Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(14):4467-4476. doi:10.7150/jca.84832 This issue Cite
Research Paper
Genomic Alterations Correlated to Trastuzumab Resistance and Clinical Outcomes in HER2+/HR- Breast Cancers of Patients Living in Northwestern China
1. Department of Surgical Oncology, Shaanxi Provincial People's Hospital, Xi'an, China.
2. Department of Pathology, Shaanxi Provincial People's Hospital, Xi'an, China.
Received 2024-3-31; Accepted 2024-5-27; Published 2024-6-17
Abstract
Anti-HER2 therapy has significantly improved the survival rates of patients with HER2+ breast cancer. However, a subset of these patients eventually experience treatment failure, and the underlying genetic mechanisms remain largely unexplored. This underscores the need to investigate the genomic heterogeneity of HER2+ breast cancer. In this study, we focus on HER2+/HR- breast cancer, as it differs from HER2+/HR+ breast cancer in terms of genetic and biological characteristics. We performed gene-targeted genome sequencing on 45 HER2+/HR- breast cancer samples and identified 650 mutations across 268 cancer-related genes. TP53 (71.1%) and PIK3CA (35.6%) were the most frequently mutated genes in our sample. Additionally, ERBB2 (77.8%), CDK12 (42.2%), and MYC (11.1%) exhibited a high frequency of copy number amplifications (CNAs). Comparative analysis with two other HER2+/HR- breast cancer cohorts revealed that our cohort had higher genetic variation rates in ARID1A, PKHD1, PTPN13, FANCA, SETD2, BRCA2, BLM, STAG2, FAT1, TOP2A, POLE, ATM, KMT2B, FGFR4, and EPAS1. Notably, in our cohort, NF1 and ATM mutations were more prevalent in trastuzumab-resistant patients (NF1, p=0.016; ATM, p=0.006) and were associated with primary trastuzumab resistance (NF1, p=0.042; ATM, p=0.021). Moreover, patients with NF1 mutations (p=0.009) and high histological grades (p=0.028) were more likely to experience early relapse. Ultimately, we identified a unique cancer-related gene mutation profile and a subset of genes associated with primary resistance to trastuzumab and RFS in patients with HER2+/HR- breast cancer in Northwest China. These findings could lay the groundwork for future studies aimed at elucidating the mechanisms of resistance to trastuzumab and improving HER2-targeted treatment strategies.
Keywords: Genomic profiling, Breast cancer, HER2 positive, Trastuzumab resistance
Introduction
Breast cancer is the most prevalent cancer among women. There are approximately 306,000 new cases of female breast cancer reported in China annually, leading to approximately 71,700 deaths [1]. Roughly a quarter of patients with breast cancer exhibit HER2 (also known as ERBB2) gene amplification and therefore require anti-HER2 therapy [2]. The introduction of anti-HER2 medications, including monoclonal antibodies, tyrosine kinase inhibitors, and antibody-drug conjugates, has significantly improved survival rates for patients with HER2-positive breast cancer [3]. However, approximately 20% of patients with HER2-positive breast cancer do not respond to trastuzumab initially, and about 70% of patients with metastatic disease who receive trastuzumab are resistant to treatment [4-7], underscoring the importance of investigating the genetic intricacies of HER2-positive breast cancer due to its heterogeneity. It is recognized that HER2-positive/hormone receptor-positive (hereinafter referred to as HER2+/HR+) and HER2-positive/hormone receptor-negative (hereinafter referred to as HER2+/HR-) breast cancers are biologically and genetically different [8]. To eliminate the effect of the estrogen receptor pathway on HER2-positive breast cancer, this study focuses on HER2+/HR- breast cancer. We employed a gene-targeted genome sequencing approach, analyzing 425 cancer genomes, to explore the relationship between known cancer-associated genes and HER2+/HR- breast cancer in northwestern China.
Materials and Methods
Patients and sample collection
Forty-five women diagnosed with HER2+/HR- breast cancer at Shaanxi Province Hospital between January 2016 and December 2018 were included in this study. Clinical and pathological data were collected from each patient, including age, menopausal status, TNM stage based on the AJCC 7th edition [9], lymph node status, estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 status, and Ki67 index. Two experienced pathologists determined the immunohistological results. When the HER2 result was 2+, FISH detection was performed to confirm the HER2 status [10]. All patients underwent surgery, followed by standard adjuvant regimens containing doxorubicin and paclitaxel. Additionally, all patients received 1-year standard trastuzumab treatment and were followed-up for a median of 36 months. This study was approved by the Ethics Committee of Shaanxi Provincial People's Hospital and was conducted in accordance with the principles of the Declaration of Helsinki.
Sequencing and bioinformatics analysis
Formalin-fixed, paraffin-embedded (FFPE) tumor tissue blocks of 45 patients with HER2+/HR- breast cancer were obtained from the Pathology Department of Shaanxi Province People's Hospital. Pathologists verified the tumor purity for each sample. The blocks were de-paraffinized using xylene and genomic DNA was extracted using the QIAamp DNA FFPE Tissue kit (Qiagen). The extracted genomic DNA was quantified with Nanodrop2000 (Thermo Fisher Scientific, Waltham, MA), and further assessed using a dsDNA HS Assay kit on a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA). Sequencing libraries were prepared with the KAPA Hyper Prep kit (KAPA Biosystems, Wilmington, MA, USA), following previously described methods [11]. Hybridization-based targeted gene capture utilized the Geneseeq pan-cancer gene panel (425 cancer-relevant genes, Supplementary Table 1), and the targeted libraries were sequenced on an Illumina HiSeq4000 NGS platform, achieving a mean coverage depth exceeding 300×. Quality trimming was performed with Trimmomatic [12] to remove bases of quality lower than 15 or N bases, followed by mapping to the hg19 genome (Human Genome version 19) using the Burrows-Wheeler Aligner. Local realignments around indels and base quality control, along with germline mutation detection, were conducted using the Genome Analysis Toolkit (GATK3.4.0). Somatic SNPs and indels were identified with VarScan2 [13] and HaplotypeCaller/UnifiedGenotyper in GATK. The mutant allele frequency (MAF) cut-off was set at 0.1% for samples, and a minimum of three unique mutant reads was needed. Common SNPs were filtered out using dbSNP (v137) and the 1000 Genome database. Gene fusions were detected with FACTERA software [14]. The copy number variations (CNVs) were analyzed using ADTEx, with a log2 ratio cut-off of 2.0 for copy number gain in samples.
Statistical methods
Categorical variables are represented by frequency and percentage, while quantitative variables are presented as mean ± SEAM. To compare categorical variables, the chi-square or Fisher's exact test was employed, and logistic regression analysis was utilized to examine potential predictive factors for trastuzumab resistance. The log-rank test and Cox regression analysis were applied to assess prognostic factors for recurrence-free survival (RFS) (SPSS 18.1.0 for Windows, SPSS Inc., Chicago, IL, USA). Survival curves were generated with GraphPad Prism8. All statistical tests were two-sided, with statistical significance established at P< 0.05.
Results
Clinical characteristics
We performed specific gene sequencing on FFPE tumor samples from 45 patients with HER2+/HR- breast cancer, whose detailed clinical characteristics are presented in Table 1. These patients were classified based on their menopausal status, TNM stage, lymph node stage, histological grade, Ki67 index, and follow-up outcomes. The median age of the cohort was 53 years with a range of 26-80 years. Among the 45 patients, 62.2% were postmenopausal, 71.1% were at TNM II stage, 75.5% had N1 lymph node involvement, 37.8% were grade 3, and 57.8% had a Ki67 index over 30%. About 33.3% (15/45) experienced recurrence within the median follow-up period of 36 months, with 66.7% (10/15) of these recurrences occurring within 24 months post-operation.
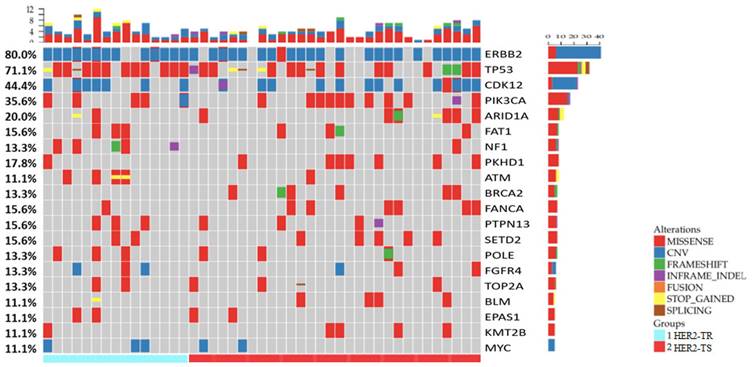
Summary of the somatic mutations and Indels of all 45 patients with HER2+/HR- breast cancer in Northwestern China. (HER2-TR = HER2+/HR- breast cancer with primary trastuzumab-resistant status, HER2-TS = HER2+/HR- breast cancer with tastuzumab sensitive status).
Clinicopathological features of the HER2+/HR- breast cancer cohort in Northwestern China.
| Clinicopathological features | Total cohort, n=45 (%) |
|---|---|
| Age (median, range) | 53.0 (26-80) |
| Menopausal status | |
| Premenopausal | 17 (37.8%) |
| Postmenopausal | 28 (62.2%) |
| pTNM stage | |
| I stage | 0 (0.0%) |
| II stage | 32 (71.1%) |
| III stage | 13 (28.9%) |
| Lymph node stage | |
| N0 | 0 (0.0%) |
| N1 | 34 (75.5%) |
| N2 | 11 (24.5%) |
| N3 | 0 (0.0%) |
| Histological grade | |
| 1 | 0 (0.0%) |
| 2 | 28 (62.2%) |
| 3 | 17 (37.8%) |
| Ki 67 index | |
| ≤30% | 19 (42.2%) |
| >30% | 26 (57.8%) |
| Recurrence | |
| Yes | 15 (33.3%) |
| RFS≤24 months | 10 (22.2%) |
| RFS>24 months | 5 (11.1%) |
| No | 30 (66.7%) |
The mutation spectrum of northwest Chinese patients with HER2+/HR- breast cancer
In total, 650 mutations were identified, including 458 single nucleotide variations (SNVs), 38 insertion-deletion variants with 25 frameshift variants (indels), 22 nonsense variants, 21 splice site variants, 96 copy number amplifications (CNA), and 13 structural variations (SV), spanning 268 genes out of the 425 genes listed. Among these 268 genes, 17 were frequently mutated, exhibiting mutation rates greater than 10%. These included: TP53 (71.1%), PIK3CA (35.6%), ARID1A (20.0%), PKHD1 (17.8%), FAT1 (15.6%), FANCA (15.6%), PTPN13 (15.6%), SETD2 (15.6%), NF1 (13.3%), POLE (13.3%), TOP2A (13.3%), BRCA2 (13.3%), ATM (11.1%), BLM (11.1%), EPAS1 (11.1%), KMT2B (11.1%), and ERBB2 (11.1%). In addition, genes with high-level CNA exceeding 10% included ERBB2 (77.8%, fold change 2-25), CDK12 (42.2%, fold change 2-9), and MYC (11.1%, fold change 1.6-24). A summary of the gene alterations in our cohort is depicted in Figure 1.
To investigate the variances in gene alterations in HER2+/HR- breast cancer among Chinese and Western women, we selected patients with HER2+/HR- breast cancer based on the pathological index from the published databases TCGA PanCancer Atlas [15] and METABRIC [16], which included 78 and 110 HER2+/HR- breast cancer cases, respectively (www.cBioportal.org). We compared the mutated gene profiles of our cohort with those from these two databases, and the 25 most frequently mutated genes across the three cohorts are depicted in Figure 2a. For the two most commonly mutated genes, TP53 and PIK3CA, there were no significant differences in mutation rates across the three cohorts (TP53: NW China 71.1% vs. TCGA 70.5% vs. METABRIC 79.1%, p>0.05; PIK3CA: NW China 35.6% vs. TCGA 33.3% vs. METABRIC 38.2%, p>0.05). Furthermore, the comparison revealed differences between our cohort and the two published cohorts. Several genes, including ARID1A, PKHD1, PTPN13, FANCA, SETD2, BRCA2, BLM, STAG2, FAT1, TOP2A, POLE, ATM, KMT2B, FGFR4, and EPAS1, were found to be more frequently mutated in our cohort than in the two published cohorts, suggesting that Chinese women with HER2-positive breast cancer exhibit a distinct gene mutation spectrum compared to Western women.
Comparison of gene mutations and CNAs between our cohort and the two published HER2+/HR- breast cancer cohorts (TCGA PanCancer Atlas and METABRIC): (a) Comparison of the top twenty mutated genes among the three cohorts (the blue, yellow, and green colors represent the top twenty mutated genes of the three cohorts individually); (b) Comparison of top ten CNAs among the three cohorts (the blue, yellow, and green colors indicate the top ten CNAs of the three cohorts individually).
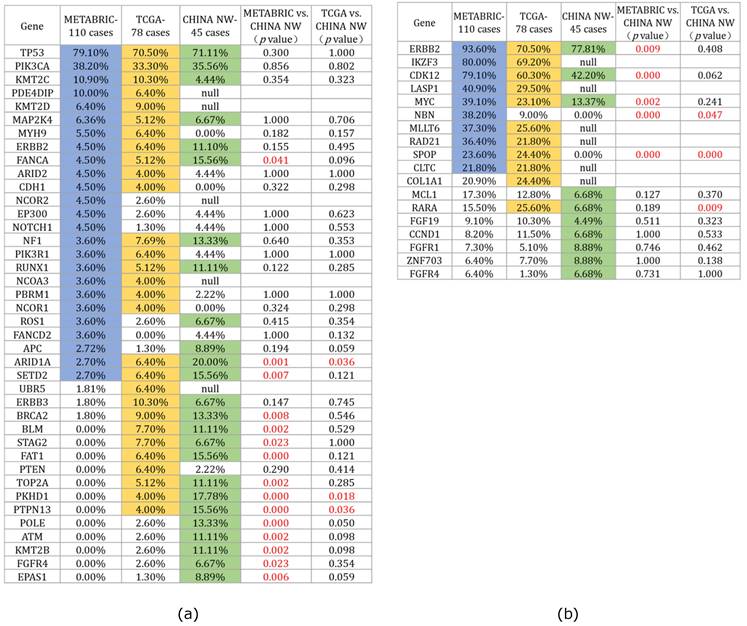
We also compared the top 10 CNAs among the three HER2-positive breast cancer cohorts (Figure 2b). Statistical results revealed that ERBB2 was the most frequently amplified gene across all three cohorts (NW China 77.81%, TCGA 70.50%, METABRIC 93.61%), although the ERBB2 amplification rate in our cohort was lower than that in the METABRIC cohort (p=0.009). Regarding the CNAs of the CDK12 and MYC genes, our cohort exhibited a lower rate compared to the METABRIC cohort (CDK12: NW China 40.4% vs. METABRIC 79.1%, p=0.000; MYC: NW China 13.37% vs. METABRIC 39.10%, p=0.002).
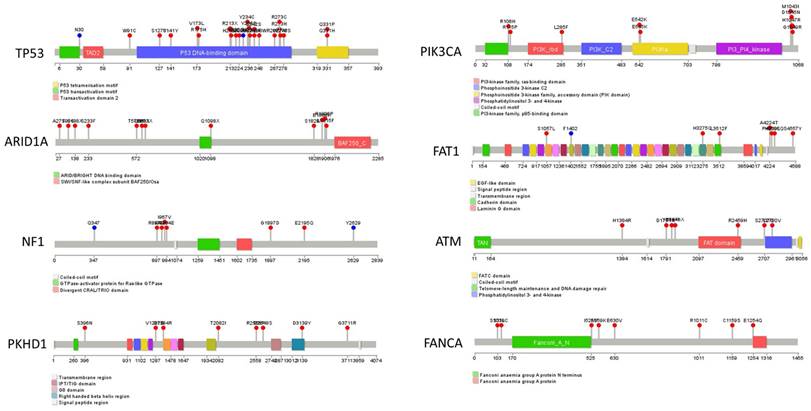
The hotspot mutations in our cohort's top eight mutated genes, including TP53, PIK3CA, ARID1A, FAT1, NF1, ATM, PKHD1, and FANCA, are illustrated as lollipops in Figure 3. Furthermore, the hotspot sites of the two most mutated genes, PIK3CA and P53, were compared between our cohort and the two previously mentioned cohorts (Supplementary Figure 1). This comparison revealed that the most frequently mutated site of PIK3CA gene in our cohort, H1047R, was also the most frequently mutated site of PIK3CA in the TCGA and METABRIC cohorts. Additionally, the mutation spectrum of the TP53 gene in our cohort showed similarities with these two cohorts.
Pathway enrichment of gene alteration and gene association and mutual exclusion analysis in HER2+/HR- breast cancer
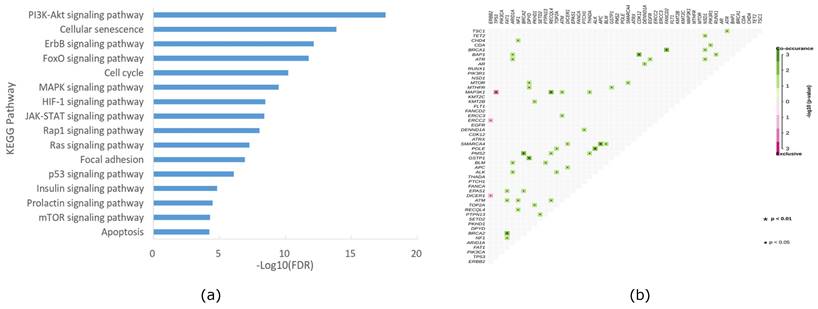
To investigate whether the altered genes in our cohort could be clustered into different pathways related to cancer, we conducted pathway analysis using KEGG. As depicted in Figure 4a, signal transduction-related pathways (including PI3K-AKT, Foxo, ErbB, MAPK, JAK-STAT, HIF-1, RAS, insulin, Hippo, AMPK, and VEGF signaling pathways), cell growth and death-related pathways (including cellular senescence, cell cycle, and apoptosis signaling pathways), and cell movement-related pathways (including adherent junction and focal adhesion signaling pathways) were the predominant enriched pathways. Notably, downstream of the HER2 pathway, PI3K-AKT signaling emerged as the most dominant enriched pathway in our cohort, aligning with previous studies [17]. Furthermore, we conducted co-occurrence and mutual exclusion analyses of altered genes in our cohort, with the results presented in Figure 4b.
Summary of hot spot mutations of eight highly mutated genes (Containing TP53, PIK3CA, ARID1A, FAT1, NF1, ATM, PKHD1, and FANCA) in the HER2+/HR- breast cancer cohort of Northwestern China. The mutations are shown in the lollipop graphs.
KEGG enrichment of mutation pathway(a) and gene association and mutual exclusion (b) in the HER2+/HR- breast cancer cohort of Northwestern China.
Correlation of altered genes with primary trastuzumab resistance in HER2+/HR- breast cancer
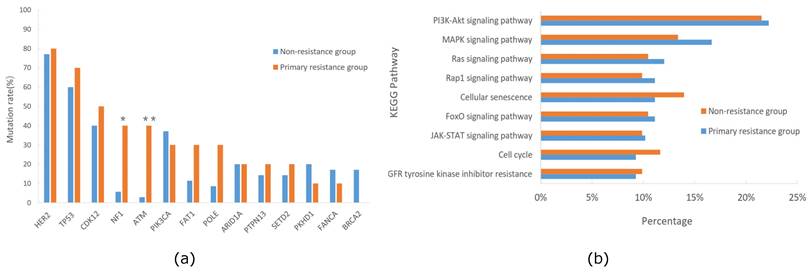
Some patients with HER2-positive breast cancer experienced relapse during or within 12 months after completing 1-year course of trastuzumab treatment, a condition deemed as primary trastuzumab resistance according to prior studies [18-20]. All patients in our cohort received standard adjuvant trastuzumab treatment for one-year post-operation, and some patients experienced relapse with a short interval. The follow-up results showed that 22.2% of patients relapsed within 24 months post-operation, suggesting primary resistance to trastuzumab. Consequently, we categorized our cohort into primary trastuzumab-resistant and non-resistant groups based on whether they relapsed within 24 months post-operation, and then compared the gene mutation profiles between these two groups. The analysis indicated significantly higher mutation frequencies of NF1 and ATM in the primary trastuzumab-resistant group compared to the non-resistant group (NF1, 40.0% vs. 5.7%, p=0.016; ATM, 40.0% vs. 2.8%, p=0.006), unlike mutations in PIK3CA or ERBB2, which were associated with trastuzumab resistance in other studies [21-23] (Figure 5a). Additionally, we conducted KEGG pathway enrichment analyses for both groups, revealing the PI3K-AKT signaling pathway as prevalent in both, while the MAPK signaling pathway was more prominent in the primary trastuzumab-resistant group, though without statistical significance (Figure 5b).
Comparison of mutation genes(a)and KEGG pathway(b) between primary trastuzumab-resistant and non-resistance groups of the HER2+/HR- breast cancer cohort in Northwestern China.
Binary logistic regression analysis of factors related to primary trastuzumab resistance in the HER2+/HR- breast cancer cohort of Northwestern China.
| Parameter | Univariable analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| TNM stage III vs.I-II | 3.375 | 0.776 to14.669 | 0.105 | |||
| Lymph node stage N2-N3 vs. N0-N1 | 4.833 | 1.057 to 22.091 | 0.042 | |||
| FAT1 gene Mutation vs. WT | 3.321 | 0.603 to 18.307 | 0.168 | |||
| NF1 gene Mutation vs. WT | 11.000 | 1.633 to 74.083 | 0.014 | 9.004 | 1.078 to 75.192 | 0.042 |
| ATM gene Mutation vs. WT | 22.667 | 2.147 to 239.32 | 0.009 | 18.919 | 1.599 to 229.630 | 0.021 |
| POLE gene Mutation vs. WT | 4.571 | 0.758 to 27.577 | 0.097 | |||
To investigate the factors responsible for primary trastuzumab resistance in our cohort, we conducted logistic regression analysis on potential factors, including six clinical characteristics and ten significantly altered genes. The univariable regression analysis revealed that three factors: lymph node stage (p=0.042), mutated NF1 (p=0.014), and mutated ATM (p=0.009), might be associated with trastuzumab resistance. Subsequently, these three factors, along with others with p-values <0.2 in the univariable analysis, such as TNM stage, mutated FAT1, and mutated POLE, were subjected to the multivariable analysis. The analysis determined that only mutated NF1 (p=0.042) and mutated ATM (p=0.021) were statistically significant (Table 2), suggesting a link between NF1 and ATM mutations and primary trastuzumab resistance in our cohort.
Analysis of the predictive factor for the recurrence-free survival of HER2+/HR- breast cancer
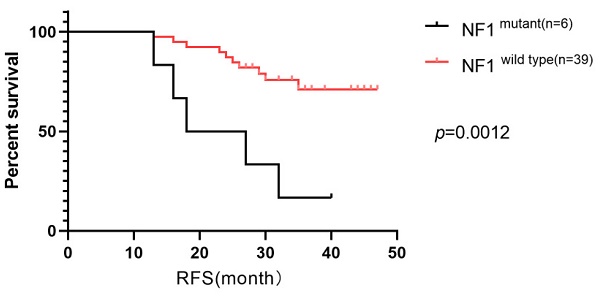
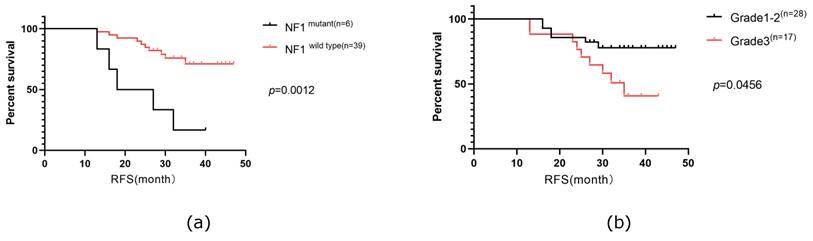
RFS in our cohort was followed up for a median time of 36 months, during which 33.3% of the patients experienced relapse or progression. Consequently, we investigated the prognostic factors for RFS. Our analysis included six clinical characteristics (age, menopausal status, TNM stage, lymph node stage, histologic grade, and Ki67 index) and ten of the most frequently altered genes (PIK3CA, TP53, ARID1A, FAT1, NF1, PKHD1, ATM, FANCA, ERBB2, and CDK12) in a Cox univariable regression analysis. This analysis identified four significant factors: histological grade (p=0.048), Ki67 index (p=0.035), mutated NF1 (p=0.001), and mutated ATM (p=0.003). These factors, along with three additional factors with p-values were <0.2 (TNM stage (p=0.136), lymph node stage (p=0.159), mutated ERBB2 (p=0.152), were further analyzed in a Cox multivariate regression analysis. Ultimately, mutated NF1 (p=0.009) and histological grade (p=0.028) emerged as independent prognostic factors for RFS in patients with HER2+/HR- breast cancer in our cohort (Table 3). Survival curves were generated using the Kaplan-Meier method (Figure 6).
Univariable and multivariable analysis of factors related to the RFS of the patients with HER2+/HR- breast cancer in Northwestern China.
| Parameter | Univariable analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| pTNM stage III vs. I-II | 2.201 | 0.780 to 6.209 | 0.136 | |||
| Lymph node stage N2-3 vs. N0-1 | 2.170 | 0.738 to 6.382 | 0.159 | |||
| Histological grade 3 vs. 1-2 | 2.835 | 1.007 to 7.980 | 0.048 | 3.613 | 1.145 to 11.394 | 0.028 |
| Ki67 index High vs. Low-Medium | 3.907 | 1.099 to 13.887 | 0.035 | |||
| NF1 gene Mutation vs. WT | 6.270 | 2.096 to 18.758 | 0.001 | 5.965 | 1.550 to 22.957 | 0.009 |
| ATM gene Mutation vs. WT | 5.961 | 1.826 to 19.455 | 0.003 | |||
| HER2 gene Mutation vs. WT | 2.534 | 0.709 to 9.054 | 0.152 | |||
The Kaplan-Meier curves of mutated NF1 gene (a)and histological grade status(b) on RFS of the patients with HER2+/HR- breast cancer in Northwestern China.
Discussion
Activation of tumor-associated signaling pathways mediated by the amplified HER2 gene is involved in 15%-20% of breast cancers [2]. Anti-HER2 therapy, including monoclonal antibodies, small molecule kinase inhibitors, and antibody-drug conjugates, has significantly improved clinical outcomes in the HER2-positive subtype of breast cancer. However, some patients with HER2-positive disease still experience progression or relapse during or after anti-HER2 therapy, underscoring the molecular complexity of cancer and highlighting the need to explore the comprehensive molecular profile of HER2-positive breast cancer. In this study, we focus on the HER2+/HR- breast cancer in Northwestern China and aim to investigate the genomic alterations and their correlation with clinical outcomes. We identified 650 genomic alterations in 268 genes in 45 primary breast cancer tissues using a gene-targeted genome sequencing method that included 425 cancer-associated genes. The most frequently mutated genes were TP53 and PIK3CA, and the most frequently amplified genes were ERBB2, CDK12, and MYC in our cohort.
Given that all breast cancer samples in our study were obtained from Chinese women of Asian ethnicity, we were curious about potential differences in the gene mutation spectrum compared to individuals of different ethnic backgrounds. Consequently, we examined the extensive breast cancer mutation data within the TCGA database, securing 78 and 110 HER2+/HR- breast cancer cases from the PanCancer Atlas and METABRIC datasets, respectively. In comparing the top 20 mutated genes across the three cohorts, we observed that TP53 and PIK3CA remained the most frequently mutated genes in all datasets, with no significant statistical difference in mutation rates. However, our cohort exhibited a notably higher mutation rate in the FANCA, ARID1A, SETD2, BRCA2, BLM, FAT1, TOP2A, PKHD1, PTPN13, POLE, ATM, and KMT2B genes compared to the other two cohorts (Figure 2). Among these genes, several are recognized as tumor-related genes critical to the onset and progression of breast cancer, including FANCA, ARID1A, BRCA2, BLM, TOP2A, ATM, and KMT2B [24-30]. Additionally, we noted that the mutation rate in these genes exceeded 10%, a rarity in other published breast cancer mutation databases [31-33]. This suggests that HER2+/HR- breast cancer in Chinese women presents a distinct mutation spectrum from that observed in Western populations.
Although the clinical application of anti-HER2 drugs has markedly improved the survival for patients with HER2-positive breast cancer, a subset of these patients still experiences treatment failure at some stage. This failure is often attributed to primary and acquired resistance for anti-HER2 therapy. Recent researches [17, 22, 23, 34-37] suggest that anti-HER2 resistance often stems from intratumor heterogeneity, specific HER2 splicing variants, activation of the PIK3CA/AKT signaling pathway, signaling pathway crosstalk, or cell cycle sensitization.
Our study focus on patients with HER2+/HR- breast cancer, identifying a subset that experienced recurrence during or within one year following trastuzumab treatment, typically classified as primary trastuzumab resistance [20]. Additionally, we investigated genetic factors that might underlie primary trastuzumab resistance in our cohort. We discovered that mutations in the ATM and NF1 tumor suppressor genes were significantly more common in the primary trastuzumab-resistant group compared to the non-resistant group (ATM, 40.0% vs. 2.8%, p=0.006; NF1, 40.0% vs. 5.7%, p=0.016). Furthermore, these mutations in ATM and NF1 were associated with primary trastuzumab resistance in both univariate and multivariate regression analyses (Table 2).
ATM, the gene mutated in ataxia-telangiectasia (AT), regulates the DNA damage response by playing a pivotal role in identifying and repairing double-strand breaks, thereby being linked to various cancer-related processes such as tumorigenesis, progression, metabolism, drug resistance, and radiosensitivity [38-40]. Moslemi et al. [41] found that somatic ATM missense mutations are associated with a 2.8- to 3-fold increased risk of sporadic breast cancer. In addition, clinical data analysis revealed that breast cancers harboring ATM mutations are highly aggressive and exhibit reduced progression-free survival and overall survival [42]. A recent study demonstrated that ATM can maintain HER2 protein stability by forming a complex with HER2 and HSP90, thus inhibiting HER2 ubiquitination and degradation in HER2-positive breast cancer cell lines [29]. In our research, ATM mutations were linked to primary trastuzumab resistance in patients with HER2+/HR- breast cancer, indicating that ATM is involved in the anti-HER2 resistance and the underlying mechanism is worth studying further in the future.
NF1, which encodes neurofibromin and negatively regulates Ras activation by converting GTP-Ras to GDP-Ras, acts as a tumor suppressor gene [43]. The loss of neurofibromin due to NF1 mutation or deletion leads to the activation of Ras-Raf-MEK-ERK/Ras-PI3K-Akt signaling pathways, playing a significant role in the initiation and progression of various tumors [44-46]. Somatic mutations of NF1 are frequently observed in many tumors, correlating with tumorigenesis and progression [44, 47-50]. In breast cancer, NF1 mutations are commonly identified through large-scale sequencing, and these mutations are associated with endocrine therapy resistance and poor outcomes [51-53]. Recent studies have shown that NF1 mutation can induce acquired resistance to anti-HER2 therapy in HER2-positive breast cancer. It has been demonstrated that the loss of NF1, mediated by mutation, enables HER2-positive breast cancer to adapt to HER2-targeted treatment by shifting from PI3K/AKT signaling pathway to MAPK signaling pathway [54-56]. In our study, we found that mutated NF1 was correlated with primary trastuzumab resistance and poor RFS in patients with HER2+/HR- breast cancer in Northwest China. Additionally, we conducted pathway enrichment analysis and identified a predominant enrichment of mutations in the MAPK signaling pathway among the primary trastuzumab-resistant patient group. Taken together, these alterations suggest a direct contribution to anti-HER2 resistance, warranting further investigation into the underlying mechanisms.
While our study has yielded some notable results, it is not devoid of limitations. First, the small number of patients enrolled limits the representativeness of the mutation spectrum of HER2+/HR- breast cancer in China. Second, our cohort's follow-up period, which spans only three years of relapse-free survival data, is insufficient for long-term analysis, limiting the prognostic value of the mutated genes identified in our cohorts. Third, the relationship between mutated genes and primary trastuzumab resistance in patients with HER2+/HR- breast cancer in China necessitates further exploration.
Conclusions
In conclusion, our study revealed a distinct mutation landscape in Northwestern Chinese patients with HER2+/HR- breast cancer, differing from the mutation profiles reported in Western HER2+/HR- breast cancer cohorts. Additionally, our study identified mutated genes associated with primary trastuzumab resistance and RSF in patients with HER2+/HR- breast cancer, which offers insights into improving the efficacy of anti-HER2 therapy for patients with HER2+/HR- breast cancer in China.
Supplementary Material
Supplementary figure and table.
Acknowledgements
Funding
This research was funded by Shaanxi Provincial Natural Science Basic Research Program (2019JM-430), Shaanxi Provincial Science and Technology Plan (2022K076), Shaanxi Provincial Key Technology Innovation Team Plan (2014KCT-24), Shaanxi Provincial Key Research and Development Plan (2019ZDLSF03-05), and Shaanxi Province Clinical Medicine Research Centre Project (S2021-0-ZC-LCZX-0002).
Author contributions
Gang Ma performed the experiments, and prepared the primary manuscript. Binliang Huo and Yanwei Shen provided technical support. Xulong Zhu and Chong Cheng analyzed the data. Wensheng Li performed and analyzed the histopathological and immunofluorescence studies. Wei Cao and Jianhui Li conceived the project, designed and supervised this study, and prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional review board statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Shaanxi Provincial People's Hospital (protocol code 2015009 and date of approval is 1 December 2015).” for studies involving humans.
Informed consent statement
Patient consent was waived as written informed consent had been obtained from the patient(s) when undergoing surgeries in the hospital.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R. et al. Cancer incidence and mortality in China, 2016. Journal of the National Cancer Center. 2022;2:1-9
2. Loibl S, Gianni L. HER2-positive breast cancer. The Lancet. 2017;389:2415-29
3. Thanopoulou E, Khader L, Caira M, Wardley A, Ettl J, Miglietta F. et al. Therapeutic strategies for the management of hormone receptor-positive, human epidermal growth factor receptor 2-Positive (HR+/HER2+) breast cancer: A review of the current literature. Cancers. 2020;12:3317
4. Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS. et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: Current knowledge, new research directions and therapeutic perspectives. Critical reviews in oncology/hematology. 2019;139:53-66
5. Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118-45
6. Harris LN, You F, Schnitt SJ, Witkiewicz A, Lu X, Sgroi D. et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin Cancer Res. 2007;13:1198-207
7. Wilken JA, Maihle NJ. Primary trastuzumab resistance: new tricks for an old drug. Ann N Y Acad Sci. 2010;1210:53-65
8. Lee HJ, Park IA, Park SY, Seo AN, Lim B, Chai Y. et al. Two histopathologically different diseases: hormone receptor-positive and hormone receptor-negative tumors in HER2-positive breast cancer. Breast Cancer Research and Treatment. 2014;145:615-23
9. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Annals of Surgical Oncology. 2010;17:1471-4
10. Muller KE, Marotti JD, Memoli VA, Wells WA, Tafe LJ. Impact of the 2013 ASCO/CAP HER2 Guideline Updates at an Academic Medical Center That Performs Primary HER2 FISH Testing: Increase in Equivocal Results and Utility of Reflex Immunohistochemistry. American journal of clinical pathology. 2015;144:247-52
11. Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X. et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018;24:3097-107
12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-20
13. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568-76
14. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M. et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390-3
15. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD. et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400-16.e11
16. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479
17. Kataoka Y, Mukohara T, Shimada H, Saijo N, Hirai M, Minami H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Annals of Oncology. 2010;21:255-62
18. Wong H, Leung R, Kwong A, Chiu J, Liang R, Swanton C. et al. Integrating molecular mechanisms and clinical evidence in the management of trastuzumab resistant or refractory HER-2⁺ metastatic breast cancer. Oncologist. 2011;16:1535-46
19. Hurvitz SA, Dalenc F, Campone M, O'Regan RM, Tjan-Heijnen VC, Gligorov J. et al. A phase 2 study of everolimus combined with trastuzumab and paclitaxel in patients with HER2-overexpressing advanced breast cancer that progressed during prior trastuzumab and taxane therapy. Breast Cancer Res Treat. 2013;141:437-46
20. Chen X, Wang J, Fan Y, Luo Y, Zhang P, Li Q. et al. Primary Trastuzumab Resistance After (Neo) adjuvant Trastuzumab-containing Treatment for Patients With HER2-positive Breast Cancer in Real-world Practice. Clinical Breast Cancer. 2021;21:191-8
21. Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M. et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18:6784-91
22. Loibl S, Majewski I, Guarneri V, Nekljudova V, Holmes E, Bria E. et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016;27:1519-25
23. Madrid-Paredes A, Cañadas-Garre M, Sánchez-Pozo A, Calleja-Hernández MÁ. De novo resistance biomarkers to anti-HER2 therapies in HER2-positive breast cancer. Pharmacogenomics. 2015;16:1411-26
24. Seal S, Barfoot R, Jayatilake H, Smith P, Renwick A, Bascombe L. et al. Evaluation of Fanconi Anemia Genes in Familial Breast Cancer Predisposition. Cancer Research. 2003;63:8596-9
25. Mamo A, Cavallone L, Tuzmen S, Chabot C, Ferrario C, Hassan S. et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2012;31:2090-100
26. Mehrgou A, Akouchekian M. The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med J Islam Repub Iran. 2016;30:369
27. Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DYH, Tothill RW. et al. Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes FANCC and BLM as Potential Breast Cancer Susceptibility Alleles. PLOS Genetics. 2012;8:e1002894
28. Järvinen T, Liu E. Topoisomerase IIα gene (TOP2A) amplification and deletion in cancer-more common than anticipated. Cytopathology. 2003;14:309-13
29. Stagni V, Manni I, Oropallo V, Mottolese M, Di Benedetto A, Piaggio G. et al. ATM kinase sustains HER2 tumorigenicity in breast cancer. Nat Commun. 2015;6:6886
30. Ghanbari M, Hosseinpour-Feizi M, Safaralizadeh R, Aghazadeh A, Montazeri V. Study of KMT2B (MLL2) gene expression changes in patients with breast cancer. Breast Cancer Management. 2019;8:BMT24
31. Ferrari A, Vincent-Salomon A, Pivot X, Sertier A-S, Thomas E, Tonon L. et al. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nature communications. 2016;7:1-9
32. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70
33. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A. et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506-19
34. Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K. et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395-402
35. Sperinde J, Jin X, Banerjee J, Penuel E, Saha A, Diedrich G. et al. Quantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res. 2010;16:4226-35
36. Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H. et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166-73
37. Hou Y, Nitta H, Wei L, Banks PM, Portier B, Parwani AV. et al. HER2 intratumoral heterogeneity is independently associated with incomplete response to anti-HER2 neoadjuvant chemotherapy in HER2-positive breast carcinoma. Breast Cancer Research and Treatment. 2017;166:447-57
38. Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351-60
39. Estiar MA, Mehdipour P. ATM in breast and brain tumors: a comprehensive review. Cancer Biol Med. 2018;15:210-27
40. Jin MH, Oh D-Y. ATM in DNA repair in cancer. Pharmacology & Therapeutics. 2019;203:107391
41. Easton DF, Pharoah PDP, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL. et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243-57
42. Bueno RC, Canevari RA, Villacis RAR, Domingues MAC, Caldeira JRF, Rocha RM. et al. ATM down-regulation is associated with poor prognosis in sporadic breast carcinomas. Annals of Oncology. 2014;25:69-75
43. Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P. et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nature genetics. 1996;12:144-8
44. Wang L-H, Wu C-F, Rajasekaran N, Shin YK. Loss of tumor suppressor gene function in human cancer: An overview. Cellular Physiology and Biochemistry. 2018;51:2647-93
45. Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Human genomics. 2017;11:1-19
46. Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170:17-33
47. Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061
48. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-75
49. Zenonos K, Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World journal of gastrointestinal oncology. 2013;5:97
50. Sangha N, Wu R, Kuick R, Powers S, Mu D, Fiander D. et al. Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations. Neoplasia. 2008;10:1362-IN9
51. Stephens P, Tarpey P, Davies H. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400-404
52. Wallace MD, Pfefferle AD, Shen L, McNairn AJ, Cerami EG, Fallon BL. et al. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192:385-96
53. Pearson A, Proszek P, Pascual J, Fribbens C, Shamsher MK, Kingston B. et al. Inactivating NF1 mutations are enriched in advanced breast cancer and contribute to endocrine therapy resistance. Clinical Cancer Research. 2020;26:608-22
54. Duso BA, Poletti C, Curigliano G, Pelicci P, Mazzarella L. Secondary mechanisms of anti-HER2 resistance in breast cancer: NF1 as an actionable target. Annals of Oncology. 2019;30:iii6-iii7
55. Wang X, Kallionpää RA, Gonzales PR, Chitale DA, Tousignant RN, Crowley JP. et al. Germline and somatic NF1 alterations are linked to increased HER2 expression in breast cancer. Cancer Prevention Research. 2018;11:655-64
56. Smith AE, Ferraro E, Safonov A, Morales CB, Lahuerta EJA, Li Q. et al. HER2+ breast cancers evade anti-HER2 therapy via a switch in driver pathway. Nature communications. 2021;12:1-10
Author contact
![]() Corresponding authors: Gang Ma (magang7010xjtu.edu.cn), Wei Cao (caowei0e1com), Jianhui Li (lijh3192com).
Corresponding authors: Gang Ma (magang7010xjtu.edu.cn), Wei Cao (caowei0e1com), Jianhui Li (lijh3192com).

 Global reach, higher impact
Global reach, higher impact