Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(16):5277-5287. doi:10.7150/jca.99351 This issue Cite
Research Paper
Association of MTHFR gene polymorphisms with non-Hodgkin lymphoma risk: Evidence from 31 articles
1. Clinical Research Institute of Zhanjiang, Central People's Hospital of Zhanjiang, Guangdong Medical University Zhanjiang Central Hospital, Zhanjiang 524045, P. R. China.
2. Department of Oncology, Central People's Hospital of Zhanjiang, Guangdong Medical University Zhanjiang Central Hospital, Zhanjiang 524045, P. R. China.
3. Department of Hematology, Central People's Hospital of Zhanjiang, Guangdong Medical University Zhanjiang Central Hospital, Zhanjiang 524045, P. R. China.
4. Zhanjiang Key Laboratory of Leukemia Pathogenesis and Targeted Therapy Research, Central People's Hospital of Zhanjiang, Guangdong Medical University Zhanjiang Central Hospital, Zhanjiang 524045, P. R. China.
Received 2024-6-6; Accepted 2024-8-2; Published 2024-8-13
Abstract
Background: Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms, particularly C677T and A1298C, have been implicated in various cancers, including non-Hodgkin lymphoma (NHL); however, their association with NHL risk remains inconclusive.
Methods: We conducted an updated meta-analysis to assess the relationship between MTHFR gene polymorphisms (C677T and A1298C) and NHL risk. Relevant studies were identified through systematic literature searches in multiple databases. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to evaluate the strength of the associations.
Results: The meta-analysis included 32 studies (8222 cases vs. 12956 controls) for MTHFR C677T and 26 studies (6930 cases vs. 11611 controls) for the A1298C polymorphism. Our meta-analysis revealed no significant associations between MTHFR gene polymorphisms (C677T and A1298C) and NHL risk. However, subgroup analysis stratified by ethnicity and NHL subtype yielded interesting findings for the C677T polymorphism. Specifically, in the subgroup analysis of Caucasians, the C677T polymorphism was significantly associated with NHL risk (heterozygous: OR=1.16, 95% CI=1.02-1.32; allele comparison: OR=1.07, 95% CI=1.01-1.13). Furthermore, in the analysis stratified by NHL subtype, the C677T polymorphism was significantly associated with increased follicular lymphoma (FL) risk (homozygous: OR=1.25, 95% CI=1.02-1.53; recessive: OR=1.28, 95% CI=1.06-1.56). False-positive result possibility (FPRP) analysis verified that the association of the MTHFR C677T polymorphism with NHL risk for Caucasians and FL subtypes was a true positive and deserves attention. We also determined that the C677T polymorphism is an expression quantitative trait locus (eQTL) since it is associated with MTHFR gene expression.
Conclusion: There was no overall association between MTHFR gene polymorphisms (C677T and A1298C) and NHL risk, but stratified analyses revealed significant associations in specific subgroups. While meta-analyses inherently build upon existing studies, our work distinguishes itself by incorporating recent data, applying rigorous analytical techniques, and providing more evidence of the MTHFR C677T polymorphism as an eQTL.
Keywords: MTHFR, C677T, A1298C, polymorphism, susceptibility
Introduction
Non-Hodgkin lymphoma (NHL), which originates in the lymphatic system, is a complex group of blood cancers with more than 50 different subtypes and is classified mainly into B-cell, T-cell, and natural killer (NK)-cell lymphomas on the basis of the type of lymphocyte affected [1]. Some of the most common types of NHL include diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), mantle cell lymphoma, and peripheral T-cell lymphoma [2]. The severity of high-grade NHL may require patients to undergo combination therapy, including chemotherapy, immunotherapy, and radiotherapy. In contrast, indolent lymphomas are usually incurable but are best managed as a lifelong, chronic disease. According to global cancer statistics from 2022, NHL ranks 10th in incidence and 11th in mortality [3]. Many lifestyle factors, environmental factors, and genetic factors, including smoking, alcohol consumption, hair dye, ultraviolet radiation, occupational exposure, immune deficiency, and micronutrients involved in one-carbon metabolism (e.g., B6, B12, methionine, and folate), have been shown to be associated with NHL risk; however, the risk may vary among NHL subtypes [2, 4-7]. Mounting evidence indicates that adequate intake of folate protects alcohol abstainers and former alcohol drinkers from developing NHL [4] and reduces the risk of DLBCL and marginal zone lymphoma [8].
Folate metabolism is closely intertwined with one-carbon metabolism. Folate, a B vitamin, is a crucial component of one-carbon metabolism. In folate metabolism, 5,10-methylenetetrahydrofolate reductase, encoded by the MTHFR gene, converts 5,10-methylenetetrahydrofolate into biologically active 5-methyltetrahydrofolate. The resulting active form of folate transfers carbon units to acceptor molecules through a series of enzymatic reactions (e.g., the conversion of homocysteine to methionine), completing the one-carbon transfer process. During one-carbon metabolism, the carbon units provided by 5-methyltetrahydrofolate are transferred and utilized to synthesize various biomolecules, including nucleic acids, amino acids, lipids, and neurotransmitters. Additionally, one-carbon metabolism is involved in methylation reactions (e.g., DNA CpG island methylation), which play critical roles in cellular differentiation, gene expression regulation, and other biological processes [9, 10]. Therefore, defects in the MTHFR gene disrupt multiple fundamental biochemical processes, including cell cycle regulation, DNA replication, and DNA and protein methylations, leading to various disorders, such as neural tube defects, cancer, and cardiovascular diseases [11-13]. Accordingly, genomic DNA methylation positively correlates with plasma folate [10]. Research has shown that two polymorphisms in the MTHFR gene, C677T and A1298C, can reduce enzyme activity [9, 10]. The MTHFR C677T variant leads to decreased intracellular methylation reactions, with the T/T genotype of MTHFR C677T dictating genomic DNA hypomethylation, a feature of most cancers [10, 14].
Polymorphisms in this gene have been studied with respect to the risk of various cancers, including NHL. Over the past few decades, many studies have investigated the associations between two MTHFR polymorphisms (C677T and A1298C) and NHL risk. However, the inconsistencies or limitations in the literature are not ignorable. Initial meta-analyses provided some insights but were limited by small sample sizes, regional biases, and variations in study quality [15]. By including a broader array of recent studies, the aim of this study was to conduct a systematic and updated meta-analysis to reassess the association between MTHFR polymorphisms and NHL risk to help elucidate the genetic underpinnings of NHL and offer potential preventative strategies.
Materials and methods
Sources
Literature searches were conducted via PubMed and EMBASE to collect studies on the association of MTHFR polymorphisms with NHL. The search employed the keywords “MTHFR or methylenetetrahydrofolate reductase”, ''polymorphism or variant or variation'', and ''non-Hodgkin lymphoma or non-Hodgkin's lymphoma or NHL”, coupled with the term “dependence” (the last search updated was on April 18, 2024). The Chinese Biomedical Literature Database (CBM) was also screened via the same search strategy to identify publications written in Chinese. We subsequently reviewed the bibliographies of the articles captured through the electronic search to find additional articles. We analysed studies that measured the association between MTHFR polymorphisms and NHL. Specifically, we were interested in studies of MTHFR polymorphisms known to affect the enzyme activity of MTHFR, C677T (Ala222Val, rs1801133) and A1298C (Glu429Ala, rs1801131).
Inclusion and exclusion criteria of studies
Studies eligible for the final meta-analysis needed to (1) investigate MTHFR C677T and/or A1298C polymorphisms in relation to NHL risk; (2) be structured as case‒control, nested case‒control, or cohort studies; (3) be published in English or Chinese; (4) be available for single nucleotide polymorphism (SNP) genotype data; (5) be distinct from other studies, with no overlapping datasets; and (6) supply adequate data to determine ORs and 95% CIs. The exclusion criteria applied to studies were the control genotype frequencies for the MTHFR C677T and A1298C polymorphisms did not adhere to Hardy‒Weinberg equilibrium (HWE) or lacked additional verification of HWE for other SNPs. Additional exclusions included case-only studies, case reports, conference abstracts, reviews, meta-analyses, and studies without adequate data. If there were two or more case‒control studies involving the same subjects, we included only the newest study or the study with the largest sample size in the final meta-analysis.
False-positive report probability analysis (FPRP)
FPRP is a statistical method that helps determine the probability that a statistically significant result is a false-positive, considering certain assumptions about prior probabilities of a true association for each finding and the observed data. We employed FPRP analysis to assess the robustness of statistically significant findings for the current genetic association studies. We calculate the FPRP for each significant finding via the observed P value, the prior probability (0.25, 0.1, 0.01, 0.001, or 0.0001), and the study's statistical power.
Expression quantitative trait locus analysis
An expression quantitative trait locus (QTL) is defined as a genetic variant that is significantly correlated with nearby gene expression alterations. The Adult Genotype Tissue Expression (GTEx) project, launched in 2010, is a large-scale research effort to understand the genetic regulation of gene expression in human tissues. GTEx collects and analyses genetic data and tissue samples from deceased adult donors across diverse populations in the United States. The project has generated extensive datasets and resources that are freely accessible to the scientific community, facilitating the study of how genetic variations influence gene expression patterns in various tissues [16]. We used this GTEx web tool (www.gtexportal.org/) to explore whether the MTHFR SNPs affect gene expression.
Statistical analysis
We calculated ORs and 95% CIs to estimate the associations between MTHFR SNPs and NHL susceptibility. We evaluated the risk of developing NHL for the assumed underlying genetic models, including the homozygous model (C677T: TT vs. CC; A1298C: CC vs. AA), heterozygous model (C677T: CT vs. CC; A1298C: AC vs. AA), recessive model (C677T: TT vs. CT+CC; A1298C: CC vs. AC+AA), and dominant model (C677T: CT +TT vs. CC; A1298C: AC+CC vs. AA). Allele comparisons were also performed to appraise the risk of mutant alleles over wild-type alleles for the two SNPs (C677T: T vs. C; A1298C: C vs. A). The goodness-of-fit chi-square test was applied to assess the departure from Hardy-Weinberg equilibrium (HWE) in the control genotypes. Significance was determined at a level of P<0.05. We examined the heterogeneity among the studies via the chi-square-based Q test. In cases where significant heterogeneity was present (Pheterogeneity<0.10), a random-effects model was selected [17], whereas a fixed-effects model (the Mantel-Haenszel method) was applied otherwise [18]. Identifying sources of heterogeneity is crucial for interpreting the overall results of a meta-analysis and can guide future research by highlighting areas where further investigation is warranted. To address heterogeneity, we examined whether the association varies across different subgroups stratified on the basis of ethnicity (Asians, Caucasians, Africans, and Mixed groups), source of control (hospital-based and population-based), and tumor subtype (FL and DLBCL). A sensitivity analysis was conducted to assess the stability of the findings, systematically excluding one study at a time and reiteratively computing the pooled ORs and 95% CIs. To investigate potential publication bias, both Begg's funnel plot [19] and Egger's linear regression test [20] were executed. All the statistical analyses were carried out via STATA software (version 11.0; Stata Corporation, College Station, TX) and SAS software (version 9.1; SAS Institute, Cary, NC). Significance was assessed via two-sided tests, with P<0.05 indicating statistical significance.
Results
Study characteristics
We initially identified 74 studies regarding the association between MTHFR polymorphisms and NHL susceptibility. After reviewing the title and abstract, we excluded 30 articles, including reviews and studies not involving the SNPs C677T and A1298C. An additional 13 articles were discarded since they overlapped with the included studies, were case-only studies, or deviated from HWE. As a result, 31 articles were selected for the final meta-analysis, with all the samples in the studies in HWE (Table 1) [9, 12, 13, 21-46]. All these studies adopted a case‒control design. Among them, 30 articles consisted of 32 studies that compared the frequency of MTHFR C677T alleles in NHL patients and controls, whereas 24 articles with 26 studies focused on the association between the MTHFR A1298C polymorphism and NHL risk (Figure 1).
Meta-analysis results
The association between the MTHFR polymorphism and the risk of NHL was recapitulated in 31 articles, consisting of 32 case‒control studies for the SNP rs1801133 (C677T) and 26 studies for the SNP rs1801131 (A1298C). The characteristics of the relevant case‒control studies evaluating the SNPs rs1801133 and rs1801131 (A1298C) are shown separately in Table 1.
The schematic diagram of the article screening process for the meta-analysis.
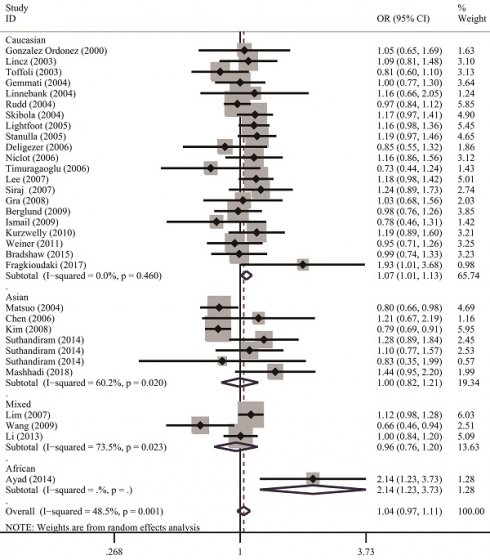
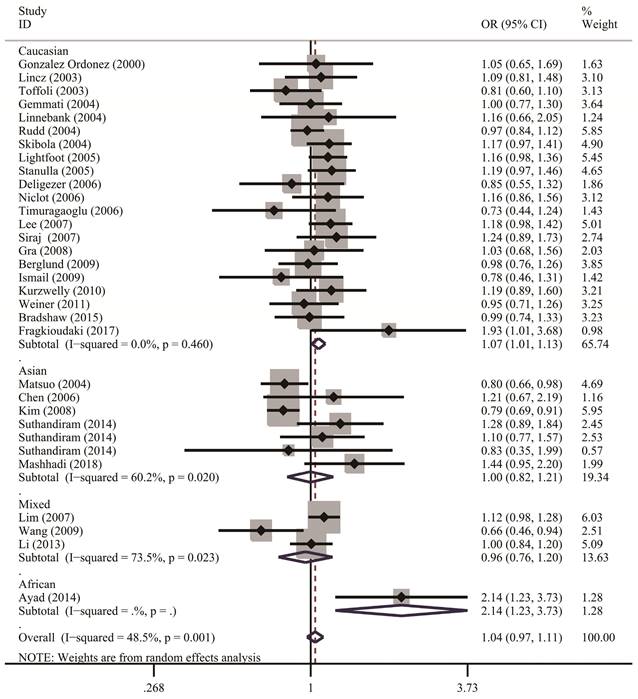
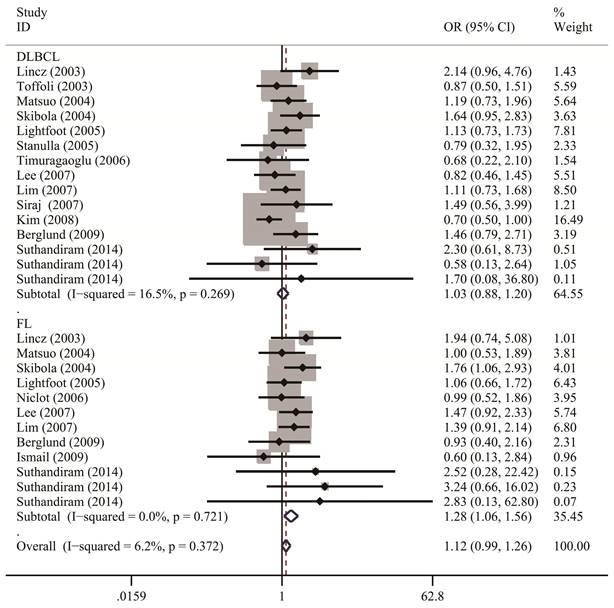
Table 2 lists the results of pooled and stratified analyses for the two SNPs. The pooled ORs and 95% CIs revealed that no association existed between the MTHFR C677T polymorphism and susceptibility to NHL across the 32 studies included in the analysis (homozygous: OR=1.10, 95% CI=0.96-1.24; heterozygous: OR=1.00, 95% CI=0.92-1.10; recessive: OR=1.06, 95% CI=0.97-1.17; dominant: OR=1.02, 95% CI=0.94-1.12; allele comparison: OR=1.04, 95% CI=0.97-1.11). Given that studies in the meta-analysis vary regarding population characteristics, tumor subtypes, and methodologies, stratified analysis may provide more informative guidance than overall analysis does and allow us to examine how these differences might affect the overall results. In the analysis stratified by ethnicity (Figure 2), the pooled OR under the homozygous model for the Caucasian subgroup was 1.16 (95% CI=1.02-1.32), with a Q statistic indicating heterogeneity (P=0.626). Allele comparison further provided evidence that the T variant allele is a risk factor for NHL in Caucasians (OR=1.07, 95% CI=1.01-1.13). Moreover, the T variant allele appeared to greatly increase the NHL risk in Africans (heterozygous: OR=2.91, 95% CI=1.34-6.32; dominant: OR=2.89, 95% CI=1.39-6.00; allele comparison: OR=2.14, 95% CI=1.23-3.73). However, this ethnic group included only one study with 49 cases and 82 controls. The source of control did not affect the significance of the association with NHL risk. In addition, stratified analysis by NHL subtype (Figure 3) revealed that carriers of the MTHFR C677T TT genotype were at significantly greater risk of developing FL (homozygous: OR=1.25, 95% CI=1.02-1.53; recessive: OR=1.28, 95% CI=1.06-1.56) than those with the CT and/or CC genotypes were (Table 2).
Like the MTHFR C677T polymorphism, in the 26-study pooled analysis, we found no significant association between the MTHFR A1298C polymorphism and overall NHL risk (homozygous: OR=1.20, 95% CI=0.99-1.47; heterozygous: OR=1.00, 95% CI=0.94-1.07; recessive: OR=1.20, 95% CI=1.00-1.44; dominant: OR=1.04, 95% CI=0.95-1.13; allele comparison: OR=1.07, 95% CI=0.98-1.17). The same applied to the stratified analysis for the A1298C polymorphism by ethnicity, source of control, and NHL subtypes (Table 2).
Heterogeneity and sensitivity analyses
The Q test revealed the presence of substantial heterogeneity in the association between the two MTHFR SNPs and NHL susceptibility, particularly in the overall analysis (Table 2). This finding suggested variability in the meta-analysis outcomes beyond what would be expected owing to chance alone. However, subgroup analyses indicated that heterogeneity was attenuated in Caucasians and in the FL subgroup (Table 2). Sensitivity analyses conducted by iteratively removing one study at a time revealed that none of the individual studies had a notable effect on the overall ORs (data not shown).
Characteristics of studies included in the final meta-analysis for the association between MTHFR C677T and A1298C polymorphisms and NHL risk
| Surname | Year | Country | Ethnicity | Source | Genotype method | Case | Control | MAF | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WW | WM | MM | All | WW | WM | MM | All | ||||||||
| C677T polymorphism | |||||||||||||||
| Gonzalez Ordonez | 2000 | Spain | Caucasian | HB | PCR-RFLP | 21 | 21 | 5 | 47 | 92 | 88 | 20 | 200 | 0.32 | 0.876 |
| Lincz | 2003 | Australia | Caucasian | HB | PCR-RFLP | 73 | 58 | 17 | 148 | 145 | 133 | 21 | 299 | 0.29 | 0.198 |
| Toffoli | 2003 | Italy | Caucasian | PB | PCR-RFLP | 44 | 49 | 18 | 111 | 147 | 233 | 85 | 465 | 0.43 | 0.662 |
| Gemmati | 2004 | Italy | Caucasian | PB | PCR-RFLP | 60 | 101 | 39 | 200 | 78 | 128 | 51 | 257 | 0.45 | 0.908 |
| Linnebank | 2004 | German | Caucasian | PB | PCR-RFLP | 13 | 12 | 6 | 31 | 66 | 52 | 24 | 142 | 0.35 | 0.019 |
| Matsuo | 2004 | Japan | Asian | HB | PCR-RFLP | 165 | 122 | 63 | 350 | 182 | 230 | 88 | 500 | 0.41 | 0.301 |
| Rudd | 2004 | UK | Caucasian | HB | Taqman | 361 | 381 | 90 | 832 | 383 | 397 | 106 | 886 | 0.34 | 0.841 |
| Skibola | 2004 | USA | Caucasian | PB | Taqman | 122 | 160 | 52 | 334 | 288 | 350 | 84 | 722 | 0.36 | 0.149 |
| Lightfoot | 2005 | UK | Caucasian | PB | Taqman | 247 | 270 | 72 | 589 | 356 | 316 | 83 | 755 | 0.32 | 0.309 |
| Stanulla | 2005 | German | Caucasian | PB | PCR-RFLP | 207 | 216 | 64 | 487 | 184 | 152 | 43 | 379 | 0.31 | 0.179 |
| Chen | 2006 | China | Asian | HB | Taqman | 11 | 13 | 4 | 28 | 72 | 66 | 19 | 157 | 0.33 | 0.522 |
| Deligezer | 2006 | Turkey | Caucasian | HB | Taqman | 31 | 30 | 5 | 66 | 66 | 72 | 16 | 154 | 0.34 | 0.574 |
| Niclot | 2006 | France | Caucasian | PB | DHPLC | 66 | 86 | 20 | 172 | 92 | 88 | 24 | 204 | 0.33 | 0.674 |
| Timuragaoglu | 2006 | Turkey | Caucasian | PB | Realtime PCR | 31 | 22 | 5 | 58 | 36 | 36 | 10 | 82 | 0.34 | 0.829 |
| Lee | 2007 | Australia | Caucasian | PB | Taqman | 253 | 227 | 74 | 554 | 256 | 190 | 57 | 503 | 0.30 | 0.019 |
| Lim | 2007 | USA | Mixed | PB | Taqman | 499 | 477 | 127 | 1103 | 443 | 396 | 86 | 925 | 0.31 | 0.853 |
| Siraj | 2007 | Saudi Arabia | Caucasian | PB | PCR-RFLP | 109 | 45 | 6 | 160 | 372 | 126 | 13 | 511 | 0.15 | 0.553 |
| Gra | 2008 | Russia | Caucasian | HB | Hybridization | 39 | 28 | 9 | 76 | 85 | 79 | 13 | 177 | 0.30 | 0.354 |
| Kim | 2008 | Korea | Asian | PB | PCR-RFLP | 223 | 286 | 75 | 584 | 540 | 863 | 297 | 1700 | 0.43 | 0.133 |
| Berglund | 2009 | Sweden | Caucasian | PB | Illumina | 154 | 85 | 24 | 263 | 241 | 157 | 32 | 430 | 0.26 | 0.363 |
| Ismail | 2009 | Jordan | Caucasian | PB | PCR-RFLP | 34 | 19 | 2 | 55 | 94 | 66 | 10 | 170 | 0.25 | 0.722 |
| Wang | 2009 | Jamaica | Mixed | PB | Taqman | 329 | 58 | 5 | 392 | 204 | 57 | 5 | 266 | 0.13 | 0.664 |
| Kurzwelly | 2010 | German | Caucasian | PB | PCR-RFLP | 78 | 81 | 26 | 185 | 96 | 96 | 20 | 212 | 0.32 | 0.568 |
| Weiner | 2011 | Russia | Caucasian | PB | Taqman | 72 | 60 | 11 | 143 | 242 | 198 | 46 | 486 | 0.30 | 0.553 |
| Li | 2013 | USA | Mixed | PB | Taqman | 202 | 206 | 72 | 480 | 236 | 246 | 82 | 564 | 0.36 | 0.173 |
| Ayad | 2014 | Egypt | African | PB | PCR-RFLP | 19 | 24 | 6 | 49 | 53 | 23 | 6 | 82 | 0.21 | 0.136 |
| Suthandiram | 2014 | Malaysia-Malay | Asian | PB | MassARRAY | 144 | 49 | 6 | 199 | 236 | 66 | 5 | 307 | 0.12 | 0.876 |
| Suthandiram | 2014 | Malaysia-Chinese | Asian | PB | MassARRAY | 67 | 48 | 6 | 121 | 155 | 98 | 12 | 265 | 0.23 | 0.479 |
| Suthandiram | 2014 | Malaysia-Indian | Asian | PB | MassARRAY | 45 | 7 | 0 | 52 | 128 | 20 | 2 | 150 | 0.08 | 0.249 |
| Bradshaw | 2015 | Australia | Caucasian | HB | PCR-RFLP | 97 | 85 | 25 | 207 | 88 | 94 | 19 | 201 | 0.33 | 0.393 |
| Fragkioudaki | 2017 | Greece | Caucasian | HB | PCR-RFLP | 4 | 10 | 5 | 19 | 235 | 291 | 74 | 600 | 0.37 | 0.268 |
| Mashhadi | 2018 | Iran | Asian | PB | TARMS-PCR | 82 | 42 | 3 | 127 | 150 | 53 | 2 | 205 | 0.14 | 0.252 |
| A1298C polymorphism | |||||||||||||||
| Lincz | 2003 | Australia | Caucasian | HB | PCR-RFLP | 64 | 68 | 13 | 145 | 124 | 139 | 31 | 294 | 0.34 | 0.385 |
| Toffoli | 2003 | Italy | Caucasian | PB | PCR-RFLP | 54 | 44 | 13 | 111 | 200 | 222 | 43 | 465 | 0.33 | 0.094 |
| Gemmati | 2004 | Italy | Caucasian | PB | PCR-RFLP | 96 | 90 | 14 | 200 | 126 | 110 | 21 | 257 | 0.30 | 0.659 |
| Linnebank | 2004 | German | Caucasian | PB | PCR-RFLP | 16 | 12 | 3 | 31 | 69 | 54 | 19 | 142 | 0.32 | 0.116 |
| Matsuo | 2004 | Japan | Asian | HB | PCR-RFLP | 209 | 122 | 19 | 350 | 327 | 150 | 23 | 500 | 0.20 | 0.282 |
| Rudd | 2004 | UK | Caucasian | HB | Taqman | 397 | 363 | 72 | 832 | 412 | 389 | 85 | 886 | 0.32 | 0.622 |
| Skibola | 2004 | USA | Caucasian | PB | Taqman | 178 | 128 | 27 | 333 | 341 | 310 | 71 | 722 | 0.31 | 0.964 |
| Lightfoot | 2005 | UK | Caucasian | PB | Taqman | 288 | 250 | 51 | 589 | 347 | 331 | 77 | 755 | 0.32 | 0.882 |
| Niclot | 2006 | France | Caucasian | PB | DHPLC | 79 | 76 | 17 | 172 | 102 | 81 | 15 | 198 | 0.28 | 0.844 |
| Lim | 2007 | USA | Mixed | PB | Taqman | 540 | 480 | 104 | 1124 | 461 | 393 | 81 | 935 | 0.30 | 0.831 |
| Siraj | 2007 | Saudi Arabia | Caucasian | PB | PCR-RFLP | 38 | 40 | 35 | 113 | 239 | 220 | 52 | 511 | 0.32 | 0.896 |
| Gra | 2008 | Russia | Caucasian | HB | Hybridization | 36 | 30 | 10 | 76 | 81 | 82 | 14 | 177 | 0.31 | 0.278 |
| Kim | 2008 | Korea | Asian | PB | Taqman | 372 | 182 | 29 | 583 | 1147 | 500 | 53 | 1700 | 0.18 | 0.868 |
| Berglund | 2009 | Sweden | Caucasian | PB | Illumina | 116 | 121 | 25 | 262 | 214 | 196 | 39 | 449 | 0.31 | 0.533 |
| Ismail | 2009 | Jordan | Caucasian | PB | PCR-RFLP | 20 | 23 | 12 | 55 | 76 | 81 | 13 | 170 | 0.31 | 0.172 |
| Wang | 2009 | Jamaica | Mixed | PB | Taqman | 277 | 98 | 15 | 390 | 201 | 65 | 9 | 275 | 0.15 | 0.198 |
| Kurzwelly | 2010 | German | Caucasian | PB | PCR-RFLP | 72 | 96 | 17 | 185 | 106 | 89 | 17 | 212 | 0.29 | 0.779 |
| Weiner | 2011 | Russia | Caucasian | PB | Taqman | 59 | 52 | 22 | 133 | 232 | 215 | 56 | 503 | 0.33 | 0.562 |
| Li | 2013 | USA | Mixed | PB | Taqman | 246 | 203 | 40 | 489 | 265 | 250 | 59 | 574 | 0.32 | 0.997 |
| Jiang | 2014 | China | Asian | HB | Taqman | 17 | 9 | 2 | 28 | 109 | 46 | 2 | 157 | 0.16 | 0.238 |
| Suthandiram | 2014 | Malaysia-Malay | Asian | PB | MassARRAY | 104 | 82 | 13 | 199 | 137 | 147 | 23 | 307 | 0.31 | 0.052 |
| Suthandiram | 2014 | Malaysia- Chinese | Asian | PB | MassARRAY | 74 | 40 | 7 | 121 | 160 | 85 | 20 | 265 | 0.24 | 0.073 |
| Suthandiram | 2014 | Malaysia-Indian | Asian | PB | MassARRAY | 11 | 27 | 14 | 52 | 57 | 75 | 18 | 150 | 0.37 | 0.375 |
| Bradshaw | 2015 | Australia | Caucasian | HB | HRM | 94 | 92 | 25 | 211 | 93 | 85 | 24 | 202 | 0.33 | 0.502 |
| Fragkioudaki | 2017 | Greece | Caucasian | HB | PCR-RFLP | 15 | 4 | 0 | 19 | 273 | 266 | 61 | 600 | 0.32 | 0.747 |
| Mashhadi | 2018 | Iran | Asian | PB | TARMS-PCR | 69 | 40 | 18 | 127 | 110 | 69 | 26 | 205 | 0.30 | 0.006 |
NHL, non-Hodgkin lymphoma; MAF, Minor allele frequency; HWE, Hardy-Weinberg equilibrium; W, wild type; M, mutant type; HB, Hospital based; PB, Population based; PCR-RFLP, Polymorphism chain reaction-restriction fragment length polymorphism; TARMS-PCR, Tetra Amplification Refractory Mutation System polymerase chain reaction; HRM, high resolution melt; DHPLC, Denaturing high performance liquid chromatography.
Meta-analysis for the association between MTHFR C677T and A1298C polymorphisms and non-Hodgkin lymphoma risk
| Variables | No. of | Homozygous | Heterozygous | Recessive | Dominant | Allele Comparing | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| studies | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | |
| C677T (rs1801133) | TT vs. CC | CT vs. CC | TT vs. (CT + CC) | (CT + TT) vs. CC | T vs. C | ||||||
| All | 32 | 1.10 (0.96-1.24) | 0.088 | 1.00 (0.92-1.10) | 0.010 | 1.06 (0.97-1.17) | 0.538 | 1.02 (0.94-1.12) | 0.001 | 1.04 (0.97-1.11) | 0.001 |
| Ethnicity | |||||||||||
| Caucasian | 21 | 1.16 (1.02-1.32) | 0.626 | 1.05 (0.97-1.14) | 0.591 | 1.12 (1.00-1.26) | 0.790 | 1.07 (0.99-1.16) | 0.479 | 1.07 (1.01-1.13) | 0.460 |
| Asian | 7 | 0.83 (0.60-1.15) | 0.242 | 0.97 (0.74-1.26) | 0.014 | 0.85 (0.69-1.04) | 0.337 | 0.98 (0.75-1.27) | 0.008 | 1.00 (0.82-1.21) | 0.020 |
| Mixed | 3 | 1.16 (0.92-1.46) | 0.364 | 0.92 (0.71-1.19) | 0.067 | 1.15 (0.93-1.43) | 0.473 | 0.93 (0.71-1.22) | 0.034 | 0.96 (0.76-1.20) | 0.023 |
| African | 1 | 2.79 (0.80-9.71) | / | 2.91 (1.34-6.32) | / | 1.77 (0.54-5.82) | / | 2.89 (1.39-6.00) | / | 2.14 (1.23-3.73) | / |
| Source of control | |||||||||||
| HB | 9 | 1.04 (0.82-1.32) | 0.299 | 0.87 (0.72-1.05) | 0.144 | 1.07 (0.89-1.29) | 0.429 | 0.91 (0.77-1.07) | 0.204 | 0.97 (0.87-1.08) | 0.312 |
| PB | 23 | 1.11 (0.95-1.29) | 0.078 | 1.05 (0.95-1.16) | 0.040 | 1.06 (0.96-1.18) | 0.487 | 1.06 (0.96-1.18) | 0.003 | 1.06 (0.97-1.11) | 0.001 |
| Subtype | |||||||||||
| DLBCL | 15 | 0.98 (0.83-1.15) | 0.077 | 0.99 (0.83-1.18) | 0.002 | 1.03 (0.88-1.20) | 0.269 | 1.01 (0.86-1.19) | 0.001 | 1.03 (0.92-1.16) | 0.011 |
| FL | 12 | 1.25 (1.02-1.53) | 0.655 | 0.91 (0.75-1.10) | 0.081 | 1.28 (1.06-1.56) | 0.721 | 0.97 (0.82-1.15) | 0.133 | 1.06 (0.95-1.17) | 0.359 |
| A1298C (rs1801131) | CC vs. AA | AC vs. AA | CC vs. (AC + AA) | (AC + CC) vs. AA | C vs. A | ||||||
| All | 26 | 1.20 (0.99-1.47) | <0.001 | 1.00 (0.94-1.07) | 0.279 | 1.20 (1.00-1.44) | <0.001 | 1.04 (0.95-1.13) | 0.011 | 1.07 (0.98-1.17) | <0.001 |
| Ethnicity | |||||||||||
| Caucasian | 16 | 1.20 (0.92-1.59) | <0.001 | 0.97 (0.89-1.07) | 0.281 | 1.21 (0.93-1.58) | <0.001 | 1.02 (0.90-1.16) | 0.022 | 1.06 (0.94-1.21) | <0.001 |
| Asian | 7 | 1.39 (0.91-2.12) | 0.047 | 1.08 (0.95-1.24) | 0.247 | 1.34 (0.95-1.89) | 0.153 | 1.11 (0.91-1.36) | 0.086 | 1.13 (0.95-1.36) | 0.032 |
| Mixed | 3 | 0.96 (0.72-1.29) | 0.293 | 1.00 (0.87-1.14) | 0.471 | 0.98 (0.77-1.24) | 0.426 | 0.99 (0.85-1.15) | 0.291 | 0.99 (0.86-1.14) | 0.199 |
| Source of control | |||||||||||
| HB | 7 | 1.04 (0.76-1.42) | 0.264 | 1.01 (0.88-1.15) | 0.192 | 1.02 (0.78-1.34) | 0.335 | 1.00 (0.81-1.24) | 0.083 | 1.02 (0.86-1.21) | 0.056 |
| PB | 19 | 1.25 (0.98-1.58) | <0.001 | 1.00 (0.93-1.08) | 0.335 | 1.24 (0.99-1.55) | <0.001 | 1.04 (0.94-1.16) | 0.019 | 1.09 (0.98-1.21) | <0.001 |
| Subtype | |||||||||||
| DLBCL | 12 | 1.21 (0.87-1.69) | 0.003 | 1.01 (0.90-1.13) | 0.738 | 1.24 (0.89-1.72) | 0.001 | 1.04 (0.92-1.17) | 0.237 | 1.07 (0.98-1.16) | 0.002 |
| FL | 11 | 1.23 (0.91-1.67) | 0.198 | 1.04 (0.90-1.20) | 0.128 | 1.19 (0.93-1.53) | 0.365 | 1.08 (0.88-1.32) | 0.056 | 1.07 (0.96-1.19) | 0.041 |
HB, Hospital based; PB, Population based; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma.
False-positive report probability analysis for the associations between MTHFR gene C677T polymorphism and non-Hodgkin lymphoma risk
| Genotype | OR (95% CI) | P a | Statistical Power b | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| TT vs. CC | ||||||||
| Caucasian | 1.16 (1.02-1.32) | 0.022 | 1.000 | 0.062 | 0.165 | 0.685 | 0.956 | 0.995 |
| FL | 1.25 (1.02-1.53) | 0.033 | 0.980 | 0.092 | 0.233 | 0.769 | 0.971 | 0.997 |
| CT vs.CC | ||||||||
| African | 2.91 (1.34-6.32) | 0.007 | 0.053 | 0.284 | 0.544 | 0.929 | 0.993 | 0.999 |
| TT vs. CT/CC | ||||||||
| FL | 1.28 (1.06-1.56) | 0.011 | 0.953 | 0.033 | 0.094 | 0.533 | 0.920 | 0.991 |
| CT/TT vs. CC | ||||||||
| African | 2.89 (1.39-6.00) | 0.005 | 0.045 | 0.250 | 0.500 | 0.917 | 0.991 | 0.999 |
| T vs. C | ||||||||
| Caucasian | 1.07 (1.01-1.13) | 0.023 | 1.000 | 0.065 | 0.171 | 0.695 | 0.958 | 0.996 |
| African | 2.14 (1.23-3.73) | 0.007 | 0.128 | 0.141 | 0.329 | 0.844 | 0.982 | 0.998 |
OR, odds ratio; CI, confidence interval; FL, follicular lymphoma.
a Chi-square test was used to calculate the genotype frequency distributions.
b Statistical power was calculated using the number of observations in the subgroup and the OR and P values in this table.
Publication bias
We checked the potential bias of the meta-analysis via Begg's funnel plot [19] and Egger's linear regression test [20]. Asymmetry was observed in the shape of the funnel plots concerning the C677T and A1298C polymorphisms (data not presented). Moreover, Egger's test did not indicate any significant publication bias for either the C677T or A1298C polymorphisms. These results suggested that this meta-analysis was not influenced by publication bias.
FPRP results
While assuming a prior probability of 0.25, low FPRP values were yielded for the significant association between the MTHFR C677T polymorphism and NHL risk among the following subgroups: Caucasian (TT vs. CC, 0.062; T vs. C, 0.065), FL subtype groups (TT vs. CC, 0.092; TT vs. CT/CC, 0.033), and African (T vs. C, 0.141) (Table 3). These findings with low FPRP values indicate a high probability that the associations are true positives and are robust against false-positives. When a stricter prior probability of 0.1 was applied, the associations for the Caucasian (TT vs. CC, 0.165; T vs. C, 0.171) and FL subtype groups (TT vs. CT/CC, 0.094) remained noteworthy (Table 3).
Genotype‒tissue expression (GTEx) analysis
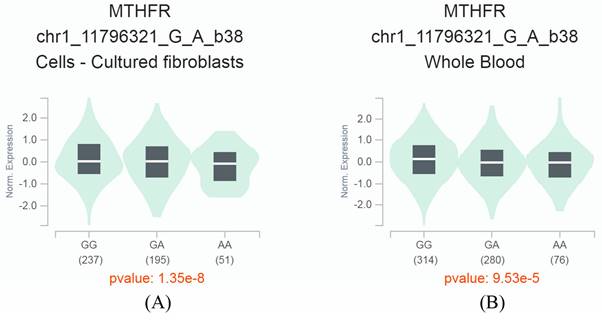
By searching, we retrieved an eQTL result from the GTEx database, indicating a significant association between the MTHFR C677T polymorphism and the expression level of the MTHFR gene. The SNP is located at position 11796321 on chromosome 1 and has two allelic variants, C (G) and T (A). In cultured fibroblasts and human blood samples, we observed significant downregulation of gene MTHFR expression in carriers of the A allele compared with carriers of the G allele (P<0.001) (Figure 4). These findings suggest that the SNP may participate in regulating the MTHFR gene, and further functional studies could help elucidate this phenomenon.
Forest plots demonstrate the association between MTHFR C677T polymorphisms and NHL risk in the stratified analysis by ethnicity regarding allele comparison. A solid diamond shape and a horizontal line on the plot visually represent the estimation of OR and its 95% CI for each study.
Discussion
This updated meta-analysis comprehensively assessed the association between MTHFR gene polymorphisms (C677T and A1298C) and susceptibility to NHL. Our overall analysis did not reveal a significant correlation between these SNPs and NHL risk. Given the significant evidence of an association of NHL risk with the two MTHFR SNPs in some studies [12, 21, 22], the contradictory findings may result from the heterogeneous nature of NHL, population ancestry, and source of controls. To control for potential confounding effects, we conducted stratified analyses to better understand the potential associations between MTHFR gene polymorphisms and NHL risk in specific subgroups. Stratified analysis revealed that the C677T polymorphism was significantly associated with increased NHL risk in Caucasians and the FL subtype but not in Asians or the DLBCL subtype. Moreover, FPRP analysis confirmed that these significant associations had low FPRP values, indicating a high probability that the associations are true positives and are robust against false-positives. These results are consistent with those of a previous meta-analysis by He et al. [15]. Notably, they also reported a significant reversal association in Asians, with 3 studies included [15], whereas the current meta-analysis with 7 studies in Asians showed no evidence of this association. The paradoxical nature of these findings indicates that they may be the result of chance. The observed association between the C677T polymorphism and increased NHL risk in Caucasians underscores the potential ethnicity-specific effects of this genetic variant on NHL pathogenesis.
The forest plots illustrate the relationship between MTHFR C677T polymorphisms and the risk of NHL under the recessive model, segmented by NHL subtypes. Each study's OR and its 95% CI are graphically presented using a solid diamond shape and a horizontal line for visual clarity.
The MTHFR C677T variant is an expression quantitative trait locus (eQTL). This eQTL modulates the expression levels of the MTHFR gene. The diagram demonstrates the impact of the MTHFR C677T variant on the MTHFR gene expression in cultured fibroblasts (A) and whole blood (B), highlighting its significance in understanding genetic regulation and its potential implications in various biological processes.
Moreover, the significant association in subgroups aligns with previous studies, which implicates the C677T polymorphism in altered folate metabolism, which may contribute to lymphomagenesis through mechanisms such as DNA methylation and nucleotide synthesis [10]. DNA methylation and synthesis rely heavily on the accessibility of one-carbon, methyl-donating nutrients, and insufficiencies in nutrients such as folate or vitamins B6 and B12 might increase the risk of gene mutations and DNA double-strand breaks [11]. A shortage of folate has been linked to several malignancies [47-49]. Consistently, several studies have revealed that genetic polymorphisms in one-carbon-metabolizing pathway genes and folate-metabolizing genes, such as thymidylate synthase, serine hydroxymethyltransferase, methionine synthase, and MTHFR, can modify NHL predisposition [9, 12, 24, 25, 30, 31, 41].
The MTHFR C677T polymorphism is known to be linked to decreased MTHFR enzyme activity and lower plasma folate levels, leading to hypomethylation [10]. It is reasonable that the MTHFR C677T polymorphism is associated with the risk of FL but not DLBCL. FL and DLBCL are the most common indolent and aggressive lymphomas, respectively; therefore, they are two distinct diseases with different environmental and genetic risk factors [2]. For example, different progressively acquired DNA alterations (e.g., gene mutation, amplification or deletion and chromosomal translocation) may contribute to the development of different subtypes of NHL, such as the causal relationship between BCL2 translocation and FL or MYC translocation and Burkitt lymphoma [50]. Studies conducted previously proposed a potential link between hypomethylation of the BCL-2 gene and the onset of this translocation [51]. One possible mechanism may be that the MTHFR C677T polymorphism results in increased global DNA hypomethylation, which in turn induces chromosomal instability through the loss of epigenetic regulation and the activation of transposable elements. Such genomic instability may predispose B lymphocytes to BCL-2 gene translocation, thereby facilitating the development of FL.
Moreover, the MTHFR A1298C polymorphism did not significantly affect NHL risk in either the overall pooled analysis or the stratified analyses of our meta-analysis. He and colleagues demonstrated that the A1298C polymorphism significantly increased NHL susceptibility among Asians in a previous meta-analysis in which 3 studies were included [15]. Intriguingly, the significance of the association disappeared in the currently updated meta-analysis, with 7 studies conducted in the Asian population. However, the potential role of the A1298C polymorphism in NHL susceptibility cannot be ruled out entirely. The lack of significance in our analysis may be attributed to the complex interplay of genetic and environmental factors influencing NHL development, highlighting the need for further investigation into the functional implications of the A1298C polymorphism in lymphomagenesis.
Overall, several unique aspects of our study may contribute to its originality and scientific value: 1) Inclusion of recent studies: The latest published meta-analysis on MTHFR SNPs and NHL susceptibility was performed in 2014. It has been ten years since then. Our meta-analysis integrated the latest available data, including several recent studies that have not been incorporated into previous analyses. This allows for an updated and comprehensive assessment of the SNPs in question, providing a more current understanding of their association with NHL. 2) Methodological Improvements: Our meta-analysis employed FPRP analysis to address possible false associations of SNPs with NHL susceptibility. Our FPRP results confirmed that the associations between the MTHFR gene C677T polymorphism and NHL risk were notable in some subgroups. These methodological enhancements increase the reliability of the conclusions drawn. 3) GTEx analysis: By exploring the GTEx database, we found that the MTHFR C677T polymorphism is an eQTL that is significantly correlated with alterations in MTHFR gene expression. This finding suggests that the SNP may participate in regulating the MTHFR gene. 4) Clinical Relevance: The findings from our updated meta-analysis have implications for personalized medicine, potentially guiding genetic screening and risk assessment strategies in clinical settings. Highlighting the translational aspect of our research underscores its practical relevance and novelty.
However, certain limitations should be acknowledged. The included studies varied in design, genotyping methods, sample size, and adjustment for confounding factors, which may have introduced heterogeneity and biased our results. Moreover, because only one study conducted with African participants was included, this meta-analysis was insufficient to estimate the risk effects of MTHRF SNPs among this population. Additionally, gene‒gene and gene‒environment interactions were not explored in this meta-analysis, warranting future research to elucidate the intricate mechanisms underlying NHL susceptibility associated with MTHFR gene polymorphisms.
In conclusion, our updated meta-analysis highlights the potential significance of the MTHFR gene C677T polymorphism in NHL risk, particularly among individuals of Caucasian ethnicity and in the FL subtype. These findings contribute to our understanding of the genetic basis of NHL and may help to foster risk stratification and personalized prevention strategies. Further large-scale studies and functional analyses are needed to validate our findings and elucidate the underlying biological mechanisms involved.
Acknowledgements
This study was supported by the Guangdong Provincial Basic and Applied Basic Research Fund Enterprise Joint Fund Project (2023A1515220173), the Medical Science and Technology Research Fund of Guangdong Province (A2024523, A2023201) and the Science and Technology Plan Project of Zhanjiang city (2021A05153).
Author contributions
Study conception and design: Zhigang Yang, Yunmiao Guo, Gang Wang; Data acquisition and formal analysis: Gang Wang, Yuluo Wu, Zuolei Jing, Ruiting Wen, Yuanrui Song, Yin Feng, Guangru Li, Xiaopeng Zou, Gaoxiang Huang, Zhirong Jia; Study supervision: Zhigang Yang, Yunmiao Guo; Writing original draft: Gang Wang, Yunmiao Guo; Reviewing & editing the article: Zhigang Yang, Yunmiao Guo, Gang Wang. All co-authors have read and approved the final manuscript.
Data availability statement
All the data are available upon request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD. et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375-90
2. Bowzyk Al-Naeeb A, Ajithkumar T, Behan S, Hodson DJ. Non-Hodgkin lymphoma. BMJ. 2018;362:k3204
3. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263
4. Muller AM, Ihorst G, Mertelsmann R, Engelhardt M. Epidemiology of non-Hodgkin's lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol. 2005;84:1-12
5. Zheng T, Mayne ST, Boyle P, Holford TR, Liu WL, Flannery J. Epidemiology of non-Hodgkin lymphoma in Connecticut. 1935-1988. Cancer. 1992;70:840-9
6. Skibola CF, Curry JD, Nieters A. Genetic susceptibility to lymphoma. Haematologica. 2007;92:960-9
7. Berndt SI, Vijai J, Benavente Y, Camp NJ, Nieters A, Wang Z, Smedby KE, Kleinstern G, Hjalgrim H, Besson C. et al. Distinct germline genetic susceptibility profiles identified for common non-Hodgkin lymphoma subtypes. Leukemia. 2022;36(12):2835-44
8. Koutros S, Zhang Y, Zhu Y, Mayne ST, Zahm SH, Holford TR, Leaderer BP, Boyle P, Zheng T. Nutrients contributing to one-carbon metabolism and risk of non-Hodgkin lymphoma subtypes. Am J Epidemiol. 2008;167:287-94
9. Lim U, Wang SS, Hartge P, Cozen W, Kelemen LE, Chanock S, Davis S, Blair A, Schenk M, Rothman N. et al. Gene-nutrient interactions among determinants of folate and one-carbon metabolism on the risk of non-Hodgkin lymphoma: NCI-SEER case-control study. Blood. 2007;109:3050-9
10. Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R. et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606-11
11. Choi SW, Kim YI, Weitzel JN, Mason JB. Folate depletion impairs DNA excision repair in the colon of the rat. Gut. 1998;43:93-9
12. Skibola CF, Forrest MS, Coppede F, Agana L, Hubbard A, Smith MT, Bracci PM, Holly EA. Polymorphisms and haplotypes in folate-metabolizing genes and risk of non-Hodgkin lymphoma. Blood. 2004;104:2155-62
13. Kurzwelly D, Knop S, Guenther M, Loeffler J, Korfel A, Thiel E, Hebart H, Simon M, Weller M, Linnebank M. et al. Genetic variants of folate and methionine metabolism and PCNSL incidence in a German patient population. J Neurooncol. 2010;100:187-92
14. Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849-53
15. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159
16. E Stranger B, E Brigham L, Hasz R. et al. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49:1664-70
17. DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45:139-45
18. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-48
19. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
20. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-34
21. Fragkioudaki S, Nezos A, Souliotis VL, Chatziandreou I, Saetta AA, Drakoulis N, Tzioufas AG, Voulgarelis M, Sfikakis PP, Koutsilieris M. et al. MTHFR gene variants and non-MALT lymphoma development in primary Sjogren's syndrome. Sci Rep. 2017;7:7354
22. Ayad MW, El Naggar AA, El Naggar M. MTHFR C677T polymorphism: association with lymphoid neoplasm and effect on methotrexate therapy. Eur J Haematol. 2014;93:63-9
23. Lincz LF, Scorgie FE, Kerridge I, Potts R, Spencer A, Enno A. Methionine synthase genetic polymorphism MS A2756G alters susceptibility to follicular but not diffuse large B-cell non-Hodgkin's lymphoma or multiple myeloma. Br J Haematol. 2003;120:1051-4
24. Li Q, Lan Q, Zhang Y, Bassig BA, Holford TR, Leaderer B, Boyle P, Zhu Y, Qin Q, Chanock S. et al. Role of one-carbon metabolizing pathway genes and gene-nutrient interaction in the risk of non-Hodgkin lymphoma. Cancer Causes Control. 2013;24:1875-84
25. Berglund M, Enblad G, Turesson I, Edman V, Thunberg U. Folate-metabolizing genes in lymphoma patients from Sweden. Scand J Immunol. 2009;70:408-10
26. Gemmati D, Ongaro A, Scapoli GL, Della Porta M, Tognazzo S, Serino ML, Di Bona E, Rodeghiero F, Gilli G, Reverberi R. et al. Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin's lymphoma in adults. Cancer Epidemiol Biomarkers Prev. 2004;13:787-94
27. Gonzalez Ordonez AJ, Fernandez Carreira JM, Fernandez Alvarez CR, Martin L, Sanchez Garcia J, Medina Rodriguez JM, Alvarez MV, Coto E. Normal frequencies of the C677T genotypes on the methylenetetrahydrofolate reductase (MTHFR) gene among lymphoproliferative disorders but not in multiple myeloma. Leuk Lymphoma. 2000;39:607-12
28. Gra OA, Glotov AS, Nikitin EA, Glotov OS, Kuznetsova VE, Chudinov AV, Sudarikov AB, Nasedkina TV. Polymorphisms in xenobiotic-metabolizing genes and the risk of chronic lymphocytic leukemia and non-Hodgkin's lymphoma in adult Russian patients. Am J Hematol. 2008;83:279-87
29. Ismail SI, Ababneh NA, Khader Y, Abu-Khader AA, Awidi A. Methylenetetrahydrofolate reductase genotype association with the risk of follicular lymphoma. Cancer Genet Cytogenet. 2009;195:120-4
30. Kim HN, Lee IK, Kim YK, Tran HT, Yang DH, Lee JJ, Shin MH, Park KS, Shin MG, Choi JS. et al. Association between folate-metabolizing pathway polymorphism and non-Hodgkin lymphoma. Br J Haematol. 2008;140:287-94
31. Lee KM, Lan Q, Kricker A, Purdue MP, Grulich AE, Vajdic CM, Turner J, Whitby D, Kang D, Chanock S. et al. One-carbon metabolism gene polymorphisms and risk of non-Hodgkin lymphoma in Australia. Hum Genet. 2007;122:525-33
32. Lightfoot TJ, Skibola CF, Willett EV, Skibola DR, Allan JM, Coppede F, Adamson PJ, Morgan GJ, Roman E, Smith MT. Risk of non-Hodgkin lymphoma associated with polymorphisms in folate-metabolizing genes. Cancer Epidemiol Biomarkers Prev. 2005;14:2999-3003
33. Linnebank M, Schmidt S, Kolsch H, Linnebank A, Heun R, Schmidt-Wolf IG, Glasmacher A, Fliessbach K, Klockgether T, Schlegel U. et al. The methionine synthase polymorphism D919G alters susceptibility to primary central nervous system lymphoma. Br J Cancer. 2004;90:1969-71
34. Matsuo K, Hamajima N, Suzuki R, Ogura M, Kagami Y, Taji H, Yasue T, Mueller NE, Nakamura S, Seto M. et al. Methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms and reduced risk of malignant lymphoma. Am J Hematol. 2004;77:351-7
35. Niclot S, Pruvot Q, Besson C, Savoy D, Macintyre E, Salles G, Brousse N, Varet B, Landais P, Taupin P. et al. Implication of the folate-methionine metabolism pathways in susceptibility to follicular lymphomas. Blood. 2006;108:278-85
36. Rudd MF, Sellick GS, Allinson R, Matutes E, Catovsky D, Houlston RS. MTHFR polymorphisms and risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev. 2004;13:2268-70
37. Siraj AK, Ibrahim M, Al-Rasheed M, Bu R, Bavi P, Jehan Z, Abubaker J, Murad W, Al-Dayel F, Ezzat A. et al. Genetic polymorphisms of methylenetetrahydrofolate reductase and promoter methylation of MGMT and FHIT genes in diffuse large B cell lymphoma risk in Middle East. Ann Hematol. 2007;86:887-95
38. Timuragaoglu A, Dizlek S, Uysalgil N, Tosun O, Yamac K. Methylenetetrahydrofolate reductase C677T polymorphism in adult patients with lymphoproliferative disorders and its effect on chemotherapy. Ann Hematol. 2006;85:863-8
39. Toffoli G, Rossi D, Gaidano G, Cecchin E, Boiocchi M, Carbone A. Methylenetetrahydrofolate reductase genotype in diffuse large B-cell lymphomas with and without hypermethylation of the DNA repair gene O6-methylguanine DNA methyltransferase. Int J Biol Markers. 2003;18:218-21
40. Wang SS, Carreon JD, Hanchard B, Chanock S, Hisada M. Common genetic variants and risk for non-Hodgkin lymphoma and adult T-cell lymphoma/leukemia in Jamaica. Int J Cancer. 2009;125:1479-82
41. Weiner AS, Beresina OV, Voronina EN, Voropaeva EN, Boyarskih UA, Pospelova TI, Filipenko ML. Polymorphisms in folate-metabolizing genes and risk of non-Hodgkin's lymphoma. Leuk Res. 2011;35:508-15
42. Mashhadi MA, Miri-Moghaddam E, Arbabi N, Bazi A, Heidari Z, Sepehri Z, Karimkoshte A, Rezvan A, Hashemi SM. C677T and A1298C polymorphisms of methylene tetrahydrofolate reductase in non-Hodgkin lymphoma: southeast Iran. Tumori. 2018;104:280-4
43. Deligezer U, Akisik EE, Yaman F, Erten N, Dalay N. MTHFR C677 T gene polymorphism in lymphoproliferative diseases. J Clin Lab Anal. 2006;20:37-41
44. Stanulla M, Seidemann K, Schnakenberg E, Book M, Mehles A, Welte K, Schrappe M, Reiter A. Methylenetetrahydrofolate reductase (MTHFR) 677C>T polymorphism and risk of pediatric non-Hodgkin lymphoma in a German study population. Blood. 2005;105:906-7
45. Chen BA, Jiang N, Ji MJ, Hou P, Lu ZH, Gao C, Ding JH, Sun YY, Wang J, Cheng J. et al. [A new method for 5, 10-methylenetetrahydrofolate reductase single nucleotide polymorphisms genotyping used to study susceptibility of hematological malignancy]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2006;14:1069-73
46. Jiang N, Zhu X, Zhang H, Wang X, Zhou X, Gu J, Chen B, Ren J. The relationship between methylenetetrahydrofolate reductase polymorphism and hematological malignancy. Clin Lab. 2014;60:767-74
47. Kwasniewska A, Tukendorf A, Semczuk M. Folate deficiency and cervical intraepithelial neoplasia. Eur J Gynaecol Oncol. 1997;18:526-30
48. Giovannucci E, Stampfer MJ, Colditz GA, Hunter DJ, Fuchs C, Rosner BA, Speizer FE, Willett WC. Multivitamin use, folate, and colon cancer in women in the Nurses' Health Study. Ann Intern Med. 1998;129:517-24
49. Stolzenberg-Solomon RZ, Albanes D, Nieto FJ, Hartman TJ, Tangrea JA, Rautalahti M, Sehlub J, Virtamo J, Taylor PR. Pancreatic cancer risk and nutrition-related methyl-group availability indicators in male smokers. J Natl Cancer Inst. 1999;91:535-41
50. Shaffer AL 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565-610
51. Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820-8
Author contact
![]() Corresponding authors: Prof. Zhigang Yang (e-mail: yangzgedu.cn) or Prof. Yunmiao Guo (e-mail: yunmiaoguocom).
Corresponding authors: Prof. Zhigang Yang (e-mail: yangzgedu.cn) or Prof. Yunmiao Guo (e-mail: yunmiaoguocom).

 Global reach, higher impact
Global reach, higher impact