3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(16):5396-5402. doi:10.7150/jca.98555 This issue Cite
Research Paper
TRMT10C gene polymorphisms confer hepatoblastoma susceptibility: evidence from a seven-center case-control study
1. Department of Pediatric Surgery, Xi'an Children's Hospital, Xi'an Jiaotong University Affiliated Children's Hospital, Xi'an 710003, Shaanxi, China.
2. Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China.
3. Department of General Pediatrics, Senior Department of Pediatrics, National Engineering Laboratory for Birth Defects Prevention and Control of Key Technology, Beijing Key Laboratory of Pediatric Organ Failure, the Seventh Medical Center of PLA General Hospital, Beijing 100000, China.
4. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
5. Department of Pediatric Surgery, Hunan Children's Hospital, Changsha 410004, Hunan, China.
6. Department of Pediatric Surgery, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning, China.
7. Department of Pediatric Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China.
8. Department of Pediatric Surgery, the Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, Shaanxi, China.
9. Kunming Key Laboratory of Children Infection and Immunity, Yunnan Key Laboratory of Children's Major Disease Research, Yunnan Institute of Pediatrics Research, Yunnan Medical Center for Pediatric Diseases, Kunming Children's Hospital, Kunming 650228, Yunnan, China.
10. Department of Pathology, Children Hospital and Women Health Center of Shanxi, Taiyuan 030013, Shannxi, China.
# These authors contributed equally.
Received 2024-5-17; Accepted 2024-8-5; Published 2024-8-19
Abstract
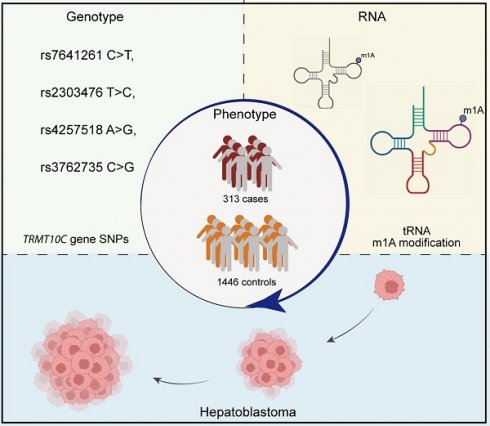
N1-methyladenosine (m1A) is a reversible epigenetic modification of RNAs. Aberrant m1A modification levels due to dysregulation of m1A regulators have been observed in multiple cancers. tRNA methyltransferase 10C (TRMT10C) can install m1A in RNAs; however, its role in hepatoblastoma remains unknown. We conducted this study to identify causal polymorphisms in the TRMT10C gene for hepatoblastoma susceptibility in a cohort of Chinese children (313 cases vs. 1446 controls). The genotypes of four potential functional polymorphisms (rs7641261 C>T, rs2303476 T>C, rs4257518 A>G, and rs3762735 C>G) were determined in participants using TaqMan real-time PCR. The associations of these polymorphisms with hepatoblastoma susceptibility were estimated by logistic regression analysis adjusted for age and sex. All four polymorphisms were significantly associated with hepatoblastoma risk. In particular, under the recessive genetic model, these polymorphisms conferred an increased risk of hepatoblastoma: rs7641261 C>T [adjusted odds ratio (OR)=1.64, 95% confidence interval (CI)=1.04-2.58, P=0.033], rs2303476 T>C (adjusted OR=1.87, 95% CI=1.16-3.02, P=0.010), rs4257518 A>G (adjusted OR=1.45, 95% CI=1.09-1.94, P=0.012), and rs3762735 C>G (adjusted OR=3.83, 95% CI=2.15-6.82, P<0.0001). Combined analysis revealed that kids had an increased risk of developing hepatoblastoma if they harbored at least one risk genotype (adjusted OR=1.94, 95% CI=1.48-2.54, P<0.0001). In addition, the combined risk effects of the four SNPs persisted across all the subgroups. We identified four hepatoblastoma susceptibility loci in the TRMT10C gene. Identifying more disease-causing loci may facilitate the development of genetic marker panels to predict individuals' hepatoblastoma predisposition.
Keywords: TRMT10C, m1A modification, polymorphism, hepatoblastoma, susceptibility
Introduction
N1-methyladenosine (m1A), a methyl group in the first nitrogen atom of adenosine, is a reversible epigenetic modification in mRNAs, tRNAs, rRNAs, and lncRNAs [1]. Positive electrostatic charge-carrying m1A modifications interfere with A:U base pairs and therefore affect the processing, structure, stability, translation, and functions of host RNAs and their interactions with other molecules [2]. Accumulating evidence indicates that dysregulation of m1A is associated with tumorigenesis, including in ovarian cancer, hepatocellular carcinoma, bladder cancer, gastric cancer, and glioma [3-7]. m1A modifications in RNAs are finely orchestrated, deposited by methyltransferases (TRMT6, TRMT10C, TRMT61A, TRMT61B, and NML) and eradicated by demethylases (ALKBH1, ALKBH3, ALKBH7, and FTO); these modifications are ultimately sensed by m1A-recognizing proteins (YTHDC1 and YTHDF1-3) [1, 5].
tRNA methyltransferase 10C (TRMT10C), also known as the mitochondrial RNase P subunit, is a bifunctional methyltransferase [8]. The TRMT10C/MRPP1-HSD17B10/MRPP2 subcomplex installs m1A and N1-methylguanine (m1G) at position 9 in tRNAs [8, 9]. TRMT10C/MRPP1 is the catalytic N1-methyltransferase subunit [8]. Moreover, this complex is also a component of mitochondrial ribonuclease P, which facilitates tRNA maturation [10]. Knowledge of the biological roles of this complex is extremely limited. Similar to other m1A methyltransferases (e.g., TRMT6 and TRMT61B) [6, 11, 12], a study suggested that TRMT10C may also be involved in carcinogenesis. Knockdown of TRMT10C suppressed the growth and migration of ovarian and cervical cancer cells [13].
Hepatoblastoma is the most frequently diagnosed malignant liver neoplasm in childhood, with an extraordinarily low incidence ranging from 1.2 to 1.5 cases per million people annually [14]. The annual incidence rate of hepatoblastoma among Chinese children is estimated to be approximately 1.4 cases per million people [15]. Despite reported environmental risk factors, such as inferior birth status, oxygen therapy, radiation, and plasticizers, hepatoblastoma is considered a disease of genetic susceptibility [16, 17]. Substantial evidence has suggested the implications of hereditary genetic variants in hepatoblastoma. Several constitutional genetic syndromes, such as trisomy 18/Edward's syndrome, Beckwith-Wiedemann syndrome (BWS), and familial adenomatous polyposis (FAP), predispose patients to hepatoblastoma. Moreover, hepatoblastoma has a lower frequency of somatic mutations than other solid pediatric tumors do [18]. In addition, hepatoblastoma susceptibility-related single-nucleotide polymorphisms (SNPs) have been reported in many genes, including genes related to RNA N6-methyladenosine (m6A) and N7-methylguanosine (m7G) modifications (e.g., WTAP, METTL3, METTL14, FTO, YTHDF1, YTHDC1, WDRT4, and METTL1) [19-26]. However, the associations between genetic polymorphisms in the TRMT10C gene and hepatoblastoma risk remain unclear. Our study aimed to identify pathogenic genetic polymorphisms for hepatoblastoma in 313 Chinese children with hepatoblastoma and 1446 age-, sex- and ethnicity-matched healthy controls.
Materials and Methods
Patients and study design
The study population included 1759 Han Chinese descendants (313 cases and 1446 controls) under 14 years of age (Table S1) [19, 27]. Hepatoblastoma was diagnosed in patients by comprehensively considering their clinical symptoms, laboratory test results, pathological reports, and imaging manifestations. Criteria for enrolling hepatoblastoma patients included: 1) Han Chinese lineage, 2) a new diagnosis with histopathological confirmation, 3) no family history of disorders or cancer, and 4) age 14 or younger. Exclusion criteria involved receiving medical intervention or not providing signed informed consent. Sufficient peripheral whole blood samples were obtained from participants for analysis. Patients and healthy controls were recruited from seven independent hospitals in the capitals of Guangdong, Yunnan, Hunan, Shaanxi, Shannxi, Henan, and Liaoning Provinces. Patients were matched to controls according to age and sex. We estimated the clinical stages of patients according to the PRETEXT classification [28]. Only children who provided informed consent and sufficient peripheral blood samples free of any treatment were included in this study. The study was evaluated and permitted by the institutional review board of Guangzhou Women and Children's Medical Center (Approval No: 202016601). This study was conducted in accordance with the Declaration of Helsinki.
Selecting and genotyping polymorphisms
Candidate SNPs in the TRMT10C gene were first selected from the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) on the basis of previously published criteria [29-31]: 1) SNPs with a minor allele frequency (MAF) of 5% in the Chinese Han population to ensure sufficient statistical power; 2) SNPs located in exonic regions, promoters and regulatory elements, splice sites, and 5' and 3' UTRs; 3) SNPs with potential functions; and 4) SNPs in low linkage disequilibrium (LD) with each other to avoid redundancy and multiple testing issues. The retrieved SNPs were subsequently entered into the SNPinfo online tool (https://snpinfo.niehs.nih.gov) to filter out SNPs without potential biological functions. For this study, we selected SNPs with low LD, specifically those with R2<0.8, according to the web tool LDlink (https://ldlink.nih.gov/?tab=ldmatrix). Four TRMT10C gene SNPs (rs7641261 C>T, rs2303476 T>C, rs4257518 A>G, and rs3762735 C>G) were eligible for the study [32]. The rs7641261 C>T, rs2303476 T>C, and rs4257518 A>G are located in the transcription factor binding site (TFBS), a region to which a specialized protein binds to increase or decrease the efficiency of gene transcription. rs3762735 C>G is a nonsynonymous SNP that results in a change in the amino acid sequence of a protein, thereby leading to changes in the function and 3D structure of the protein. We used Tiangen Blood DNA Extraction kits (Tiangen Biotechnology, Beijing, China) to recover genomic DNA from subjects' peripheral blood samples. Genotyping assays using TaqMan real-time PCR were executed on a TaqMan instrument (Applied Biosystems, Foster City, CA). For quality control, no-template controls were included in each 384-well plate to check for contamination, along with known genotype controls, to validate the assay performance. Each 384-well plate had at least four positive controls with different genotypes (e.g., homozygous for both alleles and heterozygous). We included technical replicates within the same 384-well plate to assess reproducibility, and we randomly selected 5-10% of the samples per plate for repeat genotyping to verify consistency, aiming for >99% concordance.
Statistical analysis
The Hardy‒Weinberg equilibrium (HWE) of the SNPs was assessed in the controls using a goodness-of-fit χ2 test. We performed logistic regression analysis to calculate odds ratios (ORs) and 95% confidence intervals (CIs) for the associations between selected SNPs and hepatoblastoma susceptibility with the inclusion of adjusted covariates (age and sex). The common and minor alleles of each SNP are denoted W (wild) and M (mutant), respectively. Different genetic models, including homozygous (MM vs. WW), heterozygous (MW vs. WW), dominant (MW/MM vs. WW), and recessive (MM vs. MW/WW) models, were employed to evaluate the associations between SNPs and hepatoblastoma risk in the whole study population. Association studies were also performed for subgroups classified by age, sex, and clinical stage. SAS v9.1 software (SAS Institute Inc., Cary, NC) was used for the two-tailed statistical analyses. P values < 0.05 indicated significance.
Results
Association of TRMT10C gene polymorphisms with hepatoblastoma susceptibility
Age, gender, and clinical information of participants are presented in (Table S1). We obtained genotype information on rs7641261 C>T, rs2303476 T>C, rs4257518 A>G, and rs3762735 C>G for 294 cases and 1444 controls. The goodness-of-fit chi-square test confirmed that the genotype frequencies of all the SNPs were distributed according to Hardy‒Weinberg equilibrium in the normal controls (Table 1). Compared with the CC genotype, the 7641261 CT genotype conferred a decreased risk of developing hepatoblastoma of 0.36 (adjusted OR=0.64, 95% CI=0.48-0.85, P=0.002). Interestingly, the 7641261 C>T polymorphism was associated with decreased hepatoblastoma risk under the dominant genetic model (adjusted OR=0.75, 95% CI=0.57-0.97, P=0.029) but increased risk under the recessive genetic model (adjusted OR=1.64, 95% CI=1.04-2.58, P=0.033). Similar associations with hepatoblastoma risk were found for rs4257518 A>G under the heterozygous (adjusted OR=0.59, 95% CI=0.44-0.79, P=0.0003), dominant (adjusted OR=0.74, 95% CI=0.58-0.96, P=0.025), and recessive (adjusted OR=1.45, 95% CI=1.09-1.94, P=0.012) genetic models (Table 1). The rs2303476 T>C was shown to increase hepatoblastoma risk by 1.83-fold under the homozygous model (adjusted OR=1.83, 95% CI=1.12-2.97, P=0.015) and by 1.87-fold under the recessive model (adjusted OR=1.87, 1.16-3.02, P=0.010). The opposite results were found for the rs3762735 C>G polymorphism under the heterozygous (adjusted OR=0.51, 95% CI=0.35-0.75, P=0.0005), homozygous (adjusted OR=3.42, 95%=1.91-6.11, P<0.0001), and recessive (adjusted OR=3.83, 95%=2.15-6.82, P<0.0001) genetic models. We next referred to rs7641261 TT, rs2303476 CC, rs4257518 GG, and rs3762735 GG as risk genotypes and found combined effects of the four SNPs on hepatoblastoma risk. The adjusted OR for the subpopulation of children with 1-4 risk genotypes was 1.94 (95% CI=1.48-2.54, P<0.0001), indicating the significant deleterious effects of possessing one or more risk genotypes.
Stratification analysis
Stratification analysis was conducted under the dominant genetic model. The study population was dichotomized by age, sex, and clinical stage. Surprisingly, no significant associations were detected for the rs7641261 polymorphism among all the subgroups (Table 2). Moreover, we found a significant association with the rs4257518 polymorphism in boys (OR=0.67, 95%=0.48-0.94, P=0.021). Finally, the concurrence of 1-4 risk genotypes was demonstrated to significantly increase hepatoblastoma risk among all subgroups divided by age, sex, and clinical stage. This finding suggests the robustness of the risk effects of the combined SNPs on hepatoblastoma predisposition.
Association of TRMT10C gene polymorphisms with hepatoblastoma susceptibility.
| Genotype | Cases (N=294) | Controls (N=1444) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs7641261 C>T (HWE=0.431) | |||||||
| CC | 194 (65.99) | 854 (59.14) | 1.00 | 1.00 | |||
| CT | 73 (24.83) | 506 (35.04) | 0.64 (0.48-0.85) | 0.002 | 0.64 (0.48-0.85) | 0.002 | |
| TT | 27 (9.18) | 84 (5.82) | 1.42 (0.89-2.24) | 0.140 | 1.42 (0.89-2.25) | 0.140 | |
| Additive | 0.376 | 0.91 (0.74-1.12) | 0.376 | 0.91 (0.74-1.12) | 0.376 | ||
| Dominant | 100 (34.01) | 590 (40.86) | 0.029 | 0.75 (0.57-0.97) | 0.029 | 0.75 (0.57-0.97) | 0.029 |
| Recessive | 267 (90.82) | 1360 (94.18) | 0.031 | 1.64 (1.04-2.58) | 0.033 | 1.64 (1.04-2.58) | 0.033 |
| rs2303476 T>C (HWE=0.441) | |||||||
| TT | 180 (61.43) | 908 (62.88) | 1.00 | 1.00 | |||
| TC | 88 (30.03) | 468 (32.41) | 0.94 (0.71-1.25) | 0.681 | 0.94 (0.71-1.24) | 0.664 | |
| CC | 25 (8.53) | 68 (4.71) | 1.84 (1.14-3.00) | 0.013 | 1.83 (1.12-2.97) | 0.015 | |
| Additive | 0.166 | 1.16 (0.94-1.42) | 0.166 | 1.15 (0.94-1.42) | 0.180 | ||
| Dominant | 113 (38.57) | 536 (37.12) | 0.641 | 1.06 (0.82-1.38) | 0.641 | 1.06 (0.82-1.37) | 0.666 |
| Recessive | 268 (91.47) | 1376 (95.29) | 0.008 | 1.89 (1.17-3.04) | 0.009 | 1.87 (1.16-3.02) | 0.010 |
| rs4257518 A>G (HWE=0.092) | |||||||
| AA | 117 (39.93) | 478 (33.10) | 1.00 | 1.00 | |||
| AG | 98 (33.45) | 678 (46.95) | 0.59 (0.44-0.78) | 0.0003 | 0.59 (0.44-0.79) | 0.0003 | |
| GG | 78 (26.62) | 288 (19.94) | 1.10 (0.80-1.51) | 0.572 | 1.09 (0.79-1.51) | 0.586 | |
| Additive | 0.974 | 1.00 (0.84-1.18) | 0.974 | 1.00 (0.84-1.18) | 0.957 | ||
| Dominant | 176 (60.07) | 966 (66.90) | 0.025 | 0.74 (0.58-0.96) | 0.025 | 0.74 (0.58-0.96) | 0.025 |
| Recessive | 215 (73.38) | 1156 (80.06) | 0.011 | 1.46 (1.09-1.95) | 0.011 | 1.45 (1.09-1.94) | 0.012 |
| rs3762735 C>G (HWE=0.167) | |||||||
| CC | 239 (81.29) | 1107 (76.66) | 1.00 | 1.00 | |||
| CG | 34 (11.56) | 308 (21.33) | 0.51 (0.35-0.75) | 0.0006 | 0.51 (0.35-0.75) | 0.0005 | |
| GG | 21 (7.14) | 29 (2.01) | 3.35 (1.88-5.98) | <0.0001 | 3.42 (1.91-6.11) | <0.0001 | |
| Additive | 0.874 | 1.02 (0.79-1.31) | 0.873 | 1.02 (0.80-1.31) | 0.864 | ||
| Dominant | 55 (18.71) | 337 (23.34) | 0.083 | 0.76 (0.55-1.04) | 0.084 | 0.76 (0.55-1.04) | 0.084 |
| Recessive | 273 (92.86) | 1415 (97.99) | <0.0001 | 3.75 (2.11-6.68) | <0.0001 | 3.83 (2.15-6.82) | <0.0001 |
| Risk genotypes c | |||||||
| 0 | 189 (64.29) | 1123 (77.77) | 1.00 | 1.00 | |||
| 1-4 | 105 (35.71) | 321 (22.23) | <0.0001 | 1.94 (1.49-2.54) | <0.0001 | 1.94 (1.48-2.54) | <0.0001 |
OR, odds ratio; CI, confidence interval; HWE, Hardy‒Weinberg equilibrium.
Values were in bold if the 95% CIs excluding 1.00 or the P values less than 0.05.
a χ2 test for genotype distributions between hepatoblastoma patients and cancer-free controls.
b Adjusted for age and sex.
c Risk genotypes were rs7641261 TT, rs2303476 CC, rs4257518 GG, and rs3762735 GG.
Stratification analysis of the association between TRMT10C genotypes and hepatoblastoma susceptibility.
| Variables | rs7641261 (cases/controls) | AOR (95% CI) a | P a | rs4257518 (cases/controls) | AOR (95% CI) a | P a | Risk genotypes (cases/controls) | AOR (95% CI) a | P a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT/TT | AA | AG/GG | 0 | 1-4 | |||||||
| Age, months | ||||||||||||
| <17 | 103/381 | 52/260 | 0.75 (0.52-1.08) | 0.123 | 63/213 | 91/428 | 0.72 (0.50-1.04) | 0.077 | 102/495 | 53/146 | 1.78 (1.22-2.60) | 0.003 |
| ≥17 | 91/473 | 48/330 | 0.76 (0.52-1.10) | 0.144 | 54/265 | 85/538 | 0.78 (0.54-1.13) | 0.182 | 87/628 | 52/175 | 2.13 (1.46-3.13) | 0.0001 |
| Sex | ||||||||||||
| Females | 77/344 | 43/251 | 0.77 (0.51-1.15) | 0.198 | 44/200 | 75/395 | 0.86 (0.57-1.30) | 0.482 | 76/450 | 44/145 | 1.80 (1.19-2.73) | 0.006 |
| Males | 117/510 | 57/339 | 0.73 (0.52-1.04) | 0.080 | 73/278 | 101/571 | 0.67 (0.48-0.94) | 0.021 | 113/673 | 61/176 | 2.05 (1.44-2.92) | <0.0001 |
| Clinical stages | ||||||||||||
| I+II | 100/854 | 52/590 | 0.75 (0.53-1.07) | 0.112 | 62/478 | 89/966 | 0.71 (0.51-1.00) | 0.051 | 103/1123 | 49/321 | 1.65 (1.15-2.38) | 0.007 |
| III+IV | 57/854 | 29/590 | 0.74 (0.47-1.17) | 0.197 | 37/478 | 49/966 | 0.66 (0.42-1.02) | 0.060 | 57/1123 | 29/321 | 1.80 (1.13-2.87) | 0.013 |
AOR, adjusted odds ratio; CI, confidence interval.
Values were in bold if the 95% CIs excluding 1.00 or the P values less than 0.05.
a Adjusted for age and sex, omitting the corresponding stratification factor
Discussion
Compared with adult tumors, solid pediatric tumors generally harbor far lower tumor mutation burdens; in particular, hepatoblastoma has the fewest somatic mutations [16, 33, 34]. Therefore, heritable genetic variants in cancer predisposition genes play crucial roles in the pathogenesis of hepatoblastoma.
Here, we performed a candidate gene association study to identify hepatoblastoma susceptibility loci in the TRMT10C gene. We collected genotype data for four SNPs (rs7641261 C>T, rs2303476 T>C, rs4257518 A>G, and rs3762735 C>G) from 294 cases and 1444 controls. All these SNPs independently modulated hepatoblastoma susceptibility to some extent. Moreover, combination analyses indicated that children with one or more risk genotypes of the four SNPs had a significantly greater risk of developing hepatoblastoma than noncarriers did. Stratification analysis further revealed that the existence of at least one genotype conferred hepatoblastoma susceptibility regardless of age, sex, or clinical stage. The protein product of the TRMT10C gene is a tRNA N1-methyltransferase, which, coupled with HSD17B10/MRPP2, catalyzes m1A modifications in RNAs [9]. RNA modifications, including m6A, m1A, 5-methylcytidine (m5C), m7G, and 2′-O-methylation (2′-O-Me), are garnering increasing attention in cancer research because of their ability to regulate gene expression and tumorigenesis [35]. The role of m6A RNA methylation has been investigated most often in cancer, followed by m7G [5]. Like m1A modifications, m6A and m7G RNA modifications are also dynamically controlled and recognized by regulatory enzymes categorized as writers, erasers, and readers [35]. Using the candidate gene method, we verified that these m6A and m7G regulators are susceptibility genes in several solid pediatric cancers, including glioma [36, 37], neuroblastoma [38], Wilms tumor [39-41], and hepatoblastoma [20-26].
Although less studied, m1A modifications are known to regulate diverse cellular functions in carcinogenesis, such as proliferation, invasion, cell metabolism, senescence and cell death, and the tumor microenvironment [42]. The TRM6/61 methyltransferase complex can mediate the methylation of adenosine 58 of the initiator methionine tRNA (tRNAi Met) [43]. Elevated TRM6/61 expression promotes the anchorage-independent growth and tumor sphere formation of C6 cells (rat glioma cells) by regulating the translation of relevant mRNAs [43]. ALKBH3, an m1A demethylase of tRNA, is also reported to be oncogenic [44]. ALKBH3-mediated m1A-demethylated tRNA is vulnerable to angiogenin cleavage, a process that generates apoptosis-preventing tRNA-derived small RNAs (tDRs). Therefore, ALKBH3 can accelerate cancer progression by promoting cancer cell proliferation and invasion [44]. However, the role of TRMT10C in cancer has just begun to emerge. To date, only one publication has suggested that TRMT10C is significantly upregulated in CESC, OV, and UCEC and is associated with an unfavorable prognosis in patients with gynecological cancers [13]. In vitro functional analyses revealed that depletion of TRMT10C inhibited the malignant behaviors of ovarian cancer and cervical cancer cells [13]. Consistent with previous association studies with genetic polymorphisms in m6A- and m7G-related genes, our results indicate that TRMT10C is also a hepatoblastoma susceptibility gene. Our results are biologically plausible, although the mechanisms by which TRMT10C gene SNPs affect hepatoblastoma susceptibility are unknown.
Ali and colleagues reported that mitochondrial m1A/G RNA modification levels are associated with genetic polymorphisms in the tRNA methyltransferase TRMT61B gene and the MRPP3 gene encoding mitochondrial ribonuclease P protein 3, which is responsible for mitochondrial tRNA 5'-end processing [45]. Minor alleles of the MRPP3 missense SNP rs11156878 are significantly associated with increased methylation levels of tRNA P9 [45]. Moreover, an intronic TRMT61B rs11684695 polymorphism was shown to significantly increase RNA2 methylation in blood samples [45]. More importantly, the same study demonstrated that the SNPs associated with inferred RNA methylation levels are frequently disease-causing in glaucoma, psoriasis, and breast cancer [45].
It is well documented that potential functional SNPs that may influence expression and function could modify disease susceptibility. The SNPs rs7641261 C>T, rs2303476 T>C, and rs4257518 A>G are positioned in transcription factor binding sites, where they affect TRMT10C gene transcription by enabling the binding of particular proteins. On the other hand, rs3762735 C>G is a nonsynonymous SNP that causes an amino acid substitution, impacting both the function and 3D structure of the TRMT10C protein. For instance, the nonsynonymous SNPs C677T (rs1801133) and A1298C (rs1801131) in the MTHFR gene reduce the enzyme activity of the gene-encoded protein, 5,10-methylenetetrahydrofolate reductase, consequently affecting non-Hodgkin lymphoma susceptibility [46]. Overall, it is reasonable to speculate that the studied SNPs that may alter TRMT10C expression and function could modify hepatoblastoma susceptibility by altering the m1A modification levels of target RNAs and inducing the crucial effects of m1A on fundamental cell processes.
This study has several limitations. First, this study was a retrospective case‒control study, and our conclusions should be confirmed in future prospective studies. Second, the number of cases needs to be expanded. Third, the effects of the significant SNPs on the expression levels and functions of the TRMT10C gene should be tested in clinical samples and in vitro studies. Fourth, the study cohort was exclusively composed of Han Chinese children, which ensures a homogeneous genetic background and minimizes confounding effects related to ethnic diversity. However, future research should include diverse populations to validate these findings and evaluate their broader applicability. Expanding the study to other ethnic groups will help assess the generalizability of the results and identify potential ethnic-specific genetic or environmental risk factors. Finally, the current analysis does not account for environmental influences, which are critical in disease development. Future investigations should integrate environmental factors, such as exposure to toxins, birth status, and socioeconomic conditions. This comprehensive approach could provide deeper insights into the multifactorial nature of hepatoblastoma susceptibility and enhance our understanding of the interplay between genetics and the environment.
In conclusion, we identified four potential functional SNPs in the TRMT10C gene as hepatoblastoma susceptibility loci. The discovery of TRMT10C polymorphisms as susceptibility loci may make a valuable contribution to the understanding of genetic risk factors for hepatoblastoma. Our results suggest that the role of TRMT10C in the tumorigenesis of hepatoblastoma should be explored. The discovery of new susceptibility SNPs may help improve hepatoblastoma risk assessment.
Abbreviations
m1A, N1-methyladenosine; TRMT10C, tRNA methyltransferase 10C; m1G, N1-methylguanine; BWS, Beckwith-Wiedemann syndrome; FAP, familial adenomatous polyposis; SNP, single-nucleotide polymorphism; m6A, N6-methyladenosine; m7G, N7-methylguanosine; MAF, minor allele frequency; LD, linkage disequilibrium; TFBS, transcription factor binding site; HWE, Hardy-Weinberg equilibrium; OR, odds ratio; CI, confidence interval; m5C, 5-methylcytidine; 2′-O-Me, 2′-O-methylation; tDRs, tRNA-derived small RNAs.
Supplementary Material
Supplementary table.
Acknowledgements
Funding
This study was supported by grants from the Natural Science Foundation of Shaanxi Province (No: 2023-JC-YB-807), Shaanxi Province Key R&D Program (No: 2022SF-257), and the Innovation and Cultivation Fund Project of the Seventh Medical Center, PLA General Hospital (No: QZX-2023-7).
Data availability statement
All the data are available upon request from the corresponding authors.
Ethical approval
Only children who provided informed consent and sufficient peripheral blood samples free of any treatment were included in this study. The study was evaluated and permitted by the institutional review board of Guangzhou Women and Children's Medical Center (Approval No: 202016601). This study was conducted in accordance with the Declaration of Helsinki.
Author contributions
All the authors contributed significantly to this work. Yong Li, Zhonghua Yang, Jiao Zhang, Jiwen Cheng, Li Li, Suhong Li, and Jing He performed the research study and collected the samples and information; Wenli Zhang and Jing He analyzed the data; Jun Bian and Jing He designed the research study and supervised the project; Yanfei Liu, Jinhong Zhu, Xianqiang Wang, Jing He, and Jun Bian wrote the paper; Wenli Zhang and Jing He prepared all the tables. All the authors reviewed the manuscript. In addition, all the authors have read and approved the final manuscript.
ORCID
Jing He, https://orcid.org/0000-0002-1954-2892.
Jun Bian, https://orcid.org/0000-0002-8507-5479.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li J, Zhang H, Wang H. N(1)-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J. 2022;20:6578-85
2. Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79-129
3. Ye L, Yao X, Xu B, Chen W, Lou H, Tong X. et al. RNA epigenetic modifications in ovarian cancer: The changes, chances, and challenges. Wiley Interdiscip Rev RNA. 2023;14:e1784
4. Feng Q, Wang D, Xue T, Lin C, Gao Y, Sun L. et al. The role of RNA modification in hepatocellular carcinoma. Front Pharmacol. 2022;13:984453
5. Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L. et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther. 2022;7:334
6. Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R. et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun. 2022;13:2165
7. Wang B, Niu L, Wang Z, Zhao Z. RNA m1A Methyltransferase TRMT6 Predicts Poorer Prognosis and Promotes Malignant Behavior in Glioma. Front Mol Biosci. 2021;8:692130
8. Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583-93
9. Oerum S, Roovers M, Rambo RP, Kopec J, Bailey HJ, Fitzpatrick F. et al. Structural insight into the human mitochondrial tRNA purine N1-methyltransferase and ribonuclease P complexes. J Biol Chem. 2018;293:12862-76
10. Reinhard L, Sridhara S, Hallberg BM. The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria. Nucleic Acids Res. 2017;45:12469-80
11. Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z. et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12:6314
12. Martin A, Epifano C, Vilaplana-Marti B, Hernandez I, Macias RIR, Martinez-Ramirez A. et al. Mitochondrial RNA methyltransferase TRMT61B is a new, potential biomarker and therapeutic target for highly aneuploid cancers. Cell Death Differ. 2023;30:37-53
13. Wang Q, Zhang Q, Huang Y, Zhang J. m(1)A Regulator TRMT10C Predicts Poorer Survival and Contributes to Malignant Behavior in Gynecological Cancers. DNA Cell Biol. 2020;39:1767-78
14. Czauderna P, Lopez-Terrada D, Hiyama E, Haberle B, Malogolowkin MH, Meyers RL. Hepatoblastoma state of the art: pathology, genetics, risk stratification, and chemotherapy. Curr Opin Pediatr. 2014;26:19-28
15. Bao PP, Li K, Wu CX, Huang ZZ, Wang CF, Xiang YM. et al. [Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010]. Zhonghua Er Ke Za Zhi. 2013;51:288-94
16. Capasso M, Montella A, Tirelli M, Maiorino T, Cantalupo S, Iolascon A. Genetic Predisposition to Solid Pediatric Cancers. Front Oncol. 2020;10:590033
17. Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J. et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med. 2015;373:2336-46
18. Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371-6
19. Zhuo ZJ, Hua RX, Chen Z, Zhu J, Wang M, Yang Z. et al. WTAP Gene Variants Confer Hepatoblastoma Susceptibility: A Seven-Center Case-Control Study. Mol Ther Oncolytics. 2020;18:118-25
20. Chen H, Duan F, Wang M, Zhu J, Zhang J, Cheng J. et al. Polymorphisms in METTL3 gene and hepatoblastoma risk in Chinese children: A seven-center case-control study. Gene. 2021;800:145834
21. Chen H, Chen Z, Wang M, Zhang J, Li Y, Li L. et al. METTL14 gene polymorphisms influence hepatoblastoma predisposition in Chinese children: Evidences from a seven-center case-control study. Gene. 2022;809:146050
22. Fan J, Zhuo Z, Chen G, Niu H, Yang Z, Zhang J. et al. FTO gene polymorphisms and hepatoblastoma susceptibility among Chinese children. Cell Cycle. 2022;21:1512-8
23. Luo Z, Li G, Wang M, Zhu J, Yang Z, Li Y. et al. YTHDF1 rs6090311 A>G polymorphism reduces Hepatoblastoma risk: Evidence from a seven-center case-control study. J Cancer. 2020;11:5129-34
24. Chen H, Li Y, Li L, Zhu J, Yang Z, Zhang J. et al. YTHDC1 gene polymorphisms and hepatoblastoma susceptibility in Chinese children: A seven-center case-control study. J Gene Med. 2020;22:e3249
25. He S, Zhu J, Xiao Z, Liu J, Zhang J, Li Y. et al. WDR4 gene polymorphisms increase hepatoblastoma susceptibility in girls. J Cancer. 2022;13:3342-7
26. Ge L, Zhu J, Liu J, Li L, Zhang J, Cheng J. et al. METTL1 gene polymorphisms synergistically confer hepatoblastoma susceptibility. Discov Oncol. 2022;13:77
27. Zhuo Z, Miao L, Hua W, Chen H, Yang Z, Li Y. et al. Genetic variations in nucleotide excision repair pathway genes and hepatoblastoma susceptibility. Int J Cancer. 2021;149:1649-58
28. Roebuck DJ, Aronson D, Clapuyt P, Czauderna P, de Ville de Goyet J, Gauthier F. et al. 2005 PRETEXT: a revised staging system for primary malignant liver tumours of childhood developed by the SIOPEL group. Pediatr Radiol. 2007;37:123-32 quiz 249-50
29. Zhu J, Wang M, He J, Zhu M, Wang JC, Jin L. et al. Polymorphisms in the AKT1 and AKT2 genes and oesophageal squamous cell carcinoma risk in an Eastern Chinese population. J Cell Mol Med. 2016;20:666-77
30. Chen YP, Liao YX, Zhuo ZJ, Yuan L, Lin HR, Miao L. et al. Association between genetic polymorphisms of base excision repair pathway and glioma susceptibility in Chinese children. World J Pediatr. 2022;18:632-5
31. Guan Q, Lin H, Hua W, Lin L, Liu J, Deng L. et al. Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression. Chin J Cancer Res. 2023;35:140-62
32. Liao F, Hua RX, Jia X, Liao Y, Yuan L, Ruan J. et al. Association of m1A modification gene polymorphisms with glioma risk in Chinese children. MedComm-Oncology. 2023;2:e43
33. Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA. et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321-7
34. Sweet-Cordero EA, Biegel JA. The genomic landscape of pediatric cancers: Implications for diagnosis and treatment. Science. 2019;363:1170-5
35. Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119-31
36. He J, Yuan L, Lin H, Lin A, Chen H, Luo A. et al. Genetic variants in m(6)A modification core genes are associated with glioma risk in Chinese children. Mol Ther Oncolytics. 2021;20:199-208
37. Zhu J, Liu X, Chen W, Liao Y, Liu J, Yuan L. et al. Association of RNA m(7)G Modification Gene Polymorphisms with Pediatric Glioma Risk. Biomed Res Int. 2023;2023:3678327
38. Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z. et al. METTL14 Gene Polymorphisms Confer Neuroblastoma Susceptibility: An Eight-Center Case-Control Study. Mol Ther Nucleic Acids. 2020;22:17-26
39. Zhuo Z, Hua RX, Zhang H, Lin H, Fu W, Zhu J. et al. METTL14 gene polymorphisms decrease Wilms tumor susceptibility in Chinese children. BMC Cancer. 2021;21:1294
40. Deng L, Hua RX, Deng C, Zhu J, Zhang Z, Cheng J. et al. WDR4 gene polymorphisms and Wilms tumor susceptibility in Chinese children: A five-center case-control study. J Cancer. 2023;14:1293-300
41. Deng L, Hua R, Zhang Z, Zhu J, Zhang J, Cheng J. et al. METTL1 gene polymorphisms and Wilms tumor susceptibility in Chinese children: A five-center case-control study. Chin Med J (Engl). 2023;136:1750-2
42. Ye Y, Liu M, Wu F, Ou S, Wang W, Fei J. et al. TRMT6 promotes hepatocellular carcinoma progression through the PI3K/AKT signaling pathway. Eur J Med Res. 2023;28:48
43. Macari F, El-Houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J. et al. TRM6/61 connects PKCalpha with translational control through tRNAi(Met) stabilization: impact on tumorigenesis. Oncogene. 2016;35:1785-96
44. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-45
45. Ali AT, Idaghdour Y, Hodgkinson A. Analysis of mitochondrial m1A/G RNA modification reveals links to nuclear genetic variants and associated disease processes. Commun Biol. 2020;3:147
46. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY. et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159
Author contact
![]() Corresponding authors: Jun Bian, Department of Pediatric Surgery, Xi'an Children's Hospital, Xi'an Jiaotong University Affiliated Children's Hospital, 69 Xiju Court Lane, Xi'an 710003, Shaanxi, China, Tel./Fax: (86-029) 87692108, Email: blandbirdcom or bianjun0904xjtu.edu.cn; Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (86-020) 38076560, Email: hejingorg or hejing198374com.
Corresponding authors: Jun Bian, Department of Pediatric Surgery, Xi'an Children's Hospital, Xi'an Jiaotong University Affiliated Children's Hospital, 69 Xiju Court Lane, Xi'an 710003, Shaanxi, China, Tel./Fax: (86-029) 87692108, Email: blandbirdcom or bianjun0904xjtu.edu.cn; Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (86-020) 38076560, Email: hejingorg or hejing198374com.

 Global reach, higher impact
Global reach, higher impact