3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(8):2354-2360. doi:10.7150/jca.94187 This issue Cite
Research Paper
Impact of LINC00312 gene polymorphism coupled with habitual risks on buccal mucosa cancer
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. School of Medicine, China Medical University, Taichung, Taiwan.
3. Department of Pathology, China Medical University Hospital, Taichung, Taiwan.
4. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan.
6. Department of Otolaryngology, St. Martin De Porres Hospital, Chiayi, Taiwan.
7. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan.
8. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan.
9. Department of Neurosurgery, Show Chwan Memorial Hospital, Changhua, Taiwan.
10. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
11. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
12. Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
Received 2024-1-11; Accepted 2024-2-14; Published 2024-3-4
Abstract
Oral squamous cell carcinoma (OSCC) is a prevalent and lethal malignancy with a diverse etiology. LINC00312 is a long intergenic non-coding RNA that functions as a signal hub to regulate the progression and treatment of head and neck cancer. The aim of this study was to evaluate the effect of LINC00312 single nucleotide polymorphisms (SNPs) on the development of oral cancer. Two LINC00312 SNPs, namely rs12497104 and rs164966, were investigated among 469 male patients with cancer of buccal mucosa and 1194 gender- and age-matched controls. No significant correlation was observed between these two SNPs and the occurrence of OSCC in the case and control groups. While assessing the clinicopathological features, carriers of at least one minor allele of rs164966 (GA and GG) were less prone to develop lymph node metastasis (adjusted odds ratio [AOR], 0.666; 95% confidence interval [CI], 0.447-0.991; p=0.045) in comparison with homozygous carriers of the major allele (AA). Subsequent stratifying surveys revealed that this genetic association with nodal spread was seen only in cases who habitually chewed betel quid (AOR, 0.616; 95% CI, 0.386-0.985; p=0.042) or smoked cigarettes (AOR, 0.612; 95% CI, 0.393-0.953; p=0.029), but undetected in cases free of these main behavioral risks. Our results indicate an interactivity of LINC00312 rs164966 with lifestyle-related risks on modulating OSCC progression.
Keywords: LINC00312, genetic variants, OSCC, chewed betel quid
Introduction
Oral oncogenesis, the transformation by which oral cancer develops, is a complex interplay of inherited genetic factors and environmental exposures [1]. While a number of genetic factors contribute to genome stability, cell proliferation and programmed cell death [2], various environmental factors, such as viral infections [3] and chronic use of cigarette, alcohol, and betel quid (one areca nut-related product) [4], have been identified as significant risk factors for oral cancer. Besides, additional potential contributors to oral cancer include periodontal diseases [5], and infectious and inflammatory conditions [6]. These etiologies highlight the multifaceted character of oral cancer. Oral squamous cell carcinoma (OSCC) remains the most prevalent form of oral cancer and is linked with substantial death rates [7] regardless of the presence of contemporary therapeutic approaches. Given the high heterogeneity of OSCC pathogenesis, all of the aforementioned risk factors appear to be interrelated and play a role in determining OSCC incidence and prognosis. In addition, another imperative feature of OSCC that may account for its heterogenous nature is the presence of different anatomical sites, each represented by a unique set of tissue organization and viewed as a distinct biological entity [8]. Notably, oral cancer in Taiwan occurs primarily in the buccal mucosa [9], as OSCC most commonly presents as tongue cancer in the western countries [10].
The latest breakthroughs in the understanding of long non-coding RNA (lncRNA) functions have revolutionized the scope of molecular genetics [11]. Until now, a considerable number of lncRNAs are found to be linked to a myriad of disorders [12, 13], including oral cancer [13-16]. Among these cancer-associated lncRNAs, LINC00312, long intergenic non-coding RNA 312, was reported to be significantly downregulated in nasopharyngeal carcinoma (NPC), and its levels were negatively correlated with tumor size but positively correlated with lymph node metastasis in NPC [17]. Functional assessment revealed an inhibitory effect of LINC00312 on cancer cell proliferation via induction of cell cycle arrest in NPC [18]. On the contrary, through upregulation of matrix metalloprotease-1 (MMP-1), a promotive role of LINC00312 in NPC invasion was documented [19]. In addition to the development and progression of NPC, a functional involvement of LINC00312 in NPC treatment was also noted, as this tumor suppressor was shown to improve the efficacy of radiotherapy in a xenograft mouse model of NPC by suppressing the phosphorylation of DNA-PKcs, a DNA-dependent protein kinase that is central to DNA double strand break repair [20]. These findings suggest that LINC00312, acting as a signal hub, is capable of orchestrating NPC progression and treatment.
Lately, research employing targeted gene methods has demonstrated a genetic link between single-nucleotide polymorphisms (SNPs) of LINC00312 and various aspects of NPC prognosis. Specifically, LINC00312 variants, rs15734 and rs164966, were associated with a decrease in the risks of developing NPC, as patients with rs12497104 polymorphisms showed a poorer overall survival [21, 22]. In addition, association of rs12497104 and rs15734 with chemoradiotherapy-induced hematotoxicity in NPC was recently reported [23]. As LINC00312 was found to be implicated in the development of oral submucous fibrosis [24] and head and neck cancer [25], the impact of LINC00312 SNPs together with lifestyle-related risks on OSCC is in need of clarification. In this study, we performed a case-control survey to determine the extent to which genetic variants of LINC00312 are associated with the risk of oral cancer.
Materials and Methods
Subjects
This study was approved by the institutional review board of Chung Shan Medical University Hospital, Taichung, Taiwan (CS1-21151). This study enrolled 469 male patients diagnosed with cancer of buccal mucosa and 1194 cancer-free male controls, who did not report a history of cancer or any oral precancerous conditions, to investigate the influence of LINC00312 variants on the development of OSCC. Informed written consent was collected from all participants, recruited between 2012 and 2022, during enrollment. Grading and staging of cancer were determined by using the American Joint Committee on Cancer (AJCC) TNM staging system [26]. The control group included males who did not report a history of cancer or any oral precancerous conditions like oral submucous fibrosis, leukoplakia, erythroplakia, verrucous hyperplasia, among others. Age and environmental risk information (including the use of areca nuts, tobacco, and alcohol) was gathered from all subjects.
Genotyping
Two loci located within the LINC00312 gene (rs12497104 and rs164966) were chosen for analysis in this study on the basis of their putative association with nasopharyngeal carcinoma susceptibility [21, 23]. The QIAamp DNA Blood Mini Kit (Qiagen) was used to extract genomic DNA from whole blood specimens. The allelic discrimination of these two LINC00312 SNPs including rs12497104 (assay ID: C_11757485_10), and rs164966 (assay ID: C_8753508_10) were assessed using the TaqMan assay performed with an ABI StepOne™ Real-Time PCR System (Applied Biosystems). The resulting data were processed by SDS version 3.0 software from Applied Biosystems.
Statistical analysis
Demography and environmental exposures were compared between patients and controls using Mann-Whitney U-test. Multiple logistic regression models followed by adjustment for possible confounders were used to examine the association of genotypic ratios with OSCC risk or clinical status. Calculations were performed with SAS software (v9.4, 2013; SAS Institute Inc.) The threshold for a difference or association was set at a p value of <0.05.
Results
Demographic characteristics of recruited buccal mucosa cancer patients
This study enrolled 469 male patients with squamous cell carcinoma of buccal mucosa and 1194 cancer-free males to examine the relationship between LINC00312 polymorphisms and the development of oral tumorigenesis. Demographic and clinical characteristics of both cohorts were assessed (Table 1). To exclude potential confounding factors, cancer-free controls with matched age and gender were recruited. Significant discrepancies in the proportions of cigarette smoking, alcohol consumption, and betel quid chewing were found between the case and control groups, in accordance with findings from other reports performed in Central and Southeast Asia [7, 27]. In the case group, nodal and distal metastatic disease occurred in 30.5% and 0.6% of patients, respectively. Overall, 81.9% of the tumors in our cohort were moderately or poorly differentiated.
Comparisons of clinical and demographic characteristics in male patients with cancers of the buccal mucosa (n=469) and cancer-free controls (n=1194).
| Variable | Controls (N=1194) | Patients (N=469) | p value |
|---|---|---|---|
| Age (yrs) | |||
| < 55 | 564 (47.2%) | 214 (45.6%) | p = 0.554 |
| ≥ 55 | 630 (52.8%) | 255 (54.4%) | |
| Betel quid chewing | |||
| No | 996 (83.4%) | 132 (28.1%) | |
| Yes | 198 (16.6%) | 337 (71.9%) | p < 0.001* |
| Cigarette smoking | |||
| No | 559 (46.8%) | 82 (17.5%) | |
| Yes | 635 (53.2%) | 387 (82.5%) | p < 0.001* |
| Alcohol drinking | |||
| No | 957 (80.2%) | 270 (57.6%) | |
| Yes | 237 (19.8%) | 199 (42.4%) | p < 0.001* |
| Stage | |||
| I+II | 224 (47.8%) | ||
| III+IV | 245 (52.2%) | ||
| Tumor T status | |||
| T1+T2 | 236 (50.3%) | ||
| T3+T4 | 233 (49.7%) | ||
| Lymph node status | |||
| N0 | 326 (69.5%) | ||
| N1+N2+N3 | 143 (30.5%) | ||
| Metastasis | |||
| M0 | 466 (99.4%) | ||
| M1 | 3 (0.6%) | ||
| Cell differentiation | |||
| Well differentiated | 85 (18.1%) | ||
| Moderately or poorly differentiated | 384 (81.9%) |
Mann-Whitney U test was used between controls and male patients with cancers of the buccal mucosa. * p value < 0.05 as statistically significant.
Impacts of LINC00312 genetic polymorphisms on the buccal mucosa cancer incidence
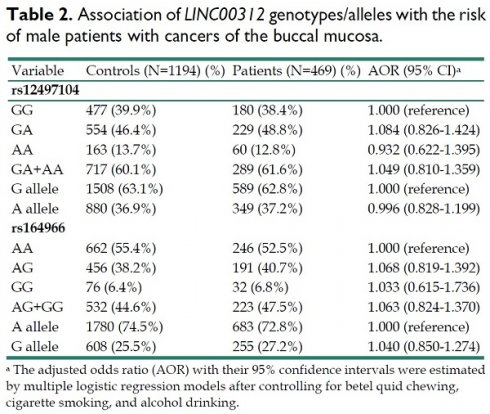
To explore the potential correlation of LINC00312 variants with the development of oral cancer, we genotyped two SNPs of LINC00312 gene, rs12497104 and rs164966, in our study groups, and no deviation from Hardy-Weinberg equilibrium was found (p>0.05) in both cohorts. Distributions of both genotype and allele frequency for individual SNP were determined in cases and controls. For these two SNPs, no significant correlations of their polymorphisms with the risk of oral cancer were observed between cases and controls (Table 2).
Association of LINC00312 genotypes/alleles with the risk of male patients with cancers of the buccal mucosa.
| Variable | Controls (N=1194) (%) | Patients (N=469) (%) | AOR (95% CI)a |
|---|---|---|---|
| rs12497104 | |||
| GG | 477 (39.9%) | 180 (38.4%) | 1.000 (reference) |
| GA | 554 (46.4%) | 229 (48.8%) | 1.084 (0.826-1.424) |
| AA | 163 (13.7%) | 60 (12.8%) | 0.932 (0.622-1.395) |
| GA+AA | 717 (60.1%) | 289 (61.6%) | 1.049 (0.810-1.359) |
| G allele | 1508 (63.1%) | 589 (62.8%) | 1.000 (reference) |
| A allele | 880 (36.9%) | 349 (37.2%) | 0.996 (0.828-1.199) |
| rs164966 | |||
| AA | 662 (55.4%) | 246 (52.5%) | 1.000 (reference) |
| AG | 456 (38.2%) | 191 (40.7%) | 1.068 (0.819-1.392) |
| GG | 76 (6.4%) | 32 (6.8%) | 1.033 (0.615-1.736) |
| AG+GG | 532 (44.6%) | 223 (47.5%) | 1.063 (0.824-1.370) |
| A allele | 1780 (74.5%) | 683 (72.8%) | 1.000 (reference) |
| G allele | 608 (25.5%) | 255 (27.2%) | 1.040 (0.850-1.274) |
a The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for betel quid chewing, cigarette smoking, and alcohol drinking.
Impacts of LINC00312 genetic polymorphisms on clinicopathologic features of the buccal mucosa cancer
In addition, we tested the relationship between genotypic ratios of LINC00312 SNPs and clinical variables of the disease cohort. We demonstrated that OSCC cases possessing at least one minor allele (G) of rs164966 (GA and GG) were less prone to develop lymph node metastasis (AOR, 0.666; 95% CI, 0.447-0.991; p=0.045) in comparison with homozygotes for the major allele (AA) (Table 3). These data indicate a putative association of LINC00312 gene variations with the metastatic potential of buccal mucosa cancer. Since a genetic correlation of LINC00312 rs164966 with nodal spread of OSCC was detected, we continued to test for a joint effect of rs164966 and three main lifestyle-related risks (cigarette smoking, betel quid chewing, and drinking alcohol) on clinical variables of buccal mucosa cancer. Among patients who habitually used areca nut-related products (betel quid chewer, n=337) or cigarettes (smoker, n=387), a significant association of LINC00312 rs164966 genotypes (GA+GG) with a decreased tendency to develop lymph node metastasis was noted, as compared to those homozygous for the major allele (AA) (AOR, 0.616; 95% CI, 0.386-0.985; p=0.042, for betel quid users; Table 4) (AOR, 0.612; 95% CI, 0.393-0.953; p=0.029, for cigarette consumers; Table 5). Nevertheless, such combined effect was not observed in patients who were not habitually exposed to betel quid chewing or cigarette smoking (Table 4-5). These results suggest that repeated exposure to carcinogens derived from lifestyle-related risks, combined with LINC00312 polymorphisms, may affect OSCC progression.
Discussion
It is widely acknowledged that the processes of oral oncogenesis are influenced by a mixture of predisposing factors, both environmental and hereditary. In this study, a link between LINC00312 rs164966 genotypes and a reduced tendency to develop lymph node metastasis was established, while no correlation was found between the genotypic variants and disease occurrence. Subsequent stratification demonstrated that the association between genotypic variants of rs164966 and decreased nodal spread was only observed in frequent consumers of betel quid or cigarettes but not in cases without these main etiological parameters. Our findings indicate that LINC00312 rs164966, coupled with lifestyle-related risks, interactively influence the progression of oral cancer.
Clinical statuses and LINC00312 rs12497104 and rs164966 genotype frequencies in male patients with cancers of the buccal mucosa.
| rs12497104 (N=469) | rs164966 (N=469) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | GG(N=180) | GA+AA(N=289) | AOR (95% CI) | p value | AA(N=246) | AG+GG(N=223) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 80 (44.4%) | 144 (49.8%) | 1.000 (reference) | 0.256 | 114 (46.3%) | 110 (49.3%) | 1.000 (reference) | 0.518 |
| Stage III+IV | 100 (55.6%) | 145 (50.2%) | 0.806 (0.554-1.170) | 132 (53.7%) | 113 (50.7%) | 0.887 (0.617-1.275) | ||
| Tumor size | ||||||||
| ≦ T2 | 88 (48.9%) | 148 (51.2%) | 1.000 (reference) | 0.625 | 121 (49.2%) | 115 (51.6%) | 1.000 (reference) | 0.606 |
| > T2 | 92 (51.1%) | 141 (48.8%) | 0.911 (0.628-1.322) | 125 (50.8%) | 108 (48.4%) | 0.909 (0.633-1.306) | ||
| Lymph node metastasis | ||||||||
| No | 123 (68.3%) | 203 (70.2%) | 1.000 (reference) | 0.662 | 161 (65.4%) | 165 (74.0%) | 1.000 (reference) | 0.045* |
| Yes | 57 (31.7%) | 86 (29.8%) | 0.914 (0.611-1.368) | 85 (34.6%) | 58 (26.0%) | 0.666 (0.447-0.991) | ||
| Metastasis | ||||||||
| M0 | 179 (99.4%) | 287 (99.3%) | 1.000 (reference) | 0.857 | 244 (99.2%) | 222 (99.6%) | 1.000 (reference) | 0.621 |
| M1 | 1 (0.6%) | 2 (0.7%) | 1.247 (0.112-13.856) | 2 (0.8%) | 1 (0.4%) | 0.550 (0.049-6.102) | ||
| Cell differentiation | ||||||||
| Well | 32 (17.8%) | 53 (18.3%) | 1.000 (reference) | 0.878 | 44 (17.9%) | 41 (18.4%) | 1.000 (reference) | 0.888 |
| Moderate or poor | 148 (82.2%) | 236 (81.7%) | 0.963 (0.593-1.563) | 202 (82.1%) | 182 (81.6%) | 0.967 (0.604-1.547) | ||
Cell differentiation grade: grade I, well differentiated; grade II, moderately differentiated; grade III, poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for cigarette smoking, and alcohol drinking. * p value < 0.05 as statistically significant.
Clinical statuses and genotypic frequencies of LINC00312 rs164966 in buccal mucosa cancer who are betel quid chewers or not betel quid chewers.
| Betel Quid Chewers (N=337) | Non-Betel Quid Chewers (N=132) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | AA(N=173) | AG+GG(N=164) | AOR (95% CI) | p value | AA(N=73) | AG+GG(N=59) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 78 (45.1%) | 81 (49.4%) | 1.000 (reference) | 0.429 | 36 (49.3%) | 29 (49.2%) | 1.000 (reference) | 0.985 |
| Stage III+IV | 95 (54.9%) | 83 (50.6%) | 0.841 (0.548-1.291) | 37 (50.7%) | 30 (50.8%) | 1.007 (0.507-1.999) | ||
| Tumor size | ||||||||
| ≦ T2 | 82 (47.4%) | 81 (49.4%) | 1.000 (reference) | 0.715 | 39 (53.4%) | 34 (57.6%) | 1.000 (reference) | 0.629 |
| > T2 | 91 (52.6%) | 83 (50.6%) | 0.923 (0.602-1.416) | 34 (46.6%) | 25 (42.4%) | 0.843 (0.422-1.684) | ||
| Lymph node metastasis | ||||||||
| No | 111 (64.2%) | 122 (74.4%) | 1.000 (reference) | 0.042* | 50 (68.5%) | 43 (72.9%) | 1.000 (reference) | 0.583 |
| Yes | 62 (35.8%) | 42 (25.6%) | 0.616 (0.386-0.985) | 23 (31.5%) | 16 (27.1%) | 0.809 (0.379-1.725) | ||
| Metastasis | ||||||||
| M0 | 172 (99.4%) | 163 (99.4%) | 1.000 (reference) | 0.970 | 72 (98.6%) | 59 (100.0%) | 1.000 (reference) | 0.367 |
| M1 | 1 (0.6%) | 1 (0.6%) | 1.055 (0.065-17.010) | 1 (1.4%) | 0 (0.0%) | --- | ||
| Cell differentiation | ||||||||
| Well | 37 (21.4%) | 34 (20.7%) | 1.000 (reference) | 0.883 | 7 (9.6%) | 7 (11.9%) | 1.000 (reference) | 0.673 |
| Moderate or poor | 136 (78.6%) | 130 (79.3%) | 1.040 (0.616-1.757) | 66 (90.4%) | 52 (88.1%) | 0.788 (0.260-2.388) | ||
Cell differentiation grade: grade I, well differentiated; grade II, moderately differentiated; grade III, poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for cigarette smoking, and alcohol drinking. * p value < 0.05 as statistically significant.
Clinical statuses and genotypic frequencies of LINC00312 rs164966 in buccal mucosa patients who are cigarette smokers or not cigarette smokers.
| Cigarette Smokers (N=387) | Non-Cigarette Smokers (N=82) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | AA(N=209) | AG+GG(N=178) | AOR (95% CI) | p value | AA(N=37) | AG+GG(N=45) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 95 (45.5%) | 92 (51.7%) | 1.000 (reference) | 0.222 | 19 (51.4%) | 18 (40.0%) | 1.000 (reference) | 0.304 |
| Stage III+IV | 114 (54.5%) | 86 (48.3%) | 0.779 (0.522-1.163) | 18 (48.6%) | 27 (60.0%) | 1.583 (0.658-3.811) | ||
| Tumor size | ||||||||
| ≦ T2 | 103 (49.3%) | 97 (54.5%) | 1.000 (reference) | 0.306 | 18 (48.6%) | 18 (40.0%) | 1.000 (reference) | 0.432 |
| > T2 | 106 (50.7%) | 81 (45.5%) | 0.811 (0.544-1.211) | 19 (51.4%) | 27 (60.0%) | 1.421 (0.590-3.420) | ||
| Lymph node metastasis | ||||||||
| No | 136 (65.1%) | 134 (75.3%) | 1.000 (reference) | 0.029* | 25 (67.6%) | 31 (68.9%) | 1.000 (reference) | 0.898 |
| Yes | 73 (34.9%) | 44 (24.7%) | 0.612 (0.393-0.953) | 12 (32.4%) | 14 (31.1%) | 0.941 (0.370-2.394) | ||
| Metastasis | ||||||||
| M0 | 207 (99.0%) | 177 (99.4%) | 1.000 (reference) | 0.659 | 37 (100.0%) | 45 (100.0%) | 1.00 (reference) | ---- |
| M1 | 2 (1.0%) | 1 (0.6%) | 0.585 (0.053-6.503) | 0 (0.0%) | 0 (0.0%) | --- | ||
| Cell differentiation | ||||||||
| Well | 38 (17.4%) | 37 (20.8%) | 1.000 (reference) | 0.518 | 6 (16.2%) | 4 (8.9%) | 1.000 (reference) | 0.313 |
| Moderate or poor | 171 (81.8%) | 141 (79.2%) | 0.847 (0.511-1.403) | 31 (83.8%) | 41 (91.1%) | 1.984 (0.515-7.641) | ||
Cell differentiation grade: grade I, well differentiated; grade II, moderately differentiated; grade III, poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for betel quid chewing, and alcohol drinking. * p value < 0.05 as statistically significant.
Dysregulation of LINC00312 has been observed in various types of malignancies. In nasopharyngeal [17], blabber [28], and hepatic tumor [29], a decrease in LINC00312 expression was previously documented. However, rather than downregulation, upregulation of LINC00312 in oral submucous fibrosis, a precancerous disorder in the oral cavity, was reported [24]. Such fluctuation of LINC00312 expression levels seems to be, in part, regulated by major habitual risks of OSCC. One piece of evidence has shown a significant induction of LINC00312 in oral cancer from patients with the history of tobacco chewing [30]. Another study demonstrated a reduction of LINC00312 expression levels in OSCC cell lines treated with arecoline, a key component of areca nut extract [25]. Chewing of areca nut-related products is a common risk factor for oral cancer among men in Taiwan (80%) [31], along with tobacco and alcohol use, but is rare in western countries [32]. As cigarette smoking is known as a major driver of carcinogenesis in head and neck cancers [33, 34], chewing of areca nut-related products has been strongly implicated in the development of OSCC through prolonged exposure of various areca nut-derived carcinogens [35]. It is reported that nitrosation of arecoline, the most abundant alkaloid in areca nuts, could generate a myriad of nitrosamines, which have high affinity with DNA, proteins or other molecules to trigger carcinogenic responses [36]. Furthermore, reactive oxygen species (ROS) are produced in great amounts in the oral cavity during areca nut chewing to mediate oxidative damages in the DNA of buccal mucosa tissues [37]. These metabolically activated reactive species of BQ-derived carcinogens collectively can induce genotoxic events in humans, such as production of Safrole-like DNA adducts [38] or chromosome breaks [39], ultimately causing oral tumorigenesis. In the present study, the genetic association of LINC00312 with OSCC progression was found only in frequent users of betel quid or cigarettes but not in those who had no access to these key etiological parameters. Our results, together with findings from other reports, suggest that chronic exposure of tobacco- and areca nut-related products amplifies the effect of LINC00312 gene polymorphisms on OSCC metastasis likely through manipulation of LINC00312 expression levels.
In addition to altered expression, functionality of LINC00312 may be also switched due to polymorphic alleles. It has been reported that polymorphic alleles of LINC00312 rs164966 were predicted to cause corresponding changes in the secondary structure of LINC00312 RNA transcripts, subsequently creating or abolishing the binding sites for miR-136-5p and miR-6729-3p, respectively [21]. Accumulative evidence has pointed out a key role of miR-136-5p in cancer prognosis and treatment [40]. As proposed as a tumor suppressor, miR-136-5p functionally interacts with a variety of target genes, such as ROCK1 [41], BCL2 [42], or SMAD3 [43], to interfere with cancer cell invasion and migration. These findings indicate that altered sponging activities of LINC00312 attributed to its polymorphic alleles may be involved in regulating the invasive potential of OSCC.
In Taiwan, there has been a five-fold increase in the incidence of OSCC in men [44]. The age-standardized incidence rate for men and the ratio of men to women are among the highest in Asia [1, 45]. To eliminate one of potential confounders, the gender, only male subjects were included in this study. Another imperative feature of tumors in the oral cavity is the presence of different anatomical sites, which are characterized by unique sets of tissue organization and considered as different biological entities [8]. The presence of high site-specific heterogeneity in molecular expression [46-48] could very likely impact the cancer development and precision medicine. It is noteworthy that oral cancer in Taiwan most commonly occurs in the buccal mucosa [9], accounting for approximately 40% of cases. However, in western countries, carcinoma of buccal mucosa is significantly less common, as OSCC primarily presents as tongue cancer in the West [10]. These regional differences and lncRNA landscape disparities highlight the long-standing notion that Asian OSCC, particularly those caused by betel quid chewing, is a distinct disease from those in the West.
The present investigation revealed a synergistic effect of LINC00312 gene polymorphisms and behavioral risk parameters on lymph node metastasis of OSCC. However, further explorations are necessary to address certain limitations of this work. One such limitation is the absence of quantitative definitions for key behavioral risks like the use of betel nuts, alcohol, and cigarettes. This could lead to an underestimation of these etiologic factors on affecting OSCC progression. Another concern is that the mechanistic roles of LINC00312 rs164966 in orchestrating nodal spread remain unclear, as the expression levels of this gene have been observed to be positively correlated with lymph node metastasis in NPC [17]. Additional investigations are needed to determine whether LINC00312 rs164966 serves as a quantitative trait locus to modulate its own expression or alters the hub activities to perturb its affinity to specific microRNAs. Furthermore, our results might only apply to certain ethnic groups and require replication in additional study cohorts.
In conclusion, our data demonstrate a correlation between LINC00312 rs164966 and the frequency of developing nodal spread in cancer of buccal mucosa. This genetic association was only found in cases who habitually consumed betel quid or cigarettes, and undetected in those who were not exposed to these significant environmental risks. These results indicate that repeated access to carcinogens derived from lifestyle-related risks, interacted with LINC00312 polymorphisms, may affect the metastatic potential of oral cancer.
Acknowledgements
We thank the Human Biobank of Chung Shan Medical University Hospital, Taichung, Taiwan for specimen preparation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Krishna Rao SV, Mejia G, Roberts-Thomson K, Logan R. Epidemiology of oral cancer in Asia in the past decade-an update (2000-2012). Asian Pacific journal of cancer prevention: APJCP. 2013;14:5567-77
2. Scully C, Field JK, Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral oncology. 2000;36:256-63
3. Gupta K, Metgud R. Evidences Suggesting Involvement of Viruses in Oral Squamous Cell Carcinoma. Pathology research international. 2013;2013:642496
4. Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral oncology. 2009;45:301-8
5. Tezal M, Sullivan MA, Hyland A, Marshall JR, Stoler D, Reid ME. et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:2406-12
6. Hooper SJ, Wilson MJ, Crean SJ. Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head & neck. 2009;31:1228-39
7. Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2010;39:299-305
8. Sarode SC, Sarode G, Patil S. Site-specific Oral Cancers are different Biological Entities. J Contemp Dent Pract. 2017;18:421-2
9. Lin NC, Hsien SI, Hsu JT, Chen MYC. Impact on patients with oral squamous cell carcinoma in different anatomical subsites: a single-center study in Taiwan. Sci Rep. 2021;11:15446
10. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
11. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
12. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cellular and molecular life sciences: CMLS. 2016;73:2491-509
13. Bozgeyik E, Bozgeyik I. Non-coding RNA variations in oral cancers: A comprehensive review. Gene. 2023;851:147012
14. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM. et al. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. Journal of dental research. 2018;97:717-24
15. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. Journal of pineal research. 2021;71:e12760
16. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL. et al. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. Journal of cellular and molecular medicine. 2023;27:3395-403
17. Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X. et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545-54
18. Tan C, Peng C, Huang YC, Zhang QH, Tang K, Li XL. et al. Effects of NPC-associated gene NAG7 on cell cycle and apoptosis in nasopharyngeal carcinoma cells. Ai Zheng. 2002;21:449-55
19. Huang C, Wu M, Tang Y, Li X, Ouyang J, Xiao L. et al. NAG7 promotes human nasopharyngeal carcinoma invasion through inhibition of estrogen receptor alpha and up-regulation of JNK2/AP-1/MMP1 pathways. J Cell Physiol. 2009;221:394-401
20. Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH, Huang WH. et al. LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death Dis. 2021;12:69
21. Guo Z, Bao MH, Fan YX, Zhang Y, Liu HY, Zhou XL. et al. Genetic Polymorphisms of Long Non-coding RNA Linc00312 Are Associated With Susceptibility and Predict Poor Survival of Nasopharyngeal Carcinoma. Front Cell Dev Biol. 2021;9:698558
22. Abdi E, Latifi-Navid S. Emerging long noncoding RNA polymorphisms as novel predictors of survival in cancer. Pathology, research and practice. 2022;239:154165
23. Guo Z, Wang YJ, He BS, Zhou J. Linc00312 Single Nucleotide Polymorphism as Biomarker for Chemoradiotherapy Induced Hematotoxicity in Nasopharyngeal Carcinoma Patients. Dis Markers. 2022;2022:6707821
24. Yu CH, Fang CY, Yu CC, Hsieh PL, Liao YW, Tsai LL. et al. LINC00312/YBX1 Axis Regulates Myofibroblast Activities in Oral Submucous Fibrosis. International journal of molecular sciences. 2020 21
25. Huang HH, You GR, Tang SJ, Chang JT, Cheng AJ. Molecular Signature of Long Non-Coding RNA Associated with Areca Nut-Induced Head and Neck Cancer. Cells. 2023 12
26. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471-4
27. Jadhav KB, Gupta N. Clinicopathological Prognostic Implicators of Oral Squamous Cell Carcinoma: Need to Understand and Revise. North American journal of medical sciences. 2013;5:671-9
28. Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu S. et al. LINC00312 inhibits the migration and invasion of bladder cancer cells by targeting miR-197-3p. Tumour Biol. 2016;37:14553-63
29. Wu J, Zhou X, Fan Y, Cheng X, Lu B, Chen Z. Long non-coding RNA 00312 downregulates cyclin B1 and inhibits hepatocellular carcinoma cell proliferation in vitro and in vivo. Biochem Biophys Res Commun. 2018;497:173-80
30. Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Arun K, Vinothkumar V, Revathidevi S. et al. Expression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancer. Tumour Biol. 2017;39:1010428317698366
31. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP. et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088-99
32. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576-82
33. South AP, den Breems NY, Richa T, Nwagu U, Zhan T, Poojan S. et al. Mutation signature analysis identifies increased mutation caused by tobacco smoke associated DNA adducts in larynx squamous cell carcinoma compared with oral cavity and oropharynx. Sci Rep. 2019;9:19256
34. Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S. et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618-22
35. Nair U, Bartsch H, Nair J. Alert for an epidemic of oral cancer due to use of the betel quid substitutes gutkha and pan masala: a review of agents and causative mechanisms. Mutagenesis. 2004;19:251-62
36. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Betel-quid and areca-nut chewing and some areca-nut derived nitrosamines. IARC Monogr Eval Carcinog Risks Hum. 2004;85:1-334
37. Nair UJ, Nair J, Friesen MD, Bartsch H, Ohshima H. Ortho- and meta-tyrosine formation from phenylalanine in human saliva as a marker of hydroxyl radical generation during betel quid chewing. Carcinogenesis. 1995;16:1195-8
38. Chen CL, Chi CW, Chang KW, Liu TY. Safrole-like DNA adducts in oral tissue from oral cancer patients with a betel quid chewing history. Carcinogenesis. 1999;20:2331-4
39. Nair U, Obe G, Nair J, Maru GB, Bhide SV, Pieper R. et al. Evaluation of frequency of micronucleated oral mucosa cells as a marker for genotoxic damage in chewers of betel quid with or without tobacco. Mutat Res. 1991;261:163-8
40. Hsu CY, Allela OQB, Mahdi SA, Doshi OP, Adil M, Ali MS. et al. miR-136-5p: A key player in human cancers with diagnostic, prognostic and therapeutic implications. Pathol Res Pract. 2023;250:154794
41. Zhang W, Shi J, Cheng C, Wang H. CircTIMELESS regulates the proliferation and invasion of lung squamous cell carcinoma cells via the miR-136-5p/ROCK1 axis. J Cell Physiol. 2020;235:5962-71
42. Geng Y, Bao Y, Zhang W, Deng L, Su D, Zheng H. Circular RNA hsa_circ_0014130 Inhibits Apoptosis in Non-Small Cell Lung Cancer by Sponging miR-136-5p and Upregulating BCL2. Molecular cancer research: MCR. 2020;18:748-56
43. Kang W, Wang Q, Dai Y, Wang H, Wang M, Wang J. et al. Hypomethylation of PlncRNA-1 promoter enhances bladder cancer progression through the miR-136-5p/Smad3 axis. Cell Death Dis. 2020;11:1038
44. Su CC, Yang HF, Huang SJ, Lian Ie B. Distinctive features of oral cancer in Changhua County: high incidence, buccal mucosa preponderance, and a close relation to betel quid chewing habit. Journal of the Formosan Medical Association = Taiwan yi zhi. 2007;106:225-33
45. Dhanuthai K, Rojanawatsirivej S, Thosaporn W, Kintarak S, Subarnbhesaj A, Darling M. et al. Oral cancer: A multicenter study. Med Oral Patol Oral Cir Bucal. 2018;23:e23-e9
46. Sathyan KM, Sailasree R, Jayasurya R, Lakshminarayanan K, Abraham T, Nalinakumari KR. et al. Carcinoma of tongue and the buccal mucosa represent different biological subentities of the oral carcinoma. J Cancer Res Clin Oncol. 2006;132:601-9
47. Trivedi TI, Tankshali RA, Goswami JV, Shukla SN, Shah PM, Shah NG. Identification of site-specific prognostic biomarkers in patients with oral squamous cell carcinoma. Neoplasma. 2011;58:217-26
48. Kannan S, Tahara H, Yokozaki H, Mathew B, Nalinakumari KR, Nair MK. et al. Telomerase activity in premalignant and malignant lesions of human oral mucosa. Cancer Epidemiol Biomarkers Prev. 1997;6:413-20
Author contact
![]() Corresponding authors: Shun-Fa Yang, Ph.D. and Chiao-Wen Lin, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); cwlinedu.tw (Chiao-Wen Lin).
Corresponding authors: Shun-Fa Yang, Ph.D. and Chiao-Wen Lin, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); cwlinedu.tw (Chiao-Wen Lin).

 Global reach, higher impact
Global reach, higher impact