Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(10):2900-2912. doi:10.7150/jca.92704 This issue Cite
Research Paper
Cell-free Nucleic Acid as Promising Diagnostic Biomarkers for Gastric Cancer: a Systematic Review
Department of Digestive Surgical Oncology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
#Qun Zhang and Zhouyuan Du contributed equally to this paper.
Received 2023-11-29; Accepted 2024-3-9; Published 2024-3-25
Abstract
Background: Gastric cancer (GC) is a common malignancy with early detection being crucial for survival. Liquid biopsy analysis using cell-free nucleic acid is a preferred method for detection. Hence, we conducted a systematic review to assess the diagnostic efficacy of cell-free nucleic acid markers for GC.
Methods: We searched PubMed and ISI Web of Science databases for articles that conformed to our inclusion and exclusion criteria from 2012 to 2022. The following information was abstracted: first author, year of publication, country/region, age, male proportion, tumor stage for cases, specimen type, measurement method, targeted markers and diagnostic related indicators (including sensitivity, specificity, AUC, P-value).
Results: Fifty-eight studies examined cell-free RNAs (cfRNAs) with a total of 62 individual circulating markers and 7 panels in serum or plasma, while 21 studies evaluated cell-free DNAs (cfDNAs) with 29 individual circulating markers and 7 panels. For individual cfRNAs, the median (range) sensitivity and specificity were 80% (21% - 98%) and 80% (54% - 99%), respectively. The median (range) sensitivity and specificity for cfRNA panels were 86% (83% - 90%) and 75% (60% - 98%), respectively. In comparison, the median (range) sensitivity and specificity reported for individual cfDNAs were 50% (18% - 96%) and 93% (57% - 100%), respectively, while cfDNA panels had a median (range) sensitivity and specificity of 85% (41% - 92%) and 73.5% (38% - 90%), respectively. The meta results indicate that cfRNA markers exhibit high sensitivity (80%) and low specificity (80%) for detecting GC, while cfDNA markers have lower sensitivity (59%) but higher specificity (92%).
Conclusions: This review has demonstrated that cell-free nucleic acids have the potential to serve as useful diagnostic markers for GC. Given that both cfRNA and cfDNA markers have shown promising diagnostic performance for GC, the combination of the two may potentially enhance diagnostic efficiency.
Keywords: gastric cancer, early detection, cfRNA, cfDNA, biomarker
1. Introduction
Gastric cancer (GC) is one of the most widespread malignant tumors globally, responsible for over a million new cases and an estimated 769,000 deaths in 2020 [1]. It ranks fifth for incidence and fourth for mortality worldwide, with Asia and Eastern Europe experiencing the highest burden of the disease, particularly in Japan and China [2]. Prognosis worsens as the tumor advances, with metastatic gastric cancer patients having a five-year survival rate of only 3.1% compared to a five-year survival rate of over 90% in patients with early gastric cancer [3]. Thus, early diagnosis and prompt treatment are crucial in improving the survival and prognosis of GC.
The notion of liquid biopsy was first introduced for circulating tumor cells (CTC) a decade ago and was subsequently extended to include circulating tumor DNA (ctDNA) and circulating cell-free RNA (cfRNA) [4]. It is widely acknowledged that the bulk of such ctDNA stems from apoptotic and necrotic tumor cells that discharge their fragmented DNA into circulation. Essentially, ctDNA bears the genetic flaws that are identical to those of the tumor cells they are derived from [5]. In addition to mutation analysis, trustworthy tests have been developed over the last few years for the assessment of epigenetic alterations, including DNA methylation [4]. Compared to ctDNA, cfRNA is unstable and has a short half-life, but its broad classification and distribution in peripheral blood and other bodily fluids make it a superior biomarker for liquid biopsy [6]. MicroRNAs (miRNAs) are typically the primary targets of cfRNA. Additionally, long noncoding RNA (lncRNA), circular RNA (circRNA), and transfer RNA (tRNA)-derived fragments (tRF) are also novel biomarkers for cancer diagnosis [7-9].
Currently, cell-free nucleic acid is widely acknowledged as a superior biomarker class for the liquid biopsy analysis of GC. The aim of this systematic review is to provide a comprehensive summary of published articles on the utility of cell-free nucleic acid as a biomarker for GC detection, with a specific emphasis on its potential for early disease detection, which is an indispensable factor in enhancing patient prognosis and management.
2. Methods
We conducted this systematic review following a predefined protocol and adhering to the reporting standards set out in the PRISMA statement [10]. Seventy-nine studies were included in the meta-analysis. Summary receiver operator characteristics (SROC) for cf-RNAs and cf-DNAs were drawn. Calculations were made by using bivariate mixed-effects model developed by von Houwelingen for treatment trial meta-analysis and then modified for synthesis of diagnostic test data, which is in order midas in STATA 17.0. As we solely used data from previously published literature, no ethical approval or patient informed consent was necessary.
2.1 Search strategy
We searched PubMed and ISI Web of Science databases for articles that conformed to our inclusion and exclusion criteria from 2012 to 2022. The combination keywords we used to separately search for DNA and RNA markers were as follows: [(gastric OR stomach) AND (cancer OR carcinoma OR neoplasm OR tumor OR adenocarcinoma OR squamous carcinoma OR malignancy) AND (cell-free RNA OR cfRNA OR mRNA fragment OR microRNA* OR miRNA* OR long noncoding RNA OR lncRNA OR circular RNA OR circRNA OR transfer RNA OR tRNA noncoding RNA OR ncRNA OR RNA) AND (detection OR diagnosis OR biomarker OR marker OR sensitivity OR specificity OR area under the curve OR AUC) AND (blood OR serum OR plasma)] for cf-RNA markers, and [(gastric OR stomach) AND (cancer OR carcinoma OR neoplasm OR tumor OR adenocarcinoma OR squamous carcinoma OR malignancy) AND (Circulating Tumor DNA OR ctDNA OR cell-free DNA OR cfDNA OR DNA) AND (detection OR diagnosis OR biomarker OR marker OR sensitivity OR specificity OR area under the curve OR AUC) AND (blood OR serum OR plasma)] for cf-DNA markers.
2.2 Eligibility criteria
In this systematic review, eligible studies were selected based on the following inclusion criteria: (1) reported at least one of the diagnostic values of cfDNA or cfRNA detection in GC patients or able to calculate it from published data; (2) samples collected from peripheral blood; (3) clearly stated the techniques and target gene used in the study; (4) studies must include negative controls. In addition, the following exclusion criteria were also applied: (1) studies published in languages other than English; (2) repeated or overlapping publications that included the same population and gene; (3) studies with a poor sample size (≤20); (4) studies that were letters, editorials, case reports or case series; (5) non-human studies; (6) experiments only based on cell lines rather than clinical samples; (7) studies using treated cases before sampling or disease controls were also excluded.
2.3 Data extraction and statistical analysis
Two investigators (QZ and ZD) independently performed data extraction of all included studies. The following information was abstracted: first author, year of publication, country/region, age, male proportion, tumor stage for cases, specimen type, measurement method, targeted markers and diagnostic indicators (including sensitivity, specificity, AUC, P-value). Consensus was obtained by discussion in case of any disagreement. The analytical software STATA 17.0 was used to analyze the diagnostic value. Sensitivity, specificity, area under curve (AUC) were calculated.
2.4 Quality assessment
The two authors independently assessed the risk of bias and applicability concerns of the included studies using Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) [11]. Any disagreement was settled by further discussion among the authors.
3. Results
3.1 Literature search result
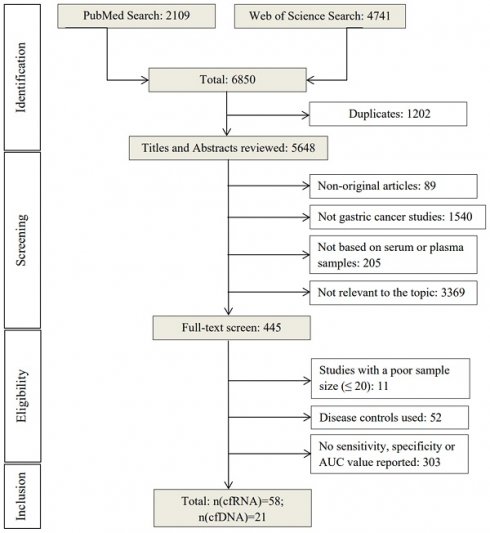
Initial electronic search retrieved 6850 articles from PubMed (2109) and Web of Science (4741) using the search terms mentioned above (Figure 1). After removing duplicates (n=1202), the remaining 5648 articles were screened by title and abstract based on the exclusion criteria. 5203 articles were excluded and 445 were selected for full-text reading. Among these, 366 were excluded for using disease controls or not reporting sensitivity, specificity, or AUC values, leaving a total of 79 studies [12-59] for evaluating the diagnostic performance of cell-free nucleic acid for GC [60-90].
3.2 Study quality and characteristics
QUADAS-2 was carried out for the 79 included studies for quality assessment (Figures S1, S2, S3 and S4). Study quality assessment was completed by two reviewers (QZ and HY) independently. Any initial inconsistencies were resolved by further discussion between the investigators. No risk of bias or applicability concern was found in the reference standard domain, and the flow and timing domain.
The two reviewers (QZ and ZD) independently carried out quality assessment of the 79 included studies (see Supplementary Figures S1, S2, S3 and S4). Any inconsistencies were resolved through further discussion. No bias or applicability concerns were found in the reference standard domain, as well as in the flow and timing domain.
All 79 included studies were case-control studies that collected blood samples after disease diagnosis. Of these, 58 studies evaluated cfRNAs and 21 studies evaluated cfDNAs. Within the cfRNA studies, 55 articles evaluated individual cfRNAs, including 34 miRNAs, 10 lncRNAs, eight circRNAs, and four tRFs (Table 1). Seven studies assessed RNA panels, five of which were miRNA panels (Table 2). Among the cfDNA studies, we reviewed 20 articles on individual cfDNAs, including three on DNA hypermethylation (Table 3). Seven studies evaluated cfDNA panels, three of which were hypermethylated panels (Table 4). Information on each study, such as the number of cases and controls, mean or median age, male proportion, specimen type, tumor stage, and diagnostic indicators, was summarized in Tables 1, 2, 3, and 4. Tables 1 and 3 also presented the p-value for testing the difference of each individual RNA between cases and controls or the statistical significance of AUC values.
Overview of the literature search process (up to 17th of November 2022).
Diagnostic performance of individual cell-free RNA markers in gastric cancer
| Case VS Control | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Author | Year | Region | Number | Age (years) | Male (%) | cfRNA marker | Specimen | Stage | SEN (%) | SPE (%) | AUC | P-value |
| 12 | Aalami AH | 2020 | Iran | 39/39 | 65/69 | 80/77 | miR-223-5p | Serum | I-III | 90 | 85 | 0.9 | <0.0001 |
| 13 | Bai SY | 2019 | china | 50/53 | / | 74/NA | miR-551b-3p | Serum | I-IV | 70 | 96 | 0.86 | <0.001 |
| 14 | Cai CC | 2019 | china | 63/29 | 63/NA | 71/NA | Exo-Lnc RNA PCSK2-2:1 | Serum | I-IV | 84 | 87 | 0.9 | / |
| 15 | Chen JL | 2020 | china | 90/45 | 65/59 | 76/47 | miR‑650 | Plasma | I-IV | 62 | 93 | 0.7 | 0.0001 |
| 16 | Chen JL | 2019 | china | 90/45 | / | / | miR-421 | Plasma | I-IV | 97 | 98 | 0.98 | <0.0001 |
| 17 | Chen SJ | 2017 | china | 104/104 | / | 72/72 | circ_0000190 | Plasma | I-IV | 72 | 68 | 0.75 | / |
| 18 | Chen X | 2020 | china | 80/80 | / | / | miR-125b-5p | Serum | I-IV | 78 | 60 | 0.68 | <0.001 |
| miR-196a5p | 70 | 70 | 0.73 | <0.001 | |||||||||
| miR-1-3p | 80 | 60 | 0.72 | <0.001 | |||||||||
| miR-149-5p | 58 | 68 | 0.66 | <0.001 | |||||||||
| 19 | Dong ZG | 2019 | china | 119/31 | / | 75/61 | Exosomal MT1-MMP mRNA | Serum | I-IV | 64 | 87 | 0.79 | / |
| 20 | Elsayed ET | 2018 | Egypt | 50/50 | / | / | lncRNA HOTAIR | Plasma | I-IV | 88 | 84 | 0.94 | / |
| 21 | Emami SS | 2019 | Iran | 30/30 | 63/67 | miR-21 | Plasma | I-IV | 87 | 72 | 0.89 | <0.0001 | |
| miR-222 | 63 | 56 | 0.75 | 0.044 | |||||||||
| 22 | Fu ZC | 2014 | china | 114/56 | / | 47/NA | miR-222 | Plasma | I-IV | 66 | 88 | 0.85 | / |
| 23 | Gong Y | 2018 | china | 42/60 | / | 64/62 | miR-199a | Serum | I-IV | 93 | 71 | 0.92 | / |
| 24 | Gu XL | 2021 | china | 130/110 | / | 72NA | tsr016141 | Serum | I-IV | 75 | 78 | 0.81 | / |
| 25 | Guo YT | 2020 | china | 90/90 | / | 67/NA | miR-296-5p | Serum | I-IV | 84 | 92 | 0.92 | <0.001 |
| miR-28-3p | 91 | 80 | 0.91 | <0.001 | |||||||||
| 26 | Han WW | 2021 | china | 146/95 | 53/46 | 66/58 | miR-135 | Serum | I-IV | 78 | 80 | 0.87 | / |
| miR-20a | 79 | 72 | 0.79 | / | |||||||||
| 27 | Hou X | 2015 | china | 80/80 | 68/67 | 58/55 | miR-106a | Plasma | I-IV | 94 | 76 | 0.89 | / |
| 29 | Huang YJ | 2021 | china | 111/89 | / | 63/NA | tRF-31-U5YKFN8DYDZDD | Serum | 60 | 81 | 0.74 | / | |
| 30 | Ji B | 2019 | china | 168/74 | / | 60/NA | lncRNA LINC00086 | Plasma | I-IV | 73 | 84 | 0.86 | / |
| miR-214 | 73 | 92 | 0.88 | / | |||||||||
| 31 | Kong S | 2019 | china | 30/30 | / | / | circ_0001821 | Plasma | I-IV | 87 | 87 | 0.87 | / |
| 32 | Kong Y | 2019 | china | 184/78 | 62/63 | 61/56 | miR-25 | Plasma | I-IV | 89 | 85 | 0.77 | <0.0001 |
| 33 | Li BH | 2017 | china | 116/85 | 61/58 | 66/55 | miR-320 | Plasma | I-IV | 82 | 76 | 0.86 | / |
| 34 | Li C | 2013 | china | 180/80 | 58/59 | 69/65 | miR-199a-3p | Plasma | I-IV | 80 | 74 | 0.84 | / |
| 35 | Li C | 2013 | china | 80/70 | 57/59 | 69/64 | miR-199a-3p | Plasma | I-IIA | 76 | 74 | 0.82 | / |
| 36 | Li FX | 2017 | china | 65/65 | 54/56 | 77/77 | miR-106 | Plasma | I-IV | 86 | 92 | 0.9 | <0.001 |
| miR-93 | 82 | 74 | 0.76 | <0.001 | |||||||||
| miR-25 | 88 | 77 | 0.82 | <0.001 | |||||||||
| 37 | Li Y | 2019 | china | 40/40 | 58/60 | 65/55 | miR-381 | Serum | I-II | 83 | 93 | 0.92 | <0.0001 |
| 38 | Liu H | 2017 | china | 137/145 | 54/54 | 62/64 | miR-217 | Plasma | I-IV | 81 | 83 | 0.89 | / |
| 39 | Liu HF | 2017 | china | 145/145 | / | 65/NA | miR-205 | Serum | I-III | 98 | 83 | 0.91 | / |
| 40 | Liu HS | 2012 | china | 40/41 | / | / | miR-378 | Serum | I-IV | 88 | 71 | 0.86 | / |
| 41 | Liu WW | 2020 | china | 89/73 | / | 71/NA | lncRNA FEZF1-AS1 | Serum | I-IV | 75 | 66 | 0.81 | <0.001 |
| lncRNA AFAP1-AS1 | 76 | 56 | 0.82 | <0.001 | |||||||||
| 42 | Liu Y | 2019 | china | 94/40 | 59/59 | 61/65 | lncRNA HOXA11-AS | Serum | I-IV | 79 | 98 | 0.92 | 0.001 |
| 43 | Park JL | 2015 | Korea | 35/35 | 52/49 | 51/51 | miR-27a | Plasma | I-IV | 77 | 56 | 0.7 | / |
| 44 | Qin SY | 2021 | china | 98/82 | / | 58/NA | LncRNA HCP5 | Serum | I-IV | 80 | 70 | 0.82 | / |
| 46 | Saliminejad K | 2022 | Iran | 97/100 | 59/56 | 64/NA | miR-18a | Plasma | I-IV | 55 | 57 | 0.67 | 0.027 |
| miR-21 | 56 | 54 | 0.65 | 0.042 | |||||||||
| miR-125b | 64 | 65 | 0.69 | 0.004 | |||||||||
| 47 | Shan LC | 2019 | china | 117/100 | 58/50 | 75/58 | lncRNA UCA1 | Serum | I-IV | 93 | 79 | 0.76 | / |
| 48 | Shao YF | 2022 | china | 42/40 | / | / | circ_0086720 | Plasma | I-II | 67 | 87 | 0.77 | <0.001 |
| 49 | Shen Y | 2020 | china | 98/40 | 58/59 | 49/63 | miR-30c | Serum | I-IV | 90 | 84 | 0.92 | / |
| 50 | Shen YJ | 2021 | china | 89/98 | / | 67/NA | tRF-19-3L7L73JD | Plasma | I-IV | 40 | 79 | 0.62 | / |
| 52 | Sun XY | 2022 | china | 71/60 | / | 69/38 | circ_0002874 | Plasma | I-IV | 87 | 77 | 0.84 | / |
| 53 | Tian WY | 2022 | china | 112/40 | 58/NA | 63/63 | miR-181 | Serum | I-II | 84 | 78 | 0.82 | / |
| miR-652 | 86 | 80 | 0.84 | / | |||||||||
| 54 | Wu DY | 2017 | china | 32/32 | / | NA/67 | miR-503 | Serum | I-IV | 97 | 79 | 0.89 | 0.006 |
| 55 | Wu JH | 2015 | china | 90/90 | / | 49/NA | miR-421 | Serum | I-IV | 90 | 86 | 0.78 | / |
| 56 | Wu JH | 2015 | china | 50/50 | / | 48/NA | miR-21 | Serum | I-IV | 88 | 80 | 0.91 | / |
| 57 | Xiao K | 2021 | china | 113/27 | / | 74/30 | EV lncRNA CCAT1 | Serum | I-IV | 80 | 93 | 0.89 | / |
| 58 | Yan JN | 2022 | china | 62/46 | / | 70/NA | circ_0001020 | plasma | I-IV | 47 | 98 | 0.74 | <0.001 |
| 59 | Yin G | 2020 | china | 80/60 | 58/53 | 56/42 | circ_0141633 | Serum | I-IV | 85 | 94 | 0.84 | / |
| 60 | Yuan RS | 2016 | china | 48/22 | / | 79/NA | miR-106a | Plasma | I-IV | 77 | 64 | 0.83 | / |
| 61 | Zeng QH | 2014 | china | 40/36 | / | 70/NA | miR-17 | Serum | I-IV | 81 | 88 | 0.88 | / |
| miR-106b | 75 | 93 | 0.86 | / | |||||||||
| 62 | Zeng WW | 2020 | china | 86/50 | 60/45 | 42/52 | miR-101-3p | Serum | I-IV | 72 | 86 | 0.87 | <0.0001 |
| 63 | Zhang WW | 2021 | china | 100/80 | / | 61/NA | circ_0007507 | Serum | I-IV | 73 | 85 | 0.83 | <0.001 |
| 64 | Zhang Y | 2022 | china | 124/119 | / | 67/NA | tRF-23-Q99P9P9NDD | Serum | I-IV | 67 | 86 | 0.78 | / |
| 65 | Zhao QF | 2018 | china | 102/105 | / | 71/NA | circular RNA 0000181 | Plasma | I-IV | 21 | 99 | 0.58 | / |
| 66 | Zheng GD | 2021 | china | 168/50 | 61/40 | 70/NA | Exosomal miR-590-5p | Serum | I-IV | 64 | 86 | 0.81 | / |
| 67 | Zhou XY | 2015 | china | 70/70 | / | 63/NA | lncRNA H19 | Plasma | I-IV | 83 | 73 | 0.84 | / |
| 68 | Zhou XY | 2015 | china | 50/50 | / | / | miR-223 | Plasma | I-IV | 70 | 80 | 0.81 | / |
| 69 | Zong W | 2019 | china | 110/84 | / | 662/NA | lncRNA CTC-497E21.4 | Serum | I-IV | 82 | 75 | 0.85 | / |
SEN: sensitivity; SEP: specificity; AUC: area under the curve; NA: not available.
Diagnostic performance of cell-free RNA panels in gastric cancer
| Case VS Control | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Author | Year | Region | Number | Age (years) | Male (%) | Panel | Specimen | Stage | SEN (%) | SPE (%) | AUC | P-value |
| 18 | Chen X | 2020 | china | 80/80 | / | / | Panel A | Serum | I-IV | 86 | 79 | 0.89 | <0.001 |
| 28 | Huang SK | 2016 | china | 62/59 | / | / | Panel B | Serum | I-IV | 86 | 98 | 0.92 | / |
| 41 | Liu WW | 2020 | china | 89/73 | / | 71/NA | Panel C | Serum | I-IV | 85 | 60 | 0.87 | <0.0001 |
| 45 | Roy S | 2022 | Japan | 102/48 | / | / | Panel D | Serum | I-IV | 89 | 62 | 0.83 | / |
| 69/48 | / | / | I-II | 90 | 60 | 0.82 | / | ||||||
| 46 | Saliminejad K | 2022 | Iran | 97/100 | 59/56 | 64/NA | Panel E | Plasma | I-IV | 86 | 85 | 0.92 | <0.001 |
| 31/100 | NA/56 | / | I-II | 83 | 75 | 0.83 | 0.001 | ||||||
| 59/100 | NA/56 | / | III-IV | 86 | 75 | 0.93 | 0.001 | ||||||
| 51 | So JBY | 2021 | Singaporean | 125/4441 | 57/57 | 61/53 | Panel F | Serum | I-IV | 87 | 68 | 0.85 | / |
| 61 | Zeng QH | 2014 | china | 40/36 | / | 70/NA | Panel G | Serum | I-IV | 83 | 87 | 0.91 | / |
SEN: sensitivity; SEP: specificity; AUC: area under the curve; NA: not available.
Panel A: miR-125b-5p, miR-196a5p, miR-1-3p, miR-149-5p; Panel B: miR-21, miR-31, miR-92a, miR-181b, miR-203; Panel C: lncRNA FEZF1-AS1, lncRNA AFAP1-AS1; Panel D: circ_0045602,circ_0008768, circ_0007380, circ_0002019, circ_0006089, circ_0034398, circ_0052001, circ_0001013; Panel E: miR-18a, miR-21, miR-125b; Panel F: miR-140, miR-183, miR- 30e, miR- 103a, miR-126, miR-93, miR-142, miR-21, miR29c, miR-424,miR-340, miR- 181a; Panel G: miR-17, miR-106b.
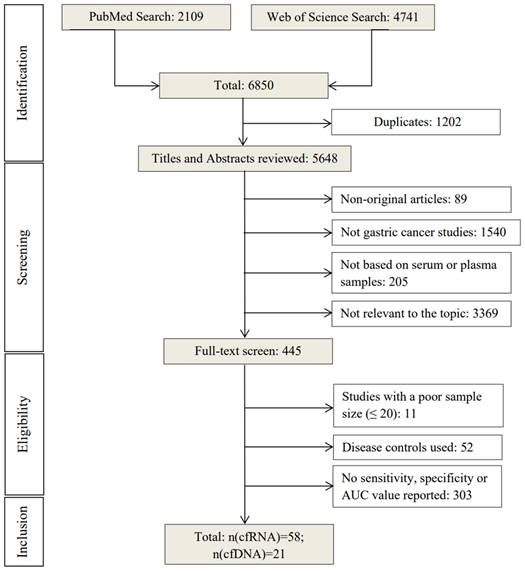
Graphical representation of sensitivity vs specificity of analyzed cfRNA markers. Sensitivity is plotted on the y-axis while on the x-axis the false-positive rate is presented (100-Specificity).
Twenty-five studies analyzed plasma samples and 33 studies analyzed serum samples for cfRNA. Overall, 58 studies evaluated 62 individual circulating cfRNA markers and seven cfRNA panels in serum or plasma. All enrolled cfRNA studies used quantitative real time polymerase chain reaction (qRT-PCR) to detect cfRNAs concentrations. The normalization methods for the expression of RNAs were not uniform. For example, miR-39, U6 snRNA, miR-16, 18S rRNA, GAPDH, β-actin, and snord47 were being used as reference standards for data normalization (Table S1). Thirteen studies analyzed plasma samples and eight studies analyzed serum samples for cfDNA. The cfDNAs were isolated by different extraction kits among the included studies (Table S2). Twenty-one studies reported 29 individual circulating cfDNA markers and seven cfDNA panels in serum or plasma. Most of 21 studies quantified methylation levels using Methylation Specific PCR (MSP). Only one study conducted by Hideura E et al [76]. additionally used digital PCR to quantify methylation level.
3.3 Diagnostic efficiency of cfRNAs and cfDNAs
The 58 included studies reported a total number of 84 cfRNAs with the diagnostic potential for GC, of which, seven miRNAs were reported in more than two studies (Table S3). The panels ranged from two to 12 miRNAs, with the smallest and largest panel sizes being two and 12 miRNAs, respectively. Figure 2 presents an overview of the diagnostic performance of all reported cfRNAs and cfRNA panels. For individual cfRNAs, the median (range) reported sensitivity and specificity were 80% (21%-98%) and 80% (54%-99%), respectively.
Diagnostic performance of individual cell-free DNA markers in gastric cancer
| Case VS Control | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Author | Year | Region | Number | Age (years) | Male (%) | Target gene | Alteration type | Method | Specimen | Stage | SEN (%) | SPE (%) | AUC | P-value |
| 70 | Anderson BW | 2018 | USA | 36/38 | / | 61/58 | ELMO1 | methylation | QuARTS | Plasma | I-IV | 96 | 78 | 0.94 | / |
| ZNF569 | 76 | 66 | 0.72 | / | |||||||||||
| C13orf18 | 73 | 58 | 0.73 | / | |||||||||||
| 71 | Ioanna B | 2013 | Greece | 73/20 | 67/64 | 70/NA | SOX17 | methylation | MSP | Serum | I-IV | 59 | 100 | / | / |
| 72 | Ioanna B | 2015 | Greece | 73/20 | 67/64 | 70/NA | APC | methylation | MSP | Serum | I-IV | 84 | 100 | / | / |
| RASSF1A | 68 | 100 | / | / | |||||||||||
| 73 | Cao CQ | 2020 | China | 74/57 | / | 64/46 | SEPT9 | methylation | MSP | Plasma | I-II | 28 | 94 | 0.62 | / |
| RFP 180 | 32 | 90 | 0.64 | / | |||||||||||
| 74 | Chen L | 2012 | China | 58/30 | 62/55 | / | FAM5C | hypermethylation | MSP | Serum | I-IV | / | / | 0.64 | / |
| MYLK | / | / | 0.82 | / | |||||||||||
| 75 | Han J | 2014 | China | 92/88 | / | 58/61 | MINT2 | methylation | MSP | Serum | I-IV | 39 | 97 | / | / |
| 76 | Hideura E | 2020 | Japan | 50/61 | 72/58 | 82/52 | RUNX3 | methylation | digital PCR | Serum | I-II | 50 | 80 | 0.7 | / |
| 77 | Lee HS | 2013 | Korea | 153/96 | / | 59/NA | SEPT9 | methylation | MSP | Plasma | I-IV | 18 | 91 | / | / |
| 78 | Li H | 2022 | China | 55/50 | 66/55 | 73/52 | KCNQ5 | methylation | MSP | Plasma | I-IV | 35 | 100 | 0.69 | / |
| C9orf50 | 51 | 98 | 0.74 | / | |||||||||||
| CLIP4 | 25 | 92 | 0.6 | / | |||||||||||
| 57/82 | 66/28 | 67/30 | KCNQ5 | 23 | 100 | 0.63 | / | ||||||||
| C9orf50 | 65 | 94 | 0.82 | / | |||||||||||
| CLIP4 | 44 | 90 | 0.68 | / | |||||||||||
| 80 | Lin ZH | 2017 | China | 131/34 | 61/57 | 67/65 | ZIC1 | methylation | MSP | Plasma | I-IV | 70 | 69 | / | / |
| HOXD10 | 48 | 80 | / | / | |||||||||||
| RUNX3 | 43 | 79 | / | / | |||||||||||
| 81 | Ling ZQ | 2013 | China | 202/88 | 59/NA | / | XAF1 | methylation | MSP | Serum | I-IV | 70 | 100 | / | / |
| 82 | Miao J | 2020 | China | 92/50 | 60/NA | 73/NA | SFRP2 | methylation | MSP | Plasma | I-IV | 61 | 86 | 0.78 | / |
| 83 | Pimson C | 2016 | Thailand | 101/202 | / | 42/NA | PCDH10 | methylation | MSP | Plasma | I-IV | 94 | 97 | / | / |
| RASSF1A | 88 | 92 | / | / | |||||||||||
| 84 | Saliminejad K | 2020 | Iran | 96/88 | / | / | P16 | methylation | MSP | Plasma | I-IV | 42 | 84 | 0.63 | <0.001 |
| RASSF1A | 33 | 100 | 0.67 | <0.001 | |||||||||||
| RPRM | 67 | 93 | 0.8 | <0.001 | |||||||||||
| RUNX3 | 58 | 95 | 0.77 | <0.001 | |||||||||||
| 34/88 | / | / | P16 | I-II | 35 | 84 | 0.6 | 0.026 | |||||||
| RASSF1A | 24 | 100 | 0.62 | <0.001 | |||||||||||
| RPRM | 47 | 93 | 0.7 | <0.001 | |||||||||||
| RUNX3 | 59 | 95 | 0.77 | <0.001 | |||||||||||
| 62/88 | / | / | P16 | III-IV | 45 | 84 | 0.65 | <0.001 | |||||||
| RASSF1A | 39 | 100 | 0.69 | <0.001 | |||||||||||
| RPRM | 77 | 93 | 0.85 | <0.001 | |||||||||||
| RUNX3 | 58 | 95 | 0.76 | <0.001 | |||||||||||
| 85 | Xu JB | 2021 | China | 151/224 | / | 75/51 | SEPT9 | methylation | MSP | Plasma | I-IV | 52 | 90 | 0.77 | / |
| RNF180 | 38 | 94 | 0.72 | / | |||||||||||
| 86 | Yang QF | 2013 | China | 40/22 | / | 83/NA | BCL6B | hypermethylation | MSP | Plasma | I-IV | 43 | 100 | / | / |
| 87 | Yu JL | 2014 | China | 92/88 | / | 59/61 | TIMP-3 | methylation | MSP | Serum | I-IV | 59 | 100 | / | / |
| 88 | Zhang H | 2014 | China | 41/21 | / | 73/NA | Spastic paraplegia-20 | hypermethylation | MSP | Plasma | I-IV | 49 | 100 | / | / |
| 89 | Zhang X | 2014 | China | 57/42 | 61/57 | 68/64 | RNF180 | methylation | MSP | Plasma | I-IV | 58 | 76 | / | / |
| DAPK1 | 49 | 71 | / | / | |||||||||||
| SFRP2 | 72 | 57 | / | / | |||||||||||
| 90 | Zhao LY | 2022 | China | 60/122 | / | 73/NA | SEPT9 | methylation | MSP | Plasma | I-IV | 42 | 96 | 0.7 | / |
SEN: sensitivity; SEP: specificity; AUC: area under the curve; NA: not available; MSP: methylation specific PCR; QuARTS: Quantitative allele-specific real-time target and signal amplification.
Diagnostic performance of cell-free DNA panels in gastric cancer
| Case VS Control | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Author | Year | Region | Number | Age (years) | Male (%) | Panel | Alteration type | Method | Specimen | Stage | SEN (%) | SPE (%) | AUC |
| 70 | Anderson BW | 2018 | USA | 36/38 | / | 61/58 | Panel A | methylation | QuARTS | Plasma | I-IV | 90 | 63 | 0.91 |
| 73 | Cao CQ | 2020 | China | 74/57 | / | 64/46 | Panel B | methylation | MSP | Plasma | I-II | 41 | 85 | 0.65 |
| 74 | Chen L | 2012 | China | 58/30 | 62/55 | / | Panel C | hypermethylation | MSP | Serum | I-IV | 90 | 58 | 0.84 |
| 78 | Li H | 2022 | China | 55/50 | 66/55 | 73/52 | Panel D | methylation | MSP | Plasma | I-IV | 66 | 90 | 0.81 |
| 57/82 | 66/28 | 67/30 | 74 | 84 | 0.85 | |||||||||
| 79 | Li WH | 2016 | China | 48/25 | 57/53 | 81/72 | Panel E | hypermethylation | MSP | Serum | I-IV | 83 | 88 | / |
| 80 | Lin ZH | 2017 | China | 131/34 | 61/57 | 67/65 | Panel F | methylation | MSP | Plasma | I-IV | 92 | 50 | / |
| 89 | Zhang X | 2014 | China | 57/42 | 61/57 | 68/64 | Panel G | methylation | MSP | Plasma | I-IV | 87 | 38 | / |
SEN: sensitivity; SEP: specificity; AUC: area under the curve; MSP: methylation-specific PCR; QuARTS: Quantitative allele-specific real-time target and signal amplification.
Panel A: ELMO1, ZNF569, C13orf18; Panel B: Septin 9, RFP 180; Panel C: FAM5C, MYLK; Panel D: KCNQ5, C9orf50, CLIP4; Panel E: OSR2, VAV3, PPFIA3; Panel F: ZIC1, HOXD10, RUNX3; Panel G: RNF180, DAPK1, SFRP2.
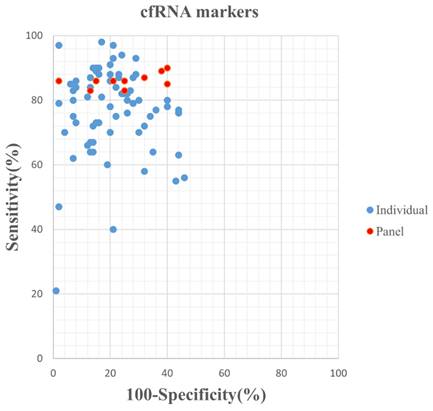
Graphical representation of sensitivity vs specificity of analyzed cfDNA markers. Sensitivity is plotted on the y-axis while on the x-axis the false-positive rate is presented (100-Specificity).
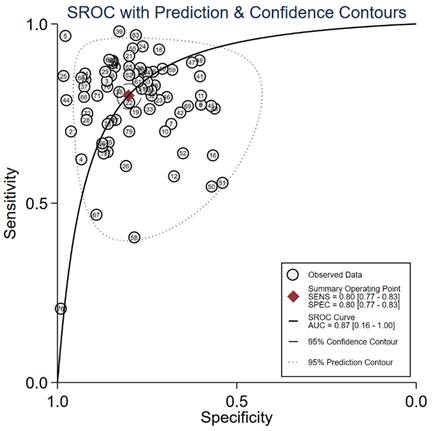
Summary of AUC of cfRNA markers for the diagnosis of gastric cancer.
Summary of AUC of cfDNA markers for the diagnosis of gastric cancer.
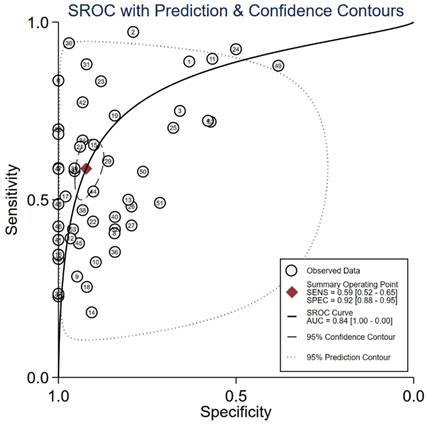
The median (range) reported sensitivity and specificity of RNA panels were 86% (83%-90%) and 75% (60%-98%), respectively. The 21 included studies reported a total number of 32 cfDNAs with the diagnostic potential for GC, of which, seven cfDNAs were reported in more than two studies (Table S4). Five reported cfDNA panels for GC diagnosis contained three number of cfDNAs and two panels contained two individual cfDNAs. An overview of the diagnostic performance of all reported cfDNAs and cfDNA panels was shown in Figure 3. The reported sensitivity and specificity for individual cfDNAs were 50% (range: 18%-96%) and 93% (range: 57%-100%), respectively. The median (range) reported sensitivity and specificity of cfDNA panels were 85% (41%-92%) and 73.5% (38%-90%), respectively. Overall, the sensitivity of cfRNA and cfDNA panels appeared to be better than that of individual cfRNAs or cfDNAs, but the specificity was lower.
Among the included studies of cfRNAs, two enrolled patients with stages I-III [12,39], three enrolled patients with earlystage disease, specifically stages I-II [35,37,48], and the remaining studies included patients with stages I-IV. Subgroup analyses were conducted in four studies [34,35,45,46]. Li et al. [35] evaluated the diagnostic efficiency of miR-199a-3p for stages I and II GC, reporting a sensitivity and specificity of 76% and 74%, respectively. They also reported the diagnostic efficiency of miR-199a-3p for stages I-IV GC, with a sensitivity of 80% and specificity of 74% [34]. Roy et al. [45] investigated an eight-circRNA panel for early GC, reporting a sensitivity of 90%, specificity of 60%, and AUC of 0.82. This panel was equally effective in diagnosing both early GC and stage I-IV GC. Saliminejad et al. [46] conducted a nested case-control study exploring the diagnostic efficacy of a three-miRNA panel in plasma for GC occurring at stages I-II, III-IV, and I-IV, reporting AUC values of 0.83, 0.93, and 0.92, respectively.
Of the included studies of cfDNAs, only two enrolled early-stage patients [73,76], and only one performed stage-specific analysis [84]. Saliminejad et al. [84] evaluated the diagnostic efficiency of four cfDNAs (P16, RASSF1A, RPRM, RUNX3) for GC and found no significant difference in AUC values among stages I-II, III-IV, and I-IV (Table 3).
Seven miRNAs were reported in at least two studies, with miRNA-21 being the most commonly reported in five studies, followed by miR-421, miR-222, miR-106a, miR-25, miR-93, and miR-199a-3p, all of which were reported in two studies (Table S3). SEPT9 was the most frequently reported cfDNA (Table S4), with sensitivity ranging from 18% to 52% (median sensitivity 41.5%), specificity ranging from 85% to 96% (median specificity 90.5%), and AUC values ranging from 0.65 to 0.77 (median AUC value = 0.7).
3.4 Overall comparison of cfRNA and cfDNA markers for GC detection
We evaluated the diagnostic efficiency of cfRNAs and cfDNAs, as shown in Figure 4 and Figure 5, respectively. The meta results showed that the diagnostic accuracy of cfRNA markers (sensitivity 0.80, specificity 0.80, and AUC 0.87) was different from that of cfDNA markers (sensitivity 0.59, specificity 0.92, and AUC 0.84). Compared to cfDNAs, the diagnostic ability of cfRNAs was better, with sensitivity increasing from 0.59 to 0.80, but worse with specificity decreasing from 0.92 to 0.80. However, differences with respect to AUCs were very limited.
4. Discussion
Our systematic review identified a total of 84 cfRNAs and 32 cfDNAs from 79 eligible studies evaluating the diagnostic performance of circulating nucleic acids for GC detection. Forty-four studies integrated individual markers into panels. Only five studies [34, 35, 45, 46, 84] conducted stage-specific analysis for the diagnostic performance of cfRNAs and cfDNAs. However, due to the lack of sufficient data, stage-specific miRNA for GC is still elusive. Overall, cfRNA and cfDNA markers show favorable diagnostic performance for GC. Compared to cfDNA markers, cfRNA markers showed better sensitivity but worse specificity for GC detection.
Cell-free RNA (cfRNA) typically includes encoded mRNAs and non-coding RNAs such as lncRNAs, miRNAs, circRNAs, and piRNAs [91, 92]. Aalami et al. [12] revealed a higher miR-223 expression (3.10-fold expression) in patients with GC compared to controls. Chen et al. [16] showed that miR-421 could achieve a satisfactory diagnostic efficiency in distinguishing GC patients from healthy controls with an AUC of 0.981 (sensitivity = 96.67% and specificity = 95.56%). In this study, they found that plasma miR-421 could well distinguish precancerous lesions of gastric cancer patients from healthy controls with an AUC of 0.872 (sensitivity = 66.29% and specificity = 95.56%). Furthermore, the diagnostic efficacy of miR-421 was markedly higher than traditional tumor markers, such as CA153, CA211, and CA50. A study by Chen et al. [15] indicated that a combination of plasma miR‑650 and CA211 was an effective and novel diagnostic biomarker panel in the diagnosis of GC. However, due to the instability of most extracellular free mRNAs and lncRNAs, they are not suitable as tumor markers. Therefore, researchers have focused more on stable small non-coding RNAs (ncRNAs), such as miRNAs, as diagnostic and prognostic markers for cancer in the past decade. Although miRNAs account for less than 2% of total ncRNAs, abnormal expression has been found in gastric tissue for pre-cancer events (such as Helicobacter pylori infection and pre-cancerous lesions including chronic atrophic gastritis and intestinal metaplasia), as well as in early and late-stage gastric cancer [93, 94]. Therefore, many studies have proposed panels with higher sensitivity and specificity, which can better diagnose tumors and determine their prognosis than single miRNAs. In recent years, with advances in small non-coding RNA sequencing technology, more and more circRNAs and piRNAs have been discovered and named, becoming a hot topic in cancer research. Similar to miRNAs, circRNAs and piRNAs are also characterized by their abundance, stability, and tissue specificity, and they are widely circulated in various body fluids and extracellular vesicles [9, 92]. Hence, circRNAs and piRNAs may become new diagnostic biomarkers for gastric cancer. Overall, small ncRNAs have certain advantages as tumor markers, as mentioned above. To better diagnose gastric cancer in its early stages, researchers should continue to explore more small non-coding RNAs and propose more accurate and specific diagnostic panels in the future.
cfDNA has become an effective biomarker for cancer detection. Pimson et al. [83] confirmed that the cfDNA level of GC patients was higher than that of healthy controls. The diagnostic sensitivity and specificity were 94% and 97% respectively. The cfDNA can also be used to distinguish benign gastric diseases (BGD) and early gastric diseases (EGC). Cao et al. [73] showed that SEPT9 was methylated in 28.4% (21/74) of (EGC) cases but in only 6.1% (6/99) of BGD cases (P<0.001), RNF180 was found to be methylated in 32.4% (24/74) of EGC cases, which was significantly higher than the 13.1% (13/99) of BGD cases (P<0.001). Lin et al. [80] discovered that the combined hypermethylated status of FAM5C and MYLK correlated with tumor size (P < 0.001), tumor invasion depth (P = 0.001) and tumor-node-metastasis (TNM) stage (P = 0.003). With the rapid decrease in sequencing costs and the emergence of more efficient library preparation techniques, researchers are able to detect cancer-related point mutations, copy number variations, and methylation markers at increasingly early disease stages [95-98]. Although these methods hold promise for cancer screening, they are fundamentally limited by the amount of tumor DNA shed into the bloodstream during cell death [99]. Small or slowly growing tumors release less DNA into circulation, resulting in decreased sensitivity for early cancer detection using cfDNA [100]. In addition, most cfDNA features, such as small nucleotide variations, are not tumor tissue-specific, making it difficult to predict the tumor origin site for positive cancer screening patients. Recently, targeted analysis of methylation markers on cfDNA has been shown to detect and locate cancer with high specificity [101]. Technological advances make cfDNA detection more specific, but recent reports indicate that aging individuals exhibit a background mutation landscape, including cancer driver genes [102, 103]. White blood cells are the main source of this background, which requires measuring the clonal hematopoiesis of indeterminate potential (CHIP) mutations and eliminating these mutations from the overall pool of cfDNA aberrations detected in individual patients [104].
Despite the increasing amount of research on cell-free nucleic acids as tumor markers in recent years and the identification of many nucleic acid markers for cancer diagnosis, personalized treatment, and prognosis, their reproducibility is low and their application in clinical practice is limited. This situation is mainly caused by several factors. Firstly, the clinical sample sizes used in most studies are too small, and the heterogeneity of tumors affects the representativeness of identified nucleic acid markers. Therefore, to ensure the reliability and effectiveness of research, it is necessary to increase the number of research samples and fully consider the impact of tumor heterogeneity. Secondly, the clinical sample types collected in most studies are single, and the identified nucleic acid markers cannot accurately distinguish between cancer patients and those with related precursor lesions. Therefore, different stages of clinical samples should be studied to obtain more accurate nucleic acid markers and improve their application value. Finally, so far, there has been no unified standard for sample collection and detection methods. Differences in sample processing, detection methods, and data processing standards can all lead to different results. Therefore, a set of standardized sample collection and detection methods needs to be developed to ensure the consistency and comparability of data.
Sample preparation is an essential pre-analytical factor that affects the identification of potential marker candidates. In 1999, the World Health Organization (WHO) observed a change made by some laboratories that indicated plasma as the preferred sample for the analysis of blood extracellular constituents as it was more representative of the in-vivo status of the patient compared to serum [105]. Plasma is recommended for metabolites in general, for circulating DNA and RNA associated with tumors in cell-free samples, and for mitochondrial RNA, while serum remains the preferred sample for proteomics and lipidology [106, 107]. Since the concentrations of cellular miRNAs and cfDNAs are relatively high compared to those in plasma and serum, it is recommended to use a second high-speed centrifugation or filtration step during blood processing [108-111]. This step serves to remove the potentially retained cells and cell debris from plasma or serum, minimizing the possibility of blood cell contamination that could lead to erroneous interpretation of results. However, only a few of the included studies applied such a high-speed centrifugation step (Table S1, S2). Hemolysis of samples is another factor that could cause variability in miRNA findings, as Pritchard et al [111]. showed that hemolysis can alter plasma miRNA biomarker levels by up to 50-fold. [110] The diverse extraction and quantification methods used in the included studies could also lead to bias in marker identification. Another important yet unresolved issue in circulating RNA investigation is normalization. Considering the limitations mentioned above, further accurate studies on the use of cell-free nucleic acid as promising diagnostic biomarkers for GC are urgently needed.
5. Conclusions
Our review concluded that cell-free nucleic acids hold the potential to serve as diagnostic markers for GC. Given that cfRNA and cfDNA markers have demonstrated favorable diagnostic performance for GC, their combination may enhance diagnostic efficiency. However, considering the differences in marker identification caused by pre-analytical factors and analytical factors, future studies should focus on standardizing sample processing procedures and detection protocols.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This work was supported by the grant from Hubei Provincial Department of Science and Technology (No. 2022BEC047).
Author contributions
Tao Liu and Haixin Yu designed the study. Xiaoxiao Wang, Fen Li and Yi Liu screened and selected the eligible studies. Jingjing Sun and Lin Zhang extracted the data. Yong Xiao and Xiaoming Lu assessed the risk of bias. Qun Zhang and Zhouyuan Du wrote the manuscript. Haixin Yu and Tao Liu revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. 2021;71:209-49
2. Jiang F, Hong F, Shah MW. et al. Circular RNAs as diagnostic biomarkers in gastric cancer: A meta-analysis review. Pathology - Research and Practice. 2019;215:152419
3. Tang L, Zhao S, Liu W. et al. Diagnostic accuracy of circulating tumor cells detection in gastric cancer: systematic review and meta-analysis. BMC Cancer. 2013;13:314
4. Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-73
5. Ye Q, Ling S, Zheng S. et al. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114
6. Ma X, Ou K, Liu X. et al. Application progress of liquid biopsy in gastric cancer. Front Oncol. 2022;12:969866
7. Pardini B, Sabo AA, Birolo G. et al. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers (Basel). 2019;11:1170
8. Qin W, Kang P, Xu Y. et al. Long non-coding RNA HOTAIR promotes tumorigenesis and forecasts a poor prognosis in cholangiocarcinoma. Sci Rep. 2018;8(1):12176
9. Shan C, Zhang Y, Hao X. et al. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019;18(1):136
10. Moher D, Liberati A, Tetzlaff J. et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097
11. Whiting PF, Rutjes AWS, Westwood ME. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-36
12. Aalami AH, Pouresmaeil V, Amirabadi A. et al. Evaluation of the Diagnostic Properties of Serum hsa-miR-223-5p in the Detection of Gastric Cancer: A Case-Control Study. Anticancer Agents Med Chem. 2020;20:800
13. Bai S, Ji R, Wei H. et al. Serum miR-551b-3p is a potential diagnostic biomarker for gastric cancer. The Turkish Journal of Gastroenterology. 2019;30:415-9
14. Cai C, Zhang H, Zhu Y. et al. Serum Exosomal Long Noncoding RNA pcsk2-2:1 As A Potential Novel Diagnostic Biomarker For Gastric Cancer. Onco Targets Ther. 2019;12:10035-41
15. Chen J, Wu L, Sun Y. et al. Diagnostic value and clinical significance of circulating miR-650 and CA211 in detecting of gastric carcinoma. Oncol Lett. 2020;20:254
16. Chen J, Wu L, Sun Y. et al. Mir-421 in plasma as a potential diagnostic biomarker for precancerous gastric lesions and early gastric cancer. PeerJ. 2019;7:e7002
17. Chen S, Li T, Zhao Q. et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167-71
18. Chen X, Li X, Peng X. et al. Use of a Four-miRNA Panel as a Biomarker for the Diagnosis of Stomach Adenocarcinoma. Dis Markers. 2020;2020:1-12
19. Dong Z, Sun X, Xu J. et al. Serum Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) mRNA Protected by Exosomes as a Potential Biomarker for Gastric Cancer. Med Sci Monit. 2019;25:7770-83
20. Elsayed ET, Salem PE, Darwish AM. et al. Plasma long non-coding RNAHOTAIR as a potential biomarker for gastric cancer. The International Journal of Biological Markers. 2018;33:528-33
21. Emami SS, Nekouian R, Akbari A. et al. Evaluation of circulating miR-21 and miR-222 as diagnostic biomarkers for gastric cancer. J Cancer Res Ther. 2019;15:115-9
22. Fu Z, Qian F, Yang X. et al. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31(9):164
23. Gong Y, Liu HB, Yuan RH. et al. Expression of miR-199a in gastric cancer tissues and serum and its clinical significance. Int J Clin Exp Med. 2018;11:7751-60
24. Gu X, Ma S, Liang B. et al. Serum hsa_tsr016141 as a Kind of tRNA-Derived Fragments Is a Novel Biomarker in Gastric Cancer. Front Oncol. 2021;11:679366
25. Guo Y, Cui X, Zhang Y. et al. Diagnostic and Prognostic Value of Serum miR-296-5p and miR-28-3p in Human Gastric Cancer. Cancer Biother Radiopharm. 2020;38(2):95-101
26. Han W, Bu X, Liu Y. et al. Clinical value of miR-135 and miR-20a combined with multi-detector computed tomography in the diagnosis of gastric cancer. World J Surg Oncol. 2021;19(1):283
27. Hou X, Zhang M, Qiao H. Diagnostic significance of miR-106a in gastric cancer. Int J Clin Exp Pathol. 2015;8:13096-101
28. Huang S, Wang J, Li J. et al. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn J Clin Oncol. 2016;46:811-8
29. Huang Y, Zhang H, Gu X. et al. Elucidating the Role of Serum tRF-31-U5YKFN8DYDZDD as a Novel Diagnostic Biomarker in Gastric Cancer (GC). Front Oncol. 2021;11:723753
30. Ji B, Huang Y, Gu T. et al. Potential diagnostic and prognostic value of plasma long noncoding RNA LINC00086 and miR-214 expression in gastric cancer. Cancer Biomark. 2019;24:249-55
31. Kong S, Yang Q, Tang C. et al. Identification of hsa_circ_0001821 as a Novel Diagnostic Biomarker in Gastric Cancer via Comprehensive Circular RNA Profiling. Front Genet. 2019;10:878
32. Kong Y, Ning L, Qiu F. et al. Clinical significance of serum miR-25 as a diagnostic and prognostic biomarker in human gastric cancer. Cancer Biomark. 2019;24:477-83
33. Li B, Zhang H. Plasma microRNA-320 is a potential diagnostic and prognostic bio-marker in gastric cancer. Int J Clin Exp Pathol. 2017;10:7356-61
34. Li C, Li JF, Cai Q. et al. MiRNA-199a-3p in Plasma as a Potential Diagnostic Biomarker for Gastric Cancer. Ann Surg Oncol. 2013;20:397-405
35. Li C, Li JF, Cai Q. et al. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108:89-92
36. Li F, Guo Y, Liu J. et al. The significance of elevated plasma expression of microRNA 106b~25 clusters in gastric cancer. PLoS One. 2017;12:e0178427
37. Li Y, Sun H, Guan J. et al. Serum microRNA-381: A Potential Marker for Early Diagnosis of Gastric Cancer. Yonsei Med J. 2019;60:720
38. Liu H, Yang Z, Zhang J. et al. MicroRNA-217 in plasma: a potential biomarker in gastric cancer. Int J Clin Exp Med. 2017;10:3313-20
39. Liu HF, Zhang XY, Liang F. et al. Downregulation of serum miR-205 as a potential biomarker for gastric cancer diagnosis, prognosis, and chemosensitivity prediction. Int J Clin Exp Med. 2017;10:13426-32
40. Liu H, Zhu L, Liu B. et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196-203
41. Liu W, Li Y, Zhang Y. et al. Circulatinglong non-coding RNA FEZF1-AS1 and AFAP1-AS1 serve as potential diagnostic biomarkers for gastric cancer. Pathology - Research and Practice. 2020;216:152757
42. Liu Y, Zhang YM, Ma FB. et al. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J Gastroenterol. 2019;25:2763-75
43. Park J, Kim M, Song K. et al. Cell-Free miR-27a, a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Genomics & Informatics. 2015;13:70
44. Qin S, Yang L, Kong S. et al. LncRNA HCP5: A Potential Biomarker for Diagnosing Gastric Cancer. Front Oncol. 2021;11:684531
45. Roy S, Kanda M, Nomura S. et al. Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol Cancer. 2022;21(1):42
46. Saliminejad K, Mahmoodzadeh H, Soleymani Fard S. et al. A Panel of Circulating microRNAs as a Potential Biomarker for the Early Detection of Gastric Cancer. Avicenna Journal of Medical Biotechnology. 2022;14(4):278-86
47. Shan L, Liu C, Ma C. High Expression of Serum UCA1 may be a Potential Biomarker for Clinical Diagnosis of Gastric Cancer. Clin Lab. 2019 65 (9)
48. Shao Y, Qi C, Yan J. et al. Biological and clinical implications of hsa_circ_0086720 in gastric cancer and its clinical application. J Clin Lab Anal. 2022 36 (5)
49. Shen Y, Xue CJ, You GL. et al. Value of serum gastric cancer-associated antigen, miR-30c, and gastrin 17 in the early diagnosis of gastric cancer. Acta Medica Mediterr. 2020;36:1393-7
50. Shen Y, Xie Y, Yu X. et al. Clinical diagnostic values of transfer RNA-derived fragment tRF-19-3L7L73JD and its effects on the growth of gastric cancer cells. J Cancer. 2021;12:3230-8
51. So JBY, Kapoor R, Zhu F. et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70:829-37
52. Sun X, Kong S, Jiang C. et al. Diagnostic Value of Circular RNA hsa_circ_0002874 Expression in Peripheral Blood of Patients with Gastric Cancer. Lab Med. 2022;53:65-70
53. Tian W, Pang X, Luan F. Diagnosis value of miR-181, miR-652, and CA72-4 for gastric cancer. J Clin Lab Anal. 2022;36(6):e24411
54. Wu D, Cao G, Huang Z. et al. Decreased miR-503 expression in gastric cancer is inversely correlated with serum carcinoembryonic antigen and acts as a potential prognostic and diagnostic biomarker. Onco Targets Ther. 2017;10:129-35
55. Wu J, Li G, Yao Y. et al. MicroRNA-421 is a new potential diagnosis biomarker with higher sensitivity and specificity than carcinoembryonic antigen and cancer antigen 125 in gastric cancer. Biomarkers. 2014;20:58-63
56. Wu J, Li G, Wang Z. et al. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis Markers. 2015;2015:1-8
57. Xiao K, Dong Z, Wang D. et al. Clinical value of lncRNA CCAT1 in serum extracellular vesicles as a potential biomarker for gastric cancer. Oncol Lett. 2021;21:447
58. Yan J, Shao Y, Lu H. et al. Hsa_circ_0001020 Serves as a Potential Biomarker for Gastric Cancer Screening and Prognosis. Dig Dis Sci. 2022;67:3753-62
59. Yin G, Zheng S, Shan Y. et al. Circulating circRNA Has_Circ_0141633 Serves as a Potential Biomarker for Gastric Cancer. Clin Lab. 2020 66(9)
60. Yuan R, Wang G, Xu Z. et al. Up-regulated Circulating miR-106a by DNA Methylation Promised a Potential Diagnostic and Prognostic Marker for Gastric Cancer. Anticancer Agents Med Chem. 2016;16:1093-100
61. Zeng Q, Jin C, Chen W. et al. Downregulation of serum miR-17 and miR-106b levels in gastric cancer and benign gastric diseases. Chin J Cancer Res. 2014;26:711-6
62. Zeng W, Zhang S, Yang L. et al. Serum miR-101-3p combined with pepsinogen contributes to the early diagnosis of gastric cancer. BMC Med Genet. 2020;21(1):28
63. Zhang W, Zheng M, Kong S. et al. Circular RNA hsa_circ_0007507 May Serve as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front Oncol. 2021;11:699625
64. Zhang Y, Gu X, Qin X. et al. Evaluation of serum tRF-23-Q99P9P9NDD as a potential biomarker for the clinical diagnosis of gastric cancer. Mol Med. 2022;28(1):63
65. Zhao Q, Chen S, Li T. et al. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333
66. Zheng G, Xu Z, Hu C. et al. Exosomal miR-590-5p in Serum as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front Mol Biosci. 2021;8:636566
67. Zhou X, Yin C, Dang Y. et al. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516
68. Zhou X, Ji G, Chen H. et al. Clinical role of circulating miR-223 as a novel biomarker in early diagnosis of cancer patients. Int J Clin Exp Med. 2015;8:16890-8
69. Zong W, Feng W, Jiang Y. et al. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clinical Chemistry and Laboratory Medicine (CCLM). 2019;57:1063-72
70. Anderson BW, Suh Y, Choi B. et al. Detection of Gastric Cancer with Novel Methylated DNA Markers: Discovery, Tissue Validation, and Pilot Testing in Plasma. Clin Cancer Res. 2018;24:5724-34
71. Balgkouranidou I, Karayiannakis A, Matthaios D. et al. Assessment of SOX17 DNA methylation in cell free DNA from patients with operable gastric cancer. Association with prognostic variables and survival. Clin Chem Lab Med. 2013;51(7):1505-10
72. Balgkouranidou I, Matthaios D, Karayiannakis A. et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2015;778:46-51
73. Cao C, Chang L, Wu Q. Circulating methylated Septin 9 and ring finger protein 180 for noninvasive diagnosis of early gastric cancer. Transl Cancer Res. 2020;9:7012-21
74. Chen L, Su L, Li J. et al. Hypermethylated FAM5C and MYLK in serum as diagnosis and pre-warning markers for gastric cancer. Dis Markers. 2012;32:195-202
75. Han J, Lv P, Yu J. et al. Circulating Methylated MINT2 Promoter DNA Is a Potential Poor Prognostic Factor in Gastric Cancer. Dig Dis Sci. 2014;59:1160-8
76. Hideura E, Suehiro Y, Nishikawa J. et al. Blood Free-Circulating DNA Testing of Methylated RUNX3 Is Useful for Diagnosing Early Gastric Cancer. Cancers (Basel). 2020;12:789
77. Lee HS, Hwang SM, Kim TS. et al. Circulating Methylated Septin 9 Nucleic Acid in the Plasma of Patients with Gastrointestinal Cancer in the Stomach and Colon. Transl Oncol. 2013;6:290-4
78. Li H, Zhao G, Guo Y. et al. Feasibility and reproducibility of a plasma-based multiplex DNA methylation assay for early detection of gastric cancer. Pathology - Research and Practice. 2022;238:154086
79. Li W, Zhou Z, Huang T. et al. Detection of OSR2, VAV3, and PPFIA3 Methylation in the Serum of Patients with Gastric Cancer. Dis Markers. 2016;2016:1-7
80. Lin Z, Luo M, Chen X. et al. Combined Detection of Plasma ZIC1, HOXD10 and RUNX3 Methylation is a Promising Strategy for Early Detection of Gastric Cancer and Precancerous Lesions. J Cancer. 2017;8:1038-44
81. Ling ZQ, Lv P, Lu XX. et al. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PLoS One. 2013;8:e67195
82. Miao J, Liu Y, Zhao G. et al. Feasibility of Plasma-Methylated SFRP2 for Early Detection of Gastric Cancer. Cancer Control. 2020;27:544332511
83. Pimson C, Ekalaksananan T, Pientong C. et al. Aberrant methylation of PCDH10 andRASSF1A genes in blood samples for non-invasive diagnosis and prognostic assessment of gastric cancer. PeerJ. 2016;4:e2112
84. Saliminejad K, Soleymani FS, Khorram KH. et al. Methylation Analysis of P16, RASSF1A, RPRM, and RUNX3 in Circulating Cell-Free DNA for Detection of Gastric Cancer: A Validation Study. Avicenna J Med Biotechnol. 2020;12:99-106
85. Xu J, Song J, Wang T. et al. A combination of methylation and protein markers is capable of detecting gastric cancer detection by combined markers. Epigenomics. 2021;13:1557-70
86. Yang Q, Gao J, Xu L. et al. Promoter hypermethylation of BCL6B gene is a potential plasma DNA biomarker for gastric cancer. Biomarkers. 2013;18:721-5
87. Yu JL, Lv P, Han J. et al. Methylated TIMP-3 DNA in body fluids is an independent prognostic factor for gastric cancer. Arch Pathol Lab Med. 2014;138:1466-73
88. Zhang H, Song Y, Xia P. et al. Detection of aberrant hypermethylated spastic paraplegia-20 as a potential biomarker and prognostic factor in gastric cancer. Med Oncol. 2014;31(2):830
89. Zhang X, Zhang X, Sun B. et al. Detection of aberrant promoter methylation of RNF180, DAPK1 and SFRP2 in plasma DNA of patients with gastric cancer. Oncol Lett. 2014;8:1745-50
90. Zhao L, Li M, Zhang S. et al. Plasma-Methylated SEPT9 for the Noninvasive Diagnosis of Gastric Cancer. J Clin Med. 2022;11(21):6399
91. Zaporozhchenko IA, Ponomaryova AA, Rykova EY. et al. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn. 2018;18:133-45
92. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-74
93. Djebali S, Davis CA, Merkel A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
94. Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol. 2018;24:3313-29
95. Newman AM, Bratman SV, To J. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-54
96. Shen SY, Singhania R, Fehringer G. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579-83
97. Bettegowda C, Sausen M, Leary RJ. et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med. 2014;6(224):224ra24
98. Chan KCA, Jiang P, Chan CWM. et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proceedings of the National Academy of Sciences. 2013;110:18761-8
99. Larson MH, Pan W, Kim HJ. et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun. 2021;12(1):2357
100. Chan KA, Jiang P, Zheng YW. et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clin Chem. 2013;59:211-24
101. Liu MC, Oxnard GR, Klein EA. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745-59
102. Razavi P, Li BT, Brown DN. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928-37
103. Genovese G, Kahler AK, Handsaker RE. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477-87
104. Chan HT, Nagayama S, Chin YM. et al. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol Oncol. 2020;14:1719-30
105. Plebani M, Banfi G, Bernardini S. et al. Serum or plasma?. An old question looking for new answers. Clin Chem Lab Med. 2020;58:178
106. Ishikawa M, Maekawa K, Saito K. et al. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects' gender and age. PLoS One. 2014;9:e91806
107. Mohamadkhani A, Poustchi H. Repository of Human Blood Derivative Biospecimens in Biobank: Technical Implications. Middle East journal of digestive diseases. 2015;7:61-8
108. Pritchard CC, Kroh E, Wood B. et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila). 2012;5:492-7
109. McDonald JS, Milosevic D, Reddi HV. et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833-40
110. Becker N, Lockwood CM. Pre-analytical variables in miRNA analysis. Clin Biochem. 2013;46:861-8
111. Greytak SR, Engel KB, Parpart-Li S. et al. Harmonizing Cell-Free DNA Collection and Processing Practices through Evidence-Based Guidance. Clin Cancer Res. 2020;26:3104-9
Author contact
![]() Corresponding authors: Dr. Tao Liu, Department of Digestive Surgical Oncology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; E-mail: uniontaoedu.cn. Haixin Yu, Department of Digestive Surgical Oncology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; E-mail: haixin.ycom.
Corresponding authors: Dr. Tao Liu, Department of Digestive Surgical Oncology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; E-mail: uniontaoedu.cn. Haixin Yu, Department of Digestive Surgical Oncology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; E-mail: haixin.ycom.

 Global reach, higher impact
Global reach, higher impact