8:79-90
50. Zhan Y, Jiang L, Jin X, Ying S, Wu Z, Wang L. et al. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed Pharmacother. 2021;133:110996
51. Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A. et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252-65
52. He J, Wei Q, Jiang R, Luan T, He S, Lu R. et al. The Core-Targeted RRM2 Gene of Berberine Hydrochloride Promotes Breast Cancer Cell Migration and Invasion via the Epithelial-Mesenchymal Transition. Pharmaceuticals (Basel). 2022 16
53. Jiang X, Li Y, Zhang N, Gao Y, Han L, Li S. et al. RRM2 silencing suppresses malignant phenotype and enhances radiosensitivity via activating cGAS/STING signaling pathway in lung adenocarcinoma. Cell Biosci. 2021;11:74
54. Samson N, Ablasser A. The cGAS-STING pathway and cancer. Nat Cancer. 2022;3:1452-63
55. Hong C, Schubert M, Tijhuis AE, Requesens M, Roorda M, van den Brink A. et al. cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature. 2022;607:366-73
56. Lv M, Chen M, Zhang R, Zhang W, Wang C, Zhang Y. et al. Manganese is critical for antitumor immune responses via cGAS-STING and improves the efficacy of clinical immunotherapy. Cell Res. 2020;30:966-79
57. Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548-69
58. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21:501-21
59. Zang N, Cui C, Guo X, Song J, Hu H, Yang M. et al. cGAS-STING activation contributes to podocyte injury in diabetic kidney disease. iScience. 2022;25:105145
60. McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 2018 359
Author contact
![]() Corresponding authors: Jianfa Li, email: lijianfayouxiangcom; Yaoting Gui, email: guiyaoting2023com.
Corresponding authors: Jianfa Li, email: lijianfayouxiangcom; Yaoting Gui, email: guiyaoting2023com.

 Global reach, higher impact
Global reach, higher impact