Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(12):3781-3793. doi:10.7150/jca.95852 This issue Cite
Research Paper
Linc00662 sponges miR-15b-5p to promote hypopharyngeal squamous cell carcinoma progression by facilitating cancer stem cell-like phenotypes
Shengli Clinical Medical College of Fujian Medical University, Fuzhou 350001, China.
Received 2024-3-1; Accepted 2024-4-28; Published 2024-5-20
Abstract
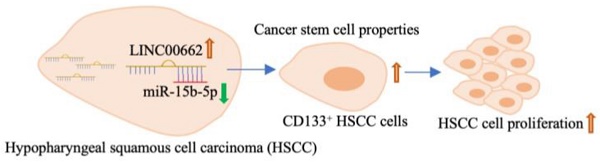
Background: Long non-coding RNAs (lncRNAs) are associated with multiple head and neck tumors and play important roles in cancer. This study explored the molecular mechanism of Linc00662 in hypopharyngeal squamous cell carcinoma (HSCC).
Methods: Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to detect gene expression in HSCC tissues. The viability and proliferation of tumor cells were measured using CCK-8 assays. HSCC cell apoptosis was measured using flow cytometry and western blotting. Cell stemness was examined using the sphere formation assay. A xenograft tumor model was established to investigate the role of Linc00662 in vivo.
Results: The expression level of Linc00662 in HSCC tissues was significantly higher than that in adjacent normal tissues. The expression of Linc00662 had no significant relationship with the tumor stage. Patients with high Linc00662 expression were found to have shorter overall survival than those with low Linc00662 expression. Linc00662 over-expression promoted cell viability and inhibited apoptosis. Using online databases and a dual luciferase reporter, miR-15b-5p was confirmed as a potential downstream sponge of Linc00662. Moreover, Linc00662 was negatively associated with miR-15b-5p in HSCC cells. Depletion of miR-15b-5p can reverse the function of Linc00662 in vivo and in vitro. Furthermore, Linc00662 promotes tumor growth, which was abolished by miR-15b-5p mimics. Importantly, the stemness of cancer stem cells was mediated by the Linc00662/miR-15b-5p axis.
Conclusion: Patients with HSCC with high Linc00662 showed poor prognosis and high Linc00662 induced stemness of tumor cells by targeting miR-15b-5p. Linc00662 may serve as a novel diagnostic and target marker for head and neck squamous cell carcinoma.
Keywords: hypopharyngeal squamous cell carcinoma, Linc00662, miR-15b-5p, cancer stem cell
Introduction
According to the 2020 Global Cancer Report, head and neck squamous cell carcinoma (HNSC) is the sixth most common cancer in the world, with approximately 700,000 cases diagnosed per year [1]. However, hypopharyngeal squamous cell carcinoma (HSCC) has the worst prognosis among all head-and-neck cancers, accounting for 0.8-1.5% of malignant tumors of HNSC, because it is generally detected at an advanced stage due to a lack of biomarkers for early diagnosis [2-4]. Treatment options for HSCC are limited, causing the incidence rate of HSCC to rise rapidly, and there has been a trend toward younger patients in recent years [3, 5, 6]. In particular, the five-year overall survival (OS) rate of patients with HSCC treated with radical radiotherapy was 25-40%, while those who received radiotherapy or chemotherapy only was 12-14% [7-9]. Therefore, it is very important to study the mechanism of the occurrence and development of HSCC and explore new therapeutic targets for improving the survival time and quality of life of patients with HSCC.
HSCC is associated with submucosal development and has a high risk of lymph node metastasis [10, 11]. Cancer stem cells (CSCs) are a small subset of tumors with the ability to regenerate and differentiate, and are considered to be one of the main drivers of tumor cell growth, migration, drug resistance, cancer recurrence, and metastasis in HNSC [12, 13]. Therefore, CSCs play a key role in the development and metastasis of HNSC, explaining the heterogeneity and resistance to cancer therapy in cancers, including head and neck cancers [14]. Therefore, targeting CSCs may be a new and effective strategy for treating HSCC.
Long non-coding RNAs (lncRNAs) have been identified as candidate tumor biomarkers that are expressed in different tissues and cancer types [15, 16]. Recent studies have demonstrated that lncRNAs play an essential role in cancer cell stemness in HSCC [17-19]. Linc00662 is a newly discovered lncRNA, which is associated with poor prognosis and radiation and chemotherapy resistance in patients with cancer [20, 21]. Xenograft experiments verified that Linc00662 promotes breast cancer tumor growth and cell stemness in vivo [22]. In particular, lncRNAs act as sponges for miRNAs to influence the progression of cancer cell stemness [23-27]. There is increasing research on the roles of lncRNAs and miR-15b-5p in the occurrence and development of cancer [28-30]. However, the role and function of Linc00662 and miR-15b-5p in HSCC and their effects on tumor cell stemness have not been completely elucidated.
This study aimed to investigate the effect of Linc00662 on cancer stemness and the underlying mechanism by which Linc00662 regulates cancer stemness in HSCC by sponging miR-15b-5p. Our results may lead to the development of novel therapeutic strategies targeting CSCs in HSCC.
Materials and Methods
Data collection and analysis
The differential Linc00662 expression in normal tissues of human hypopharyngeal carcinoma and at different tumor stages in patients with HSCC was analyzed using Gene Expression Profiling Interactive Analysis (GEPIA). Kaplan-Meier curves and relapse-free survival were used to test the association of Linc00662 with OS and relapse-free survival. The binding of miR-15b-5p to Linc00662 was monitored using miRDB, the Encyclopedia of RNA Interactomes (ENCORI), and the LncRNASNP2 online databases. MiR-15b-5p levels in HSCC were determined using the ENCORI database.
Individuals
Seventeen Patients with HSCC were enrolled in The First Affiliated Hospital of Xiamen University (Xiamen, China), and all specimens were pathologically proven to have HSCC. Basic clinical information of the clinical samples is shown in Table 1. The tumor and surrounding tissues were frozen with liquid nitrogen after resection, and the samples were immediately stored at -80°C. All experiments were approved by the Ethics Committee of the First Affiliated Hospital of Xiamen University (XMYY-2021KYSB289) and Shengli Clinical Medical College of Fujian Medical University. The patients were fully briefed on the course of the study prior to inclusion, and written informed consent was obtained from all enrolled patients.
The basic clinical information of 17 cases squamous cell carcinoma patients.
| Patient number | Differentiation degree | Tumor stage | Year | Gender | Lymph node metastasis |
|---|---|---|---|---|---|
| 932859 | Medium-low differentiation | T2N0M0 | 68 | Male | Yes |
| 930603 | High-medium differentiation | T4N2M0 | 60 | Male | No |
| 931192 | Moderately differentiation | T2N0M0 | 54 | Male | No |
| 931376 | Moderately differentiation | T4aN2cM0 | 62 | Male | No |
| 922354 | High-medium differentiation | T4aN1M0 | 65 | Male | No |
| 925424 | Medium-low differentiation | T4aN1M0 | 57 | Male | Yes |
| 926619 | Moderately differentiation | T3N1M0 | 59 | Male | No |
| 908026 | Poorly differentiation | T4N0M0 | 54 | Male | Yes |
| 871489 | Poorly differentiation | T4N0M0 | 50 | Male | Yes |
| 922673 | Moderately differentiation | T3N2M0 | 53 | Male | No |
| 447883 | High-medium differentiation | T4aN0M0 | 59 | Male | No |
| 933192 | Moderately differentiation | T2N0M0 | 59 | Male | No |
| 933196 | High-medium differentiation | T2N0M0 | 53 | Male | No |
| 930720 | Medium-low differentiation | T4N0M0 | 56 | Male | Yes |
| 943755 | Moderately differentiation | T3N0M0 | 66 | Male | Yes |
| 940629 | High-medium differentiation | T4aN2bM0 | 62 | Male | No |
| 942006 | Moderately differentiation | T4aN2bM0 | 59 | Male | No |
Cell culture, transfection, and treatment
Human oral epithelial keratinocytes (HOK) and human head and neck cancer cell lines (FaDu, SCC-4, SCC-9, Hep-2, and CAL-27) were obtained from the American Type Culture Collection (Manassas, USA) and cultured in DMEM. These cells were supplemented with 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37°C. shRNA targeting Linc00662 (ccggGGCTGATCTCACCTTGTAATTggatccAATTACAAGGTGAGATCAGCCtttttg) was subcloned into the pSin vector. The miR-15b-5p mimics and corresponding negative control miRNA were purchased from GenePharma Co., Ltd. (Shanghai, China). The pcDNA 3.1-LINC00662 overexpression plasmid and corresponding empty vector were obtained from RiboBio (Guangzhou, China). For cell transfection, the Linc00662 overexpression plasmid or Linc00662 shRNA was transfected into FaDu and SCC-9 cells for 48 h, or a miRNA inhibitor of miR-15b-5p mimic was transfected into FaDu cells for 48 h, as instructed by the manufacturer. After 48 h, the FaDu and SCC-9 cells were collected. The experiments were repeated thrice.
Real‐time quantitative reverse transcription‐polymerase chain reaction (qRT-PCR)
Total RNA was extracted from HNSC and mouse tissues treated with different groups. cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara, RR047A). Approximately 1 μg of RNA from tissue was reverse transcribed using the Prime-Script miRNA cDNA Synthesis Kit (TaKaRa) and qRT-PCR was performed to detect the expression levels of Linc00662, miR-15b-5p, CD44, Sox2, Nanog, and Oct4 usinf a SYBR Green Premix Ex Taq kit (Takara RR820A) in triplicate using synthesized primers (Tsingke, China). GAPDH and U6 served as internal controls. Primers used are listed in Table 2. All PCR assays were repeated thrice.
Cell counting kit-8 (CCK-8)
A CCK-8 assay was performed according to the manufacturer's protocol (Dojindo, Japan) to evaluate the proliferation of FaDu and SCC-9 cells. Both cell types were seeded into 96-well plates at a density of 1 × 103 cells/well. DMEM containing 10% CCK8 solution was added to each well and incubated for 24, 48, 72, and 96 h in a 37°C incubator. The absorbance of each well was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, USA) to assess the efficiency of cell proliferation.
Flow cytometry
An Annexin V-allophycocyanin apoptosis detection kit (eBioscience, San Diego, CA, USA) was used to detect apoptosis. In brief, FaDu and SCC-9 cells (1 × 106 cells/mL) were stained with 5 μL Annexin V-FITC for 15 min and 10 μL PI in a staining buffer at 4°C with light avoidance for 5 min after being subjected to different treatments with phosphate-buffered saline (PBS), followed by flow cytometry (BD Biosciences, San Diego, CA, USA). For the detection of CD133 positive cells, cells were collected with a scraper, blocked with 3% BSA, and subsequently incubated with a CD133 antibody (CL488-66666, Proteintech) for 30 min on ice in staining buffer, according to the manufacturer's protocol. The data were analyzed three times and the mean value was calculated.
Western blotting
Proteins (20 μg) were electro-transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Darmstadt, Germany) after separation by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a semi-dry blotting apparatus (Bio-Rad, Hercules, California, USA) and then PVDF were blocked with 5% nonfat milk at room temperature for 1 h and incubated with primary antibodies Bcl-2 (ab32124, Abcam, UK) and BAX (ab32503, Abcam) overnight at 4°C. On the second day, the membranes were incubated with appropriate secondary antibodies for 90 min and developed with enhancement. The protein bands were visualized using enhanced chemiluminescence (ECL Plus). The gray values of the protein bands were analyzed using ImageJ (National Institutes of Health, Bethesda, Maryland, USA). The experiment was repeated three times and the mean value was calculated.
miRNA screening
Through miRDB, ENCORI, and LncRNASNP2 databases screening, candidate miRNAs were identified, and subsequently, these miRNAs were analyzed and compared based on the ENCORI database.
Double luciferase reporter assay
Dual-luciferase reporter gene assay was performed to confirm the relationship between miR-15b-5p and Linc00662. Well-grown 293T cells (8 x104 per well in a 24-well plate) were co-transfected with mutated Linc00662 (Linc00662-MUT) and wild-type Linc00662 (Linc00662-WT) together with the miR-15b-5p mimic or negative control (NC) using Lipofectamine 2000 according to the manufacturer's protocol (Thermo Fisher Scientific, USA). After transfection for 48 h, cells were collected and luciferase activity was measured using the dual-luciferase reporter assay system (Promega). All assays were performed independently in triplicate.
Xenograft
The BALB/c nude mice (six weeks old, n=6/7) were purchased from Xiamen University and reared in plastic cages at 22-25 °C and a humidity of 40-70% under a 12 h/12 h light-dark cycle, with free access to water and food. After an adaptation period of one week, the mice were randomly divided into the following four groups: control, Linc00662 model, Linc00662+NC mimic, and Linc00662+miR-15b-5p mimic. Tumor volume was measured every three d after the transfection of FaDu cells (2×107) with Linc00662 or Linc00662 + miR-15b-5p mimic and their NC. Tumor volume was calculated using the following formula:
Tumor volume = 0.5×length×width2
Primer sets used for quantitative reverse transcription-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Linc00662 | 5′-ACACGCTTCTGAAACTGG-3′ | 5′-GTCAACATGGTGAAACCC-3′ |
| Sox2 | 5′-TGCACCGCTACGACGTGAGC-3′ | 5′-GCCCTGGAGTGGGAGGAAGA-3′ |
| CD44 | 5′-TGAGCATCGGATTTGAGAC-3′ | 5′-CATACTGGGAGGTGTTGGA-3′ |
| Oct4 | 5′-AACGATCAAGCAGTGACTATTC-3′ | 5′-GAGTACAGGGTGGTGAAGTGAGG-3′ |
| Nanog | 5′-AGTTGGACAGGGAGATGGC-3′ | 5′-AACCTTCCTTGCTTCCACG-3′ |
| GAPDH | 5′-ACAACTTTGGTATCGTGGAAGG-3′ | 5′-GCCATCACGCCACAGTTTC-3′ |
| miR-15b-5p | 5'-UAGCAGCACAUCAUGGUUUACA-3' | 5'-CTCAACTGGTGTCGTGGA-3' |
| U6 | 55'-CTCGCTTCGGCAGCACAT-3' | 5'-ACGCTTCACGAATTTGCGT-3' |
| NC mimic | 5'-UUCUCCGAACGUGUCACGUTT-3' | 5'-ACGUGACACGUUCGGAGAATT-3' |
| miR-15b-5p mimic | 5'-UAGCAGCACAUCAUGGUUUACA-3' | 5'-UAAACCAUGAUGUGCUGCUAUU-3' |
On day 22 after inoculation, the mice were euthanized by CO2 inhalation (CO2 flow rate: 10% of cage volume), and the death of the animals was confirmed by the cessation of the heartbeat. The tumor xenografts were harvested, photographed, and weighed. The experiments were carried out in accordance with the protocol approved by the Animal Welfare Ethics Committee of The First Affiliated Hospital of Xiamen University (SYXK (Min)2018-0009).
Tumorsphere assay
The protocol for the formation of tumorspheres has been described previously [31]. Briefly, FaDu cells (5000 cells/well) with Linc00662 or Linc00662 + miR-15b-5p and a relative NC were seeded on six-well ultra-low attachment plates (Corning, New York, NY, USA) and cultured in DMEM/F12 without serum, with 2% B27, EGF 20 ng/mL, and FGF 20 ng/mL. The tumor spheres were recorded and counted on the 14th day after seeding using a microscope, and the number and size of the tumor spheres were analyzed.
Statistical analysis
All data are presented as mean ± SD from at least three independent experiments. Statistical analyses were performed using IBM SPSS version 18.0. Student's t-test (unpaired and two-tailed) or one-way ANOVA was used to measure differences between two or more groups. The relationship between Linc00662 and miR-15b-5p was measured using Spearman's rank test. Differences were considered statistically significant at P < 0.05.
Results
Linc00662 is highly expressed in HSCC
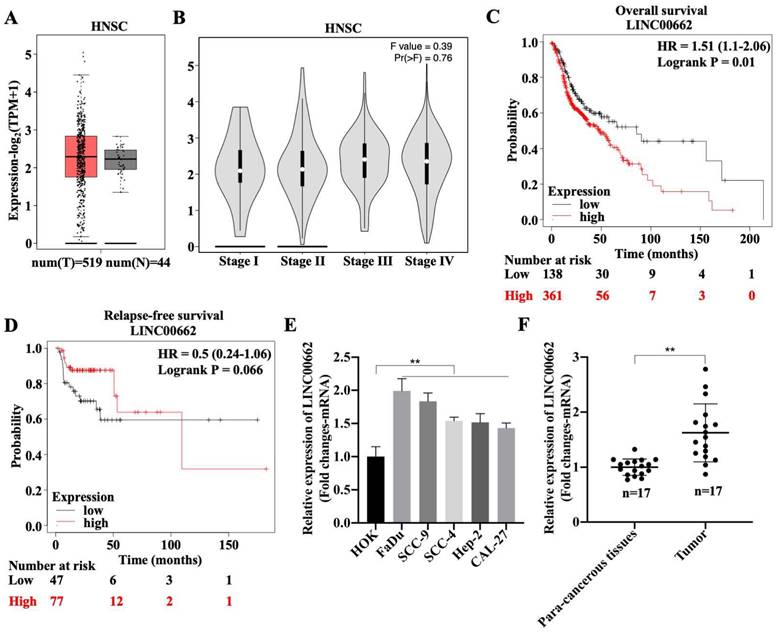
To explore the role of Linc00662 in the development of HSCC, the expression pattern of Linc00662 was determined using the online database, GEPIA2. There was no differential expression of Linc00662 between healthy individuals (n = 44) and patients with HNSC (n = 519), but Linc00662 was highly expressed in patients with HNSC compared with that in healthy individuals (Fig. 1A). Although we found that Linc00662 expression was not correlated with tumor stage or relapse-free survival in patients with HNSC (Fig. 1B-D), patients with high Linc00662 expression survived shorter than those with low Linc00662 expression (Fig. 1C). Subsequently, FaDu, SCC-9, SCC-4, Hep-2, and CAL-27 cells and normal head and neck HOK cells were cultured; the level of Linc00662 was substantially higher in HNSC cell lines than that in normal head and neck cells. There were more Linc00662 levels in FaDu and SCC-9 cells than the other two cell lines, which implied that Linc00662 may have important roles in HSCC (Fig. 1E). Additionally, Linc00662 was highly expressed in fresh HSCC tissues (n = 17), in contrast to the corresponding para-carcinoma tissues (Fig. 1F). Taken together, these results indicated that Linc00662 may be involved in HSCC progression.
Linc00662 promotes cell activity and inhibits cell apoptosis in FaDu and SCC-9 cells
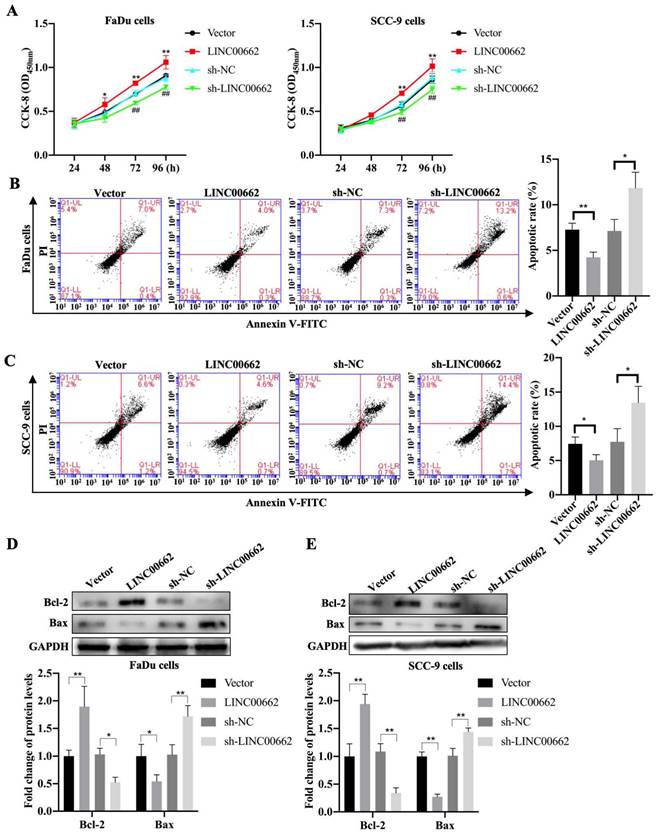
To further investigate whether Linc00662 impacted cell proliferation, we performed CCK-8 assays to detect the viability of FaDu and SCC-9 cells after Linc00662 transfection. Linc00662 significantly prompted the growth rate of FaDu and SCC-9 cells at 72 and 96 h compared to that in the control group. Knockdown of Linc00662 contributed to a conspicuous reduction in cell proliferation in FaDu and SCC-9 cells compared with that in cells transfected with scrambled shRNA (Fig. 2A, B). Flow cytometry analysis suggested that Linc00662 markedly reduced cell apoptosis and Linc00662 silencing notably repressed apoptosis in FaDu and SCC-9 cells (Fig. 2C, D). Additionally, western blot analysis showed that Bcl-2 levels increased and Bax levels decreased in the Linc00662-overexpressing group while Linc00662 silencing caused a reduction in Bcl-2 levels and increased Bax levels (Fig. 2E, F). In summary, our data reveal that Linc00662 is a regulatory factor that promotes cell proliferation and inhibits apoptosis in HSCC cells.
Linc00662 sponges miR-15b-5p in HSCC
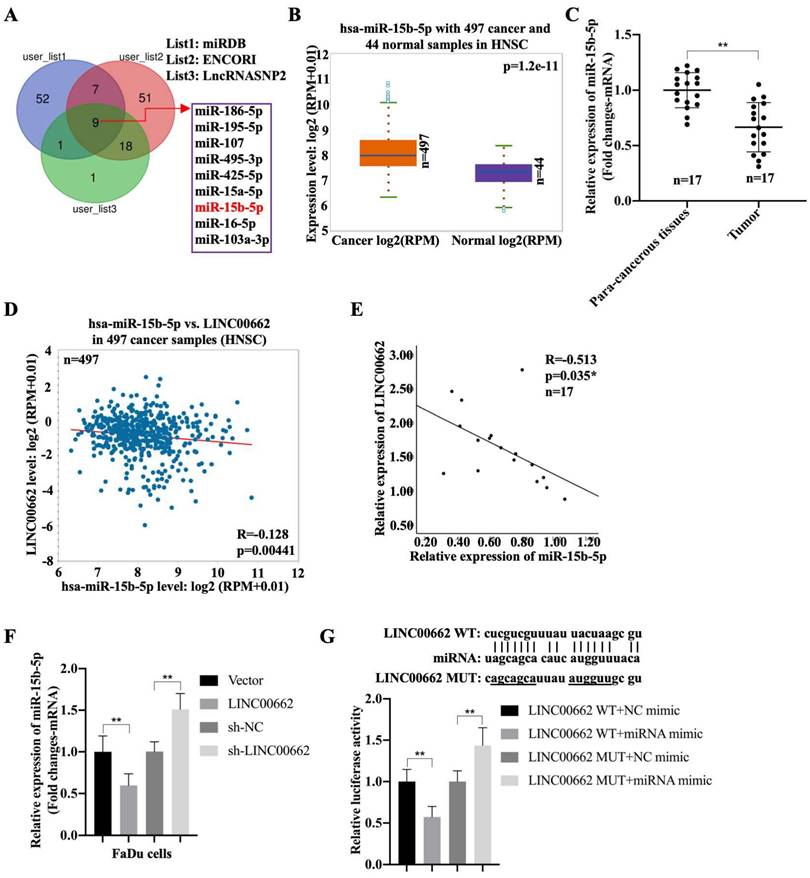
To better understand the mechanism of Linc00662 in regulating the progression of HSCC, night candidate miRNAs, including hsa-miR-15b-5p, were identified using the miRDB, ENCORI, and LncRNASNP2 databases (Fig. 3A). The results showed that the negative correlation between miR-16-5p or miR-107 with LINC00662 in patients with HNSC, but both associations were lower than the association between miR-15b-5p with LINC00662. Moreover, high expression of miRNA-15b was associated with poor prognosis, whereas low expression was associated with good prognosis (Fig. S1). Therefore, we selected miR-15b-5p as a candidate gene. In addition, miR-15b-5p was significantly expressed in patients with HNSC compared to that in the normal groups (Fig. 3B). Furthermore, miR-15b-5p was highly expressed in fresh HSCC tissues than that in para-carcinoma tissues (Fig. 3C). MiR-15b-5p also appeared to be negatively associated with Linc00662 in HNSC, and a significant negative correlation was observed between miR-15b-5p and Linc00662 in patients with HSCC (Fig. 3D, E). FaDu cells were transfected with Linc00662 and a combination of Linc00662 and miR-15b-5p mimic. qRT-PCR assays illustrated that Linc00662 silencing increased miR-15b-5p expression, whereas Linc00662 overexpression decreased miR-15b-5p expression in FaDu cells (Fig. 3F). Additionally, the relative luciferase activity of the Linc00662-WT reporter, rather than Linc00662-MUT reporter, was decreased by miR-15b-5p overexpression (Fig. 3G). These data confirm the direct binding of miR-15b-5p to Linc00662 in HSCC.
Expression and clinical value of lncRNA Linc00662 in head and neck squamous cell carcinoma (HNSC). (A) Differential expressions of Linc00662 in HNSC (n = 519) and not HNSC (n = 44) specimens were determined by using the GEPIA2 database. (B) The correlation between Linc00662 and tumor stage in patients with HNSC expression. (C, D) The Kaplan-Meier method was used to determine the relationship between Linc00662 expression and overall survival (C) or Relapse-free survival (D) of patients with HNSC. Differences between survival curves were tested by the log-rank test. Data are shown as the mean ± SD. Assays were performed in triplicate. (E) qRT-PCR analysis for Linc00662 level in normal HOK and HNSC cell lines (FaDu, SCC-9, SCC-4, Hep-2, and CAL-27). (F) The expression of Linc00662 in para-carcinoma and hypopharyngeal squamous cell carcinoma (HSCC) tissues. **P < 0.01.
Linc00662 promotes cell activity and inhibits cell apoptosis through sponging miR-15b-5p
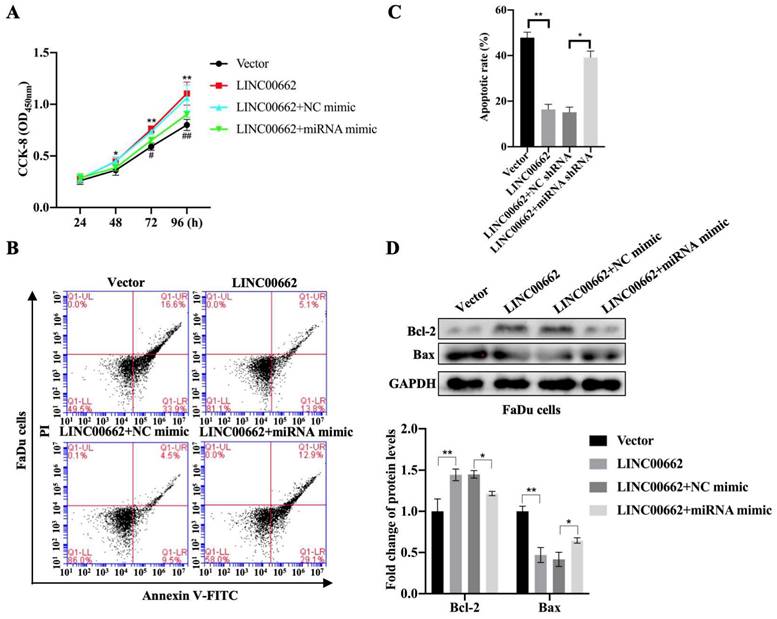
To determine whether Linc00662 could regulate the progression of HSCC by targeting miR-15b-5p, FaDu cells were transfected with Linc00662 alone or a combination of Linc00662 and miR-15b-5p mimic. The CCK-8 assay showed that Linc00662 overexpression significantly promoted FaDu cell proliferation compared with that in the control, whereas the miR-15b-5p mimic reversed the effect of Linc00662 (Fig. 4A). Flow cytometry analysis further demonstrated that overexpression of Linc00662 inhibited apoptosis. However, after transfection with the miR-15b-5p mimic, the effect of Linc00662 on cell apoptosis was significantly reversed (Fig. 4B, C). Furthermore, Linc00662 overexpression induced an increase in Bcl-2, which was prevented by treatment with the miR-15b-5p mimic, whereas the pro-apoptotic Bax protein exhibited the opposite results (Fig. 4D). These data demonstrated that the Linc00662/miR-15b-5p axis can decelerate HSCC progression.
Linc00662 affects the proliferation and apoptosis of FaDu and SCC-9 cells. FaDu cells were transfected with si-Linc00662 or Linc00662 and control groups. (A, B) CCK-8 assay was used to test proliferation of FaDu (A) and SCC-9 cells (B) at 24, 48, 72, and 96 h, respectively. (C, D) Flow cytometry shows the apoptotic rate in FaDu (C) and SCC-9 cells (D) in Linc00662 overexpression, si-Linc00662, and their control groups, respectively. (E, F) Western blot analysis was conducted to detect the expression levels of Bcl-2 and Bax in FaDu (E) and SCC-9 cells (F). All data were expressed as the mean ± SD, *P < 0.05, **P < 0.01.
Linc00662 directly binds to miR-15b-5p in FaDu and 293T cells (A) The candidate miRNAs which could potentially target Linc00662 were listed according to miRDB, ENCORI and LncRNASNP2. (B) Expression of miR-15b-5p in 497 patients with HNSC and 44 normal samples. (C) The expression of miR-15b-5p in para-carcinoma and HSCC cancer tissues. (D) The association between miR-15b-5p and Linc00662 in HNSC was analyzed using ENCORI Starbase. (E) The association between miR-15b-5p and Linc00662 in HSCC tissues. (F) The expression of miR-15b-5p mRNA were assessed by qRT-PCR in FaDu cells. (G) Luciferase reporter assay was performed to verify the interaction of miR-15b-5p and Linc00662 in 293T cells. **P < 0.01.
The Linc00662/miR-15b-5p pathway promotes tumor growth of HSCC
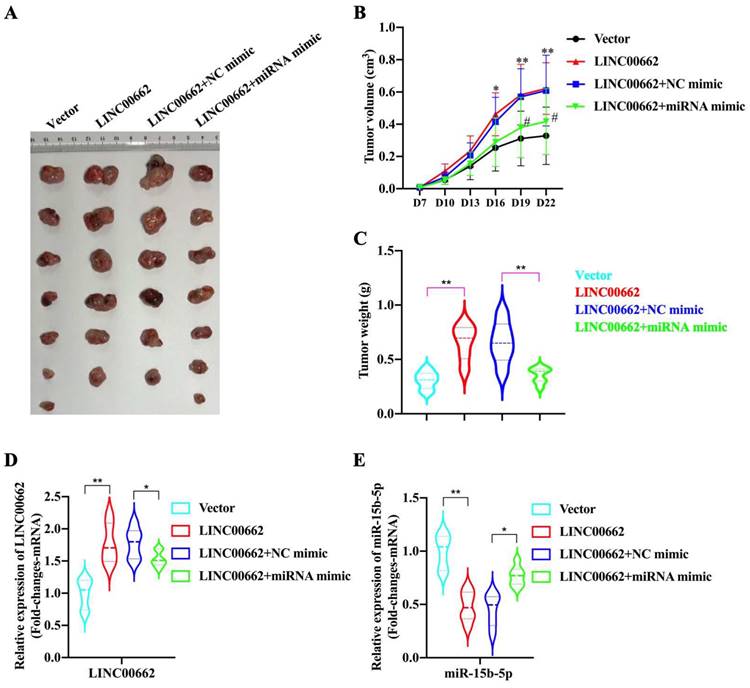
In the tumor growth xenograft model, FaDu cells were subcutaneously implanted into nude mice to induce tumor development (n = 6/7 for each group). After 22 d of injection, the mice were sacrificed, and tumors were obtained (Fig. 5A). We found that the promoting effect of Linc00662 on tumor volume was significantly reversed after transfection with miR-15b-5p mimics compared with that in the control group (Fig. 5B, C). Additionally, the weight of tumors transfected with Linc00662 significantly increased compared with that of the control group; however, when co-transfected with Linc00662 and miR-15b-5p mimic, the stimulatory effect induced by Linc00662 was rescued (Fig. 5C). The qRT-PCR results further suggested that the miR-15b-5p mimic transfection group can significantly reduce the expression of Linc00662 compared with that in the control group (Fig. 5D). Similarly, the expression of miR-15b-5p in the miR-15b-5p mimic transfection group was significantly higher than that in the LINC000662 overexpression alone (Fig. 5E). Overall, these data indicate that Linc00662 promotes HSCC growth in the xenograft model, while the miR-15b-5p mimic could rescue this result.
Cancer cell stemness induced by Linc00662 in HSCC cells is dependent on miR-15b-5p
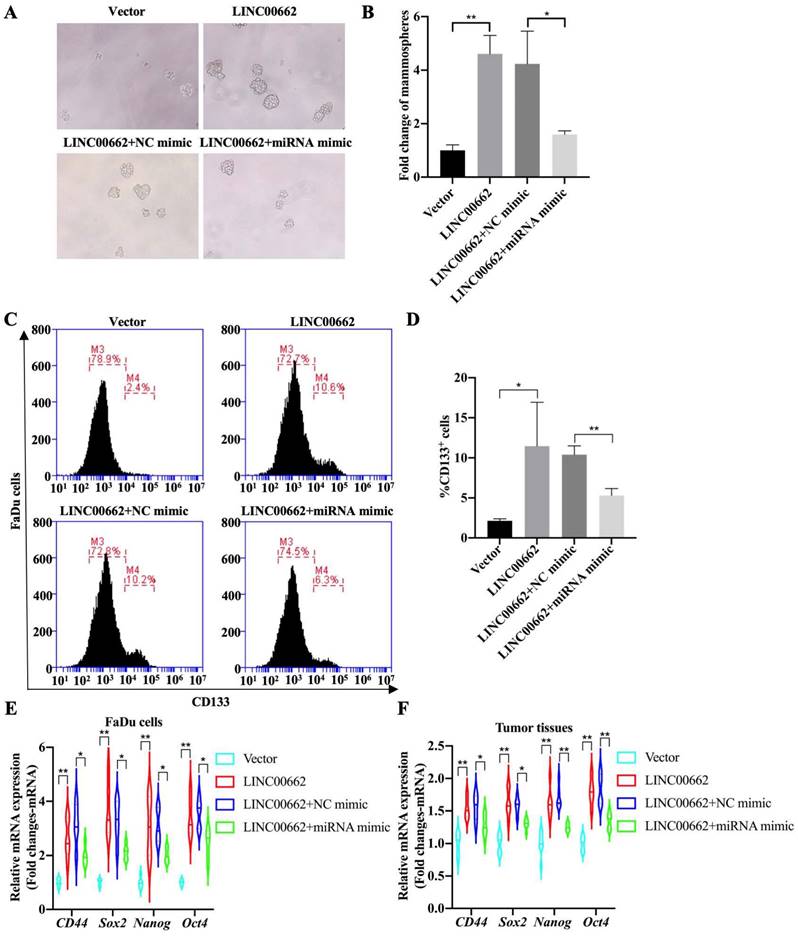
Tumor cell stemness is one of the main factors determining the progression of HSCC, therefore, whether the Linc00662/miR-15b-5p axis regulates the stemness of pharyngeal cancer cells was further explored and tumorsphere assays were evaluated. The results suggested that overexpression of Linc00662 promotes pheroidization of FaDu cells, but transfection with miR-15b-5p mimics significantly reversed the Linc00662-mediated promoting effect of cancer cell stemness (Fig. 5A, B). Flow cytometry analysis showed that Linc00662 increased the proportion of CD133-positive cells, and miR-15b-5p mimic transfection significantly abolished the promotive effects of Linc00662 (Fig. 5C, D). Next, the expression of other CSC markers was detected in FaDu cells. qRT-PCR analysis showed that the relative mRNA expression of CD44, Sox2, Nanog, and Oct4 increased after treatment with Linc00662 compared to those in the control group, which was reversed by the miR-15b-5p mimic (Fig. 6E). In addition, CSC markers in tumor tissues in vivo showed similar results (Fig. 6F). Taken together, these results indicate that Linc00662/miR-15b-5p mediates the development of HSCC tumors by influencing tumor stem cells.
Linc00662 can suppress miR-15b-5p expression. FaDu cells were transfected with Linc00662, Linc00662 + miR-15b-5p mimic or their corresponding control. (A) Cell proliferation was determined with the CCK-8 assay at the indicated time-points. (B) Flow cytometric analysis of the cell cycle in FaDu cells stained with propidium iodide. (C) The apoptotic results were evaluated by flow cytometry. (D) Apoptosis-marker Bcl-2 and Bax were determined by western blotting in FaDu cells. *P<0.05, **P<0.01.
Linc00662/miR-15b-5p modulates HSCC growth in vivo. (A) Representative images of tumor tissues from the FaDu cells transfected with Linc00662 or the combination of Linc00662 and miR-15b-5p mimic for 72 h were obtained on day 7. Vector (n=7): FaDu cell transfected with the vector; LINC00662 (n=6): FaDu cell transfected with LINC00662 overexpression plasmid; LINC00662+NC mimic (n=6): FaDu cell transfected with LINC00662 overexpression plasmid with negative control mimic; LINC00662+miRNA mimic (n=7): FaDu cell transfected with LINC00662 overexpression plasmid with miR-15b-5p mimic. (B) Tumor volume was calculated every 3 d after injection. (C) Weight of the mouse body was measured at the end of modeling. (D) The expression level of Linc000662 in tumor tissue as determined using qRT-PCR. (E) The expression level of miR-15b-5p in tumor tissue as determined using qRT-PCR. *P < 0.05, **P < 0.01.
Discussion
CSCs can initiate tumorigenesis and exist in many different tumor types. Targeting CSCs by suppressing unique molecular determinants of CSCs provides effective therapeutic intervention [32]. However, the limited knowledge of CSCs in HSCC may provide new clinical targets for metastatic or recurrent HSCC. The recurrence and metastasis of HSCC may also be attributed to the persistent presence of CSCs. In this study, we proved that Linc00662 is expressed in HSCC and could regulate the proliferation and apoptosis of HSCC cells by sponging miR-15b-5p, which in turn facilitates the appearance of a CSC-like phenotype. Therefore, this study provides an experimental and theoretical basis for the treatment of HSCC.
Linc00662 is upregulated in malignant tumors [20], such as cervical cancer [33], gastric cancer [34], and osteosarcoma [35], which is also closely related to poor prognosis and chemotherapy resistance in patients with cancer [36, 37]. In this study, we revealed that Linc00662 expression is higher in patients with HNSC, and predicted poor prognosis and shorter survival. Notably, the expression of Linc00662 was significantly higher in HSCC tissues and FaDu/SCC-9 cells. Possibly, Linc00662 may play an important role in the development and occurrence of HSCC. Linc00662 upregulation can affect the proliferation, invasion, and apoptosis of cervical cancer cells [38]. In addition, overexpression of Linc00662 facilitates the proliferation, migration, and invasion of oral squamous cell carcinoma cells [39] and promotes cell viability and metastasis in esophageal squamous cell carcinoma [40]. Our data showed that highly expressed Linc00662 promoted proliferation and inhibited apoptosis in HSCC cells. In contrast, knockdown of Linc00662 restrained proliferation and promoted apoptosis. Therefore, Linc00662 may be a positive regulator of HSCC development.
Linc00662/miR-15b-5p regulates cancer stem cell properties in HSCC. (A) Tumorsphere formation assays of Linc00662, Linc00662 + miR-15b-5p mimic or their corresponding control (B) Quantitation analysis of tumorsphere assay. (C) Flow cytometry analysis shows the proportion of CD133-positive cells in the FaDu cells co-transfected with Linc00662 and miR-15b-5p mimic and the corresponding controls. (D) Quantitative result for CD133-positive cell proportion. (E, F) The expression of CD44, Sox2, Nanog, and Oct4 in FaDu cells and tumor tissues was determined by qRT-PCR. *P < 0.05, **P < 0.01.
MiR-15b-5p has carcinogenic or tumor-inhibitory functions in different types of cancer, and its expression is upregulated in liver and breast cancers, and downregulated in castration-resistant prostate cancer cells [41-43]. Low expression of miR-15b-5p is also associated with poor prognosis in patients with HCC [28] and miR-15b-5p can predict local recurrence in patients with head and neck cancer treated with intensity-modulated radiotherapy [44]. In our data, the expression of miR-15b-5p was significantly lower in patients with HSCC than that in the normal population. Previous studies have indicated that the lncRNA TRPM2-AS/miR-15b-5p/PPM1D axis can promote malignant tumors in osteosarcoma cells [45] or Linc00662 can act as a sponge for miR-15b-5p, facilitating the progression of osteoarthritis [46]. Additionally, lncRNA-MEG8 accelerates non-small cell lung cancer progression by regulating the miR-15b-5p/PSAT1 axis [47]. In this study, our results showed that the Linc00662/miR-15b-5p axis affects HSCC cells and tumors in vivo. There is evidence indicating that the lncRNA FENDRR/miR-15b-5p/TUBA1A axis suppresses cervical cancer cell proliferation and invasion [48], and the lncRNA TTN-AS1 suppresses ovarian cancer cell proliferation and invasion by targeting miR-15b-5p and regulating FBXW7 expression [49]. Therefore, these studies indicate that miR-15b-5p exhibits substantial potential in regulating tumor cell proliferation and tumor growth. In our study, we proved that Linc00662 promoted the proliferation and inhibited the apoptosis of HSCC cells, which was reversed by the miR-15b-5p mimic. Therefore, activation of the Linc00662/miR-15b-5p signaling axis can accelerate the proliferation of HSCC cells, thereby leading to the growth of HSCC in vivo.
CSCs have a high degree of plasticity and are embedded in tumors, making them difficult to identify and eradicate [50, 51]. They are usually identified by the expression of cell surface markers, such as CD133, which is one of the most well-characterized biomarkers used to isolate CSCs [52]; CD44 is usually a highly expressed surface marker in CSC [53], and Sex2, Nanog, and Oct4 are transcription factors necessary for maintaining the phenotype of pluripotent embryonic stem cells [54, 55]. CSCs in oral/oropharyngeal squamous cell carcinoma enhance the tumorigenicity and self-renewal abilities of cancer cells in vivo [56, 57]. Moreover, many studies have reported that Linc00662 enhances cell stemness in breast cancer/osteosarcoma by sponging miR-144-3p/miR-16-5p [22, 35], and that lncRNA CERS6-AS1 acts as an oncogene that facilitates xenograft tumor growth by binding to miR-15b-5p [29]. Consistent with previous findings, we demonstrate that the Linc00662/miR-15b-5p axis affects tumor cell growth stemness. Thus, the Linc00662/miR-15b-5p axis provides new insights into the targeted mechanisms in HSCC.
Conclusion
In summary, Linc00662 acts as an oncogene that promotes stemness of HSCC cells and facilitates xenograft tumor growth by binding to miR-15b-5p. These findings may shed new light on the underlying mechanisms and treatments for HSCC. However, this study has some limitations. First, a larger clinical sample size of hypopharyngeal cancer is still needed to confirm the findings of this study. Second, 3D cell culture technology needs to be employed to confirm the role of the Linc00662/miR-15b-5p axis in tumor cell stemness.
Abbreviations
HSCC: hypopharyngeal squamous cell carcinoma; HNSC: head and neck squamous cell carcinoma; qRT-PCR: Real‐time quantitative reverse transcription‐polymerase chain reaction; CSCs: Cancer stem cells; lncRNAs: Long non-coding RNAs; PVDF: polyvinylidene fluoride; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; FCM: Flow cytometry.
Supplementary Material
Supplementary figure.
Acknowledgements
Funding
The present study was supported by Xiamen Municipal Bureau of Science and Technology (3502Z20209265).
Ethics approval and consent to participate
The experiment was carried out in accordance with the protocol approved by the Animal Welfare Ethics Committee of The First Affiliated Hospital of Xiamen University and Shengli Clinical Medical College of Fujian Medical University.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Author contributions
Y.Q. and Z.B. conceived and designed the experiments; Z.B. performed the experiments; Y.Q. and Z.B. analyzed the data; Z.B. contributed reagents and materials. All authors edited and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
2. Patel RS, Goldstein DP, Brown D, Irish J, Gullane PJ, Gilbert RW. Circumferential pharyngeal reconstruction: history, critical analysis of techniques, and current therapeutic recommendations. Head Neck. 2010;32:109-20
3. Ye J, Wang Z, Chen X, Jiang X, Dong Z, Hu S. et al. YTHDF1-enhanced iron metabolism depends on TFRC m(6)A methylation. Theranostics. 2020;10:12072-89
4. Li Y, Pan M, Lu T, Yu D, Liu C, Wang Z. et al. RAF1 promotes lymphatic metastasis of hypopharyngeal carcinoma via regulating LAGE1: an experimental research. J Transl Med. 2022;20:255
5. Cai Z, Chen L, Chen S, Fang R, Chen X, Lei W. Single-cell RNA sequencing reveals pro-invasive cancer-associated fibroblasts in hypopharyngeal squamous cell carcinoma. Cell Commun Signal. 2023;21:292
6. Ahn D, Kim JH, Sohn JH, Sin CM, Lee JE. Laryngeal preservation in stage III/IV resectable laryngo-hypopharyngeal squamous cell carcinoma following concurrent chemoradiotherapy with capecitabine/cisplatin. Mol Clin Oncol. 2013;1:685-91
7. Piazza C, Paderno A, Ravanelli M, Pessina C. Clinical and Radiological Evaluation of Hypopharyngeal Carcinoma. Adv Otorhinolaryngol. 2019;83:35-46
8. Li XY, Shen Y, Zhang L, Guo X, Wu J. Understanding initiation and progression of hepatocellular carcinoma through single cell sequencing. Biochim Biophys Acta Rev Cancer. 2022;1877:188720
9. Tang F, Li Y, Pan M, Wang Z, Lu T, Liu C. et al. HSP90AA1 promotes lymphatic metastasis of hypopharyngeal squamous cell carcinoma by regulating epithelial-mesenchymal transition. Oncol Res. 2023;31:787-803
10. Park YM, Kim DH, Kang MS, Lim JY, Koh YW, Kim SH. Transoral robotic surgery in patients with stage III/IV hypopharyngeal squamous cell carcinoma: treatment outcome and prognostic factor. Acta Otolaryngol. 2019;139:816-22
11. Zhang X, Zhang Y, Yu X, Sun Y, Miao S, Liu S. et al. Different Primary Sites of Hypopharyngeal Cancer Have Different Lymph Node Metastasis Patterns: A Retrospective Analysis From Multi-Center Data. Front Oncol. 2021;11:727991
12. Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauss A. et al. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front Immunol. 2020;11:1280
13. Jang TH, Huang WC, Tung SL, Lin SC, Chen PM, Cho CY. et al. MicroRNA-485-5p targets keratin 17 to regulate oral cancer stemness and chemoresistance via the integrin/FAK/Src/ERK/beta-catenin pathway. J Biomed Sci. 2022;29:42
14. Saito S, Ozawa H, Imanishi Y, Sekimizu M, Watanabe Y, Ito F. et al. Cyclooxygenase-2 expression is associated with chemoresistance through cancer stemness property in hypopharyngeal carcinoma. Oncol Lett. 2021;22:533
15. Sassenberg M, Droop J, Schulz WA, Dietrich D, Loick SM, Wiek C. et al. Upregulation of the long non-coding RNA CASC9 as a biomarker for squamous cell carcinoma. BMC Cancer. 2019;19:806
16. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
17. Luo Y, Huang S, Wei J, Zhou H, Wang W, Yang J. et al. Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/beta-catenin-TFE3 feedback loop signalling. Clin Transl Med. 2022;12:e752
18. Xu Z, Zhang M, Guo Z, Chen L, Yang X, Li X. et al. Stemness-related lncRNAs signature as a biologic prognostic model for head and neck squamous cell carcinoma. Apoptosis. 2023;28:860-80
19. Chen Z, Hu Z, Sui Q, Huang Y, Zhao M, Li M. et al. LncRNA FAM83A-AS1 facilitates tumor proliferation and the migration via the HIF-1alpha/ glycolysis axis in lung adenocarcinoma. Int J Biol Sci. 2022;18:522-35
20. He Y, Xu Y, Yu X, Sun Z, Guo W. The Vital Roles of LINC00662 in Human Cancers. Front Cell Dev Biol. 2021;9:711352
21. Zhong L, Hao P, Zhang Q, Jiang T, Li H, Xiao J. et al. Revised International Staging System (R-ISS) stage-dependent analysis uncovers oncogenes and potential immunotherapeutic targets in multiple myeloma (MM). Elife. 2022 11
22. An C, Hu Z, Li Y, Zhao P, Liu R, Zhang Q. et al. LINC00662 enhances cell progression and stemness in breast cancer by MiR-144-3p/SOX2 axis. Cancer Cell Int. 2022;22:184
23. Liang B, Wang X, Song X, Bai R, Yang H, Yang Z. et al. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:929-38
24. Liu X, Zhao W, Wang X. Inhibition of long non-coding RNA MALAT1 elevates microRNA-429 to suppress the progression of hypopharyngeal squamous cell carcinoma by reducing ZEB1. Life Sci. 2020;262:118480
25. Zhuang J, Shen L, Li M, Sun J, Hao J, Li J. et al. Cancer-Associated Fibroblast-Derived miR-146a-5p Generates a Niche That Promotes Bladder Cancer Stemness and Chemoresistance. Cancer Res. 2023;83:1611-27
26. Munoz-Galvan S, Felipe-Abrio B, Verdugo-Sivianes EM, Perez M, Jimenez-Garcia MP, Suarez-Martinez E. et al. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the Hippo pathway and increasing the stemness. Mol Cancer. 2020;19:7
27. Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond). 2021;41:199-217
28. Lin Z, Liu J. lncRNA DQ786243 promotes hepatocellular carcinoma cell invasion and proliferation by regulating the miR-15b-5p/Wnt3A axis. Mol Med Rep. 2021 23
29. Zhao SY, Wang Z, Wu XB, Zhang S, Chen Q, Wang DD. et al. CERS6-AS1 contributes to the malignant phenotypes of colorectal cancer cells by interacting with miR-15b-5p to regulate SPTBN2. Kaohsiung J Med Sci. 2022;38:403-14
30. Xu J, Zhang J, Shan F, Wen J, Wang Y. SSTR5-AS1 functions as a ceRNA to regulate CA2 by sponging miR-15b-5p for the development and prognosis of HBV-related hepatocellular carcinoma. Mol Med Rep. 2019;20:5021-31
31. Zou Y, Lin X, Bu J, Lin Z, Chen Y, Qiu Y. et al. Timeless-Stimulated miR-5188-FOXO1/beta-Catenin-c-Jun Feedback Loop Promotes Stemness via Ubiquitination of beta-Catenin in Breast Cancer. Mol Ther. 2020;28:313-27
32. Shin KH, Kim RH. An Updated Review of Oral Cancer Stem Cells and Their Stemness Regulation. Crit Rev Oncog. 2018;23:189-200
33. Wei J, Wang L, Sun Y, Bao Y. LINC00662 contributes to the progression and the radioresistance of cervical cancer by regulating miR-497-5p and CDC25A. Cell Biochem Funct. 2020;38:1139-51
34. Liu Z, Yao Y, Huang S, Li L, Jiang B, Guo H. et al. LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505:843-9
35. Yu M, Lu W, Cao Z, Xuan T. LncRNA LINC00662 Exerts an Oncogenic Effect on Osteosarcoma by the miR-16-5p/ITPR1 Axis. J Oncol. 2021;2021:8493431
36. Yuan L, Zhang P, Lu Y, Zhang A, Chen X. LINC00662 Promotes Proliferation and Invasion and Inhibits Apoptosis of Glioma Cells Through miR-483-3p/SOX3 Axis. Appl Biochem Biotechnol. 2022;194:2857-71
37. Jing L, Lan L, Mingxin Z, Zhaofeng Z. METTL3/LINC00662/miR-186-5p feedback loop regulates docetaxel resistance in triple negative breast cancer. Sci Rep. 2022;12:16715
38. Liu Y, Qiu S, Zheng X, Qiu Y, Yao S, Ge Y. et al. LINC00662 modulates cervical cancer cell proliferation, invasion, and apoptosis via sponging miR-103a-3p and upregulating PDK4. Mol Carcinog. 2021;60:365-76
39. Yao Y, Liu Y, Jin F, Meng Z. LINC00662 Promotes Oral Squamous Cell Carcinoma Cell Growth and Metastasis through miR-144-3p/EZH2 Axis. Yonsei Med J. 2021;62:640-9
40. Zhang Z, Liang X, Ren L, Zhang S, Li S, Wan T. et al. LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR-340-5p and upregulating HOXB2. Thorac Cancer. 2020;11:2306-15
41. Dong Y, Zhang N, Zhao S, Chen X, Li F, Tao X. miR-221-3p and miR-15b-5p promote cell proliferation and invasion by targeting Axin2 in liver cancer. Oncol Lett. 2019;18:6491-500
42. Wu B, Liu G, Jin Y, Yang T, Zhang D, Ding L. et al. miR-15b-5p Promotes Growth and Metastasis in Breast Cancer by Targeting HPSE2. Front Oncol. 2020;10:108
43. Asai S, Goto Y, Tanigawa K, Tomioka Y, Kato M, Mizuno K. et al. MiR-15b-5p inhibits castration-resistant growth of prostate cancer cells by targeting the muscarinic cholinergic receptor CHRM3. FEBS Lett. 2023;597:1164-75
44. Ahmad P, Sana J, Slavik M, Gurin D, Radova L, Gablo NA. et al. MicroRNA-15b-5p Predicts Locoregional Relapse in Head and Neck Carcinoma Patients Treated With Intensity-modulated Radiotherapy. Cancer Genomics Proteomics. 2019;16:139-46
45. Cai Y, Yang Y, Zhang X, Ma Q, Li M. TRPM2-AS promotes the malignancy of osteosarcoma cells by targeting miR-15b-5p/PPM1D axis. Cell Cycle. 2022;21:835-50
46. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Jamal HH, Taheri M, Hajiesmaeili M. A Comprehensive Review on Function of miR-15b-5p in Malignant and Non-Malignant Disorders. Front Oncol. 2022;12:870996
47. Guo K, Qi D, Huang B. LncRNA MEG8 promotes NSCLC progression by modulating the miR-15a-5p-miR-15b-5p/PSAT1 axis. Cancer Cell Int. 2021;21:84
48. Zhu Y, Zhang X, Wang L, Zhu X, Xia Z, Xu L. et al. FENDRR suppresses cervical cancer proliferation and invasion by targeting miR-15a/b-5p and regulating TUBA1A expression. Cancer Cell Int. 2020;20:152
49. Miao S, Wang J, Xuan L, Liu X. LncRNA TTN-AS1 acts as sponge for miR-15b-5p to regulate FBXW7 expression in ovarian cancer. Biofactors. 2020;46:600-7
50. Paul R, Dorsey JF, Fan Y. Cell plasticity, senescence, and quiescence in cancer stem cells: Biological and therapeutic implications. Pharmacol Ther. 2022;231:107985
51. Papadaki S, Magklara A. Regulation of Metabolic Plasticity in Cancer Stem Cells and Implications in Cancer Therapy. Cancers (Basel). 2022 14
52. Barzegar Behrooz A, Syahir A, Ahmad S. CD133: beyond a cancer stem cell biomarker. J Drug Target. 2019;27:257-69
53. Hassn Mesrati M, Syafruddin SE, Mohtar MA, Syahir A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules. 2021 11
54. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947-56
55. Lee S, Wottrich S, Bonavida B. Crosstalks between Raf-kinase inhibitor protein and cancer stem cell transcription factors (Oct4, KLF4, Sox2, Nanog). Tumour Biol. 2017;39:1010428317692253
56. Martin CE, Nguyen A, Kang MK, Kim RH, Park NH, Shin KH. DYRK1A is required for maintenance of cancer stemness, contributing to tumorigenic potential in oral/oropharyngeal squamous cell carcinoma. Exp Cell Res. 2021;405:112656
57. Lee SH, Kieu C, Martin CE, Han J, Chen W, Kim JS. et al. NFATc3 plays an oncogenic role in oral/oropharyngeal squamous cell carcinomas by promoting cancer stemness via expression of OCT4. Oncotarget. 2019;10:2306-19
Author contact
![]() Corresponding author: Qing Ye, Shengli Clinical Medical College of Fujian Medical University, Fuzhou 350001, China. Email: zbhedu.cn.
Corresponding author: Qing Ye, Shengli Clinical Medical College of Fujian Medical University, Fuzhou 350001, China. Email: zbhedu.cn.

 Global reach, higher impact
Global reach, higher impact