3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(8):2147-2159. doi:10.7150/jca.93364 This issue Cite
Research Paper
The Analysis of ceRNA Networks and Tumor Microenvironment in Endometrial Cancer
1. Clinical and Translational Research Center, Shanghai First Maternity and Infant Hospital, Tongji University School of Medicine, Shanghai, China.
2. The International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200030, China.
3. Shanghai Key Laboratory of Embryo Original Diseases, Shanghai 200030, China.
4. Blood Transfusion Department, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
5. Department of Gynecology, Affiliated Hospital of Jiangsu University, Zhenjiang, Jiangsu, China.
Abstract
Background: Endometrial carcinoma is a life-threatening and aggressive tumor that affects women worldwide. ceRNAs and carcinoma-infiltrating immunocytes can be associated with tumor formation and progression. Therefore, investigating the unique mechanisms underlying endometrial carcinoma is crucial.
Methods: Prognostic nomograms were constructed based on the differentially expressed genes between normal and tumor tissues. Twenty types of tumor immune infiltrating cells in uterine corpus endometrial carcinoma (UCEC) were examined using CIBERSORT. To identify the potential signaling pathways, the associations among essential ceRNA network genes and important immunocytes were investigated using the co-expression assay.
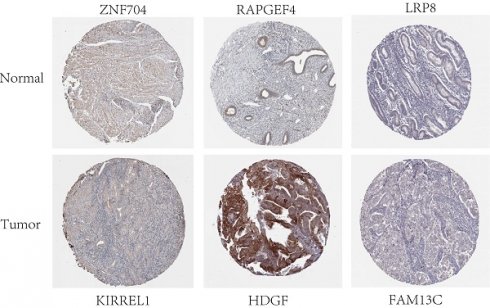
Results: Differential analysis identified 3636 mRNAs, 249 miRNAs, and 252 lncRNAs unique to UCEC. The ceRNA network was constructed using the interplays between 19 lncRNA-miRNA pairs and 434 miRNA-mRNA pairs. Furthermore, CIBERSORT and ceRNA integration analysis revealed that immune cells, including dendritic cells and natural killer cells, and associated ceRNAs such as LRP8, HDGF, PPARGC1B, and TEAD1 can appropriately predict prognosis. A receiver operating characteristic curve was constructed to predict patient outcomes.
Conclusions: Using a nomogram, we predicted the outcomes of patients with UCEC Furthermore, we revealed its significance in improving clinical management.
Keywords: Endometrial carcinoma, ceRNA, Prognosis, Immune cells infiltration, Machine learning

 Global reach, higher impact
Global reach, higher impact