Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(16):5403-5414. doi:10.7150/jca.98656 This issue Cite
Review
Oncogenic-tsRNA: A novel diagnostic and therapeutic molecule for cancer clinic
1. The First Clinical College of Hunan University of Chinese Medicine & Hunan Cancer Hospital, Changsha, 410007, China.
2. Hunan Key Laboratory of Oncotarget Gene and Clinical Laboratory of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha 410013, China.
Received 2024-5-20; Accepted 2024-7-29; Published 2024-8-19
Abstract
tsRNA (tRNA-derived small RNA) is derived from mature tRNA or precursor tRNA (pre-tRNAs). It is lately found that tsRNA's aberrant expression is associated with tumor occurrence and development, it may be used a molecule of diagnosis and therapy. Based on the cleavage position of pre-tRNAs or mature tRNAs, tsRNAs are classified into two categories: tRNA-derived fragments (tRFs) and tRNA halves (also named tiRNAs or tRHs). tsRNAs display more stability within cells, tissues, and peripheral blood than other small non-coding RNAs (sncRNAs), and play a role of stable entities that function in various biological contexts, thus, they may serve as functional molecules in human disease. Recently, tsRNAs have been found in a large number of tumors including such as lung cancer, breast cancer, gastric cancer, colorectal cancer, liver cancer, and prostate cancer. Although the biological function of tsRNAs is still poorly understood, increasing evidences have indicated that tsRNAs have a great significance and potential in early tumor screening and diagnosis, therapeutic targets and application, and prognosis. In the present review, we mainly describe tsRNAs in tumors and their potential clinical value in early screening and diagnosis, therapeutic targets and application, and prognosis, it provides theoretical support and guidance for further revealing the therapeutic potential of tsRNAs in tumor.
Keywords: tsRNAs, tumor, diagnosis, targeted therapy, biomarker
Introduction
tRNA-derived small RNAs (tsRNAs) are a novel class of functional RNA molecules that are derived from mature tRNAs or precursor tRNAs (pre-tRNAs), and are aberrantly expressed under various conditions, such as ultraviolet radiation, arsenite, heat shock, hypoxia, oxidative damage or viral infection [1-4]. tRNAs are initially transcribed by RNA polymerase III in the nucleus in the form of pre-tRNAs [5]. Mature tRNAs are usually 73-93 nucleotides (nts) in length and are characterized by a cloverleaf-shaped secondary structure with five arms designated as amino acid, including D, anticodon, variable and the TψC arms, respectively. Any additional nucleotides beyond nucleotide 73 are accommodated in the variable-loop or in the D-loop. Two of the tRNA arms are critical for its functions as an adaptor: the amino acid arm carrying a specific amino acid and the anticodon arm containing an anticodon that recognizes the codon in template mRNA. Moreover, the cloverleaf secondary structure is folded into an L-shaped tertiary structure with an amino acid arm and an anticodon arm located at separate ends [5]. With rapid advances in high-throughput sequencing technologies, many studies have reported that tsRNAs participate in the cell biology including gene regulation, transposon repression, and disease onset and progression [6-9]. Since tsRNAs are derived from tRNAs with heavy modifications and characteristic structures, they display more stability in cells, tissues, and peripheral blood than other small non-coding RNAs (sncRNAs) [10]. The heterogeneous population of tsRNAs represents distinct and stable entities that function in various biological contexts, including stress responses, tumorigenesis, stem cell biology, and epigenetic inheritance [11]. Thus, tsRNA may serve as marker molecules in human disease. Accumulating evidence has demonstrated that tsRNAs play a critical role in human cancer [3, 4, 12-19], with its potential as a biomarker for early screening, diagnosis and prognosis of tumors, but also as an important target for tumor therapy.
tsRNA belongs to one species of sncRNA. sncRNAs include microRNA (miRNA), small interfering RNA (siRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), PIWI-interacting RNA (piRNA), and RNA-derived small RNA (tsRNA). Short or small interfering RNAs (siRNAs) and microRNAs (miRNAs) are molecules similar in size [20]. The expression of some snoRNAs is cell type specific and their biogenesis is dynamically regulated during body development. snoRNA is stable, which occurs mainly by binding with proteins to form a complex that enhances stability, however, the specific mechanism of the binding remains unclear [21]. The biogenesis of snRNA is regulated by specific developmental phenomenas (such as being regulated by certain cellular stress conditions, or closely related to RNA polymerase II) and is largely conserved in its expression [21]. The piRNA has permanently been considered germline-specific. With further studies, the tissue specificity of somatic piRNA has been confirmed. Mature piRNA stability is mainly achieved by methylation methyltransferase 2'-o-methylated, and while the potential mechanisms of the biogenesis (such as ethylation modification or other modifications) have been less studied [21]. The tsRNAs have different functions from the other sncRNAs (siRNA, miRNA, snoRNA, snRNA, and piRNA). However, tsRNAs regulate gene expression mainly at post-transcription suppression, post-transcriptional, translation and post-translational, and reverse transcription regulation. siRNAs and miRNAs are complementary to its corresponding mRNA sequences, which can inhibit gene expression, inducing the degradation of the transcript or the inhibition of their translation. siRNAs bind specifcally to a single gene location by sequence complementarity and regulate gene expression by specifcally targeting transcription units to silence osttranscriptional gene. miRNAs can regulate the expression of diferent gene targets through their imperfect base pairing. siRNAs and miRNAs have been extensively studied in the past decades and their contribution to the development of various pathologenses are now well established for the treatment of different diseases including cancer [20]. snoRNAs guide post-transcriptional covalent modifications that alter RNA biophysical properties and can also modify each other's modification. In addition, selective splicing can also be regulated [21]. piRNAs can interact with PIWI protein to silence transposons. After translation, piRNAs can regulate the stability of its interacting proteins by binding to proteins [21]. Some tsRNAs can also act as a piRNA to interact with PIWI protein to regulate gene expression at the transcriptional level. tsRNA can interact with Argonaute protein to form RNA-induced silencing complex and inhibit gene expression through RNA interference. Besides, tsRNA can be used as a protein bait to affect the stability of mRNA. tsRNA can inhibit translation initiation either through binding to translation initiation complex or inhibiting transcription initiation. During retroviral cycle, cellular tRNAs serve as primers for reverse transcription in the synthesis of synthesise minus-strand cDNA, and are placed onto the site of the viral RNA. These indicate that tsRNA plays a double-edged sword in reverse transcription regulation. In the present review, we mainly describe various tsRNA in tumor and their function mechanism in tumorigenesis. The potential clinical significance of tsRNAs were evaluated in cancer early-screening and diagnosis, targeted therapy and application, and prognosis. This article provides a theoretical support and guidance for further revealing the clinical potential of tsRNAs in tumors.
Classification and biogenesis of tsRNA
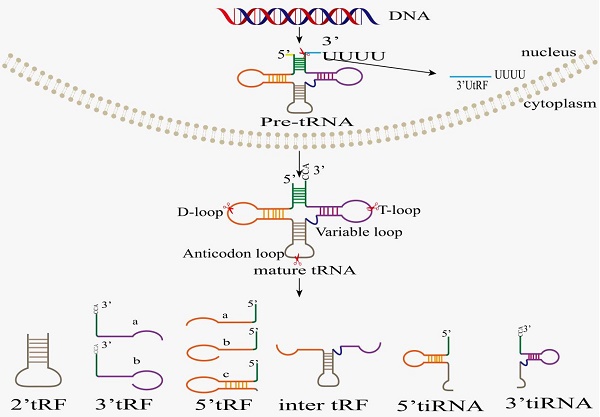
It is worthwhile to note that tsRNAs are neither remnants of tRNA maturation, nor are they random tRNA degradation products. Conversely, tsRNAs are generated by cleavage of pre-tRNAs or mature tRNAs at specific sites, and their appearance is not accompanied by a significant depletion of their cognate tRNAs [22, 23]. Based on the cleavage position of pre-tRNAs or mature tRNAs, tsRNAs are classified into two categories, tRNA-derived fragments (tRFs) and tRNA halves (also named tiRNAs or tRHs) (Figure 1). Based on their mapping positions on pre-tRNAs and mature tRNAs, tRFs and tiRNAs are divided into several subclasses [24]. tRFs are non-coding RNAs with approximately 14-30 nt in length, and they are grouped into 5 subclasses: tRF-1s (1'tRFs), tRF-3s (3'tRFs), tRF-5s (5'tRFs), tRF-2s (2'tRFs), and i-tRFs (inter tRFs, i'tRFs)[25]. tRF-1s (also named 3′U-tRFs) are derived from the 3'-end of pre-tRNA mainly through endonuclease Z (RNase Z/ELAC2) digestion [24, 26-30], which results in the presence of poly-U residues at the 3'end of tRF-1s [31]. tRF-2s only contain anticodon stem-loop sequences and are induced in hypoxic conditions [3]. tRF-3s come from TψC-loop cleavage of mature tRNAs by Dicer or other unknown nucleases [32-34], and they are classified into two subtypes, tRF-3a and tRF-3b, with lengths of 18 and 22 nucleotides, respectively. The 18-nucleotide tRF-3a is generated by the cleavage between 58th and 59th nucleotides on the TΨC-loop. The 22-nucleotide tRF-3b is produced by cleavage between 54th and 55th nucleotides on the TΨC-loop. The end of TΨC-loop sequence contains CCA sequence, which is added to TΨC-loop during mature tRNA processing. tRF-5s minimally extend to D-loop and maximally to anticodon stem, and they are classified into tRF-5a (14-16 nt), tRF-5b (22-24 nt), and tRF-5c (28-30 nt) subtypes [35] based on the cleavage by Dicer, RNase T2, or RNase A, respectively [32, 33, 35]. The i-tRFs originate from the internal body of mature tRNAs, straddling anticodon region due to cleavage by unknown nucleases [26] and including anticodon-loop and part of D/T-loop. Its production may be related to hypoxia stress stimulation, but the specific mechanism remains unclear. tiRNAs are non-coding small RNAs with 31-40 nucleotides in length, which can be classified into 3'-tiRNAs and 5'-tiRNAs [24, 26-30]. As the earliest discovered tsRNAs, tiRNAs are produced by cleavage specifically in the anticodon-loop of mature tRNAs [36]. Despite being known as stress-induced fragments, tiRNAs also exist in non-stressed conditions [37]. 5'tiRNA replication starts from the 5'end of mature tRNAs and end at the anticodon-loop. Similarly, 3'tiRNA replication starts from the anticodon-loop and end at the 3'end of mature tRNAs. 5'tRNA halves (31-40 nt) are produced by specific cleavage within the anticodon-loop of mature tRNA under various stress conditions, such as heat shock, hypoxia, UV irradiation, oxidative stress, amino acid/glucose starvation, and viral infection. Therefore, these tsRNAs are also called tiRNA-5s. 3'tRNA halves (31-40 nt), also known as tiRNA-3s, are generated by specific cleavage within anticodon-loop during stress responses [22, 38-41]. Angiogenin (ANG), RNase T2, and RNase L are responsible for tiRNA biogenesis [22, 37-43]. The biogenesis of tsRNA is enzymatically modified: the biogenesis of tsRNA is affected by modifying enzymes, but the specific mechanisms by which these modification enzymes recognize tRNA targets and induce modification to produce the corresponding tsRNA remain unclear.
tsRNAs in tumors
tsRNA belongs to a species of small non-coding RNA, a derivative of pre-tRNA or mature tRNA. Some studies have found that tsRNA is abnormally expressed in many tumor tissues, cells, or peripheral blood (Table 1).
Classification and biogenesis of tsRNAs. After being transcribed by RNA polymerase III in nucleus, pre-tRNA undergoes 5'leader, 3'tailer, and intron sequence removing, as well as 3'CCA plus and modification before tRNA maturation. Ribonuclease cleavage in specific region of pre-/mature tRNA produces different types of tsRNA, including 3'UtRF(1-tRF), 5'Trf, 3'tRF, 2'tRF, i'tRF, 5'tRH (5'tiRNA), and 3'tRH (3'tiRNA).
tsRNAs in tumors
| Tumor type | tsRNA | tsRNA type | Specimen | Refs |
|---|---|---|---|---|
| Non-small cell lung cancer (NSCLC) | tRF-Leu-CAG | tiRNAs | NSCLC tissues, cell lines, and sera | [44] |
| Lung adenocarcinoma (LUAD) | tsRNA-5001a | - | LUAD tissues | [45] |
| tRF-16-L85J3KE | i-tRF | LUAD tissues and cell lines | [46] | |
| tRF-21-RK9P4P9L0 and tRF-16-PSQP4PE | tRF-5 | LUAD tissues and cell lines | [46] | |
| Breast cancer (BC) | tsRNA-26576 | - | BC tissues | [47] |
| ts-112 | tRFs | BC cell lines | [48] | |
| tRF-Arg-CCT-017, tRF-Gly-CCC-001, and tiRNA-Phe-GAA-003 | i-tRF and 5'tiRNA | plasmas | [49] | |
| Triple-negative breast cancer (TNBC) | tDR-0009 and tDR-7336 | - | Cell lines | [50] |
| Notriple-negative breast cancer (Non-TNBC) | tDR-7816 | tRFs | Blood samples | [51] |
| Gastric cancer (GC) | tRF-33-P4R8YP9LON4VDP | - | peripheral blood samples | [52] |
| tiRNA-Val-CAC-001 | tRF-5 | GC tissues and cells | [53] | |
| tRF-Val-CAC-016 | - | GC tissues | [54] | |
| tRF-3019a | tRF-3 | GC tissues and cells | [55] | |
| tRF-31-U5YKFN8DYDZDD | i-tRF | GC tumor tissues, sera, and cell lines | [56] | |
| tRF-5026a (tRF-18-79MP0P04) | tRFs | GC tissues and plasma samples | [57] | |
| tRF-19-3L7L73JD | tRFs | plasma and cell lines | [58] | |
| Has-tsr016141 | tRFs | GC tissues and sera | [59] | |
| Hepatocellular carcinoma (HCC) | ts-N102, ts-N59 and ts-N41 | - | liver primary tumor tissues | [60] |
| tRF-Gln-TTG-006 | - | HCC sera | [61] | |
| Liver cancer | tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5 | - | plasma exosomes | [62] |
| Colorectal cancer (CRC) | tRF-22-WB86Q3P92, tRF-22-WE8SPOX52, tRF-22-WE8S68L52 and tRF-18-8R1546D2 | - | Sequence Read Archives (SRA) public repository | [63] |
| tiRNA-1:33-Pro-TGG-1 (5′tiRNA-Pro-TGG) | 5'tiRNA | Sessile serrated lesions (SSLs) tissues | [64] | |
| 5'-tiRNA-Val | 5'tiRNA | CRC tumor tissues | [65] | |
| 5'tiRNA-His-GTG | 5'tiRNA | CRC tissues | [66] | |
| Pancreatic ductal adenocarcinoma (PDAC) | tsRNA-ValTAC-41, tsRNA-MetCAT-37 and tsRNA-ThrTGT-23 | tRF-3 and i-tRF | PDAC sera and tissue | [67] |
| Pancreatic cancer (PC) | tRF-Pro-AGG-004, and tRF-Leu-CAG-002 | - | PC sera | [68] |
| Clear cell renal cell carcinoma (ccRCC) | 5'-tRNA-Arg-CCT, 5'-tRNA-Glu-CTC and 5'-tRNA-Lys-TTT | 5'tiRNA | cancerous tissues and sera | [69] |
| Prostate cancer (CaP) | 5'-tRNA-Asp-GUC-half and 3'-tRNA-Asp-GUC-half | 5'tiRNA and 3'tiRNA | prostate tissues | [70] |
| Muscle-invasive bladder cancer (MIBC) | tiRNA-1:33-Gly-GCC-1, tRF-1:32-Gly-GCC-1 | 5'tiRNA and tRF-5c | MIBC tissues | [71] |
| tRF-+1:T20-Ser-TGA-1 | tRF-1 | MIBC tissues | [71] | |
| Endometrial carcinoma (EC) | tRF-20-S998LO9D | tRF-5 | EC tissues and serumal exosomes | [72] |
| High-grade serous ovarian cancer (HGSOC) | tRF-03357 and tRF-03358 | tRFs | HGSOC sera and cells | [73] |
| Gliomas | tRFdb-3003a and tRFdb-3003b | tRF-3 | tRF explorer and tRFdb databases | [74] |
| Glioblastomas (GBMs) | tRF-1-32-chrM.Lys-TTT, tiRNA-1-33-Gly-GCC-1, tiRNA-1-33-Gly-CCC-1, tRF-1-31-His-GTG-1, and tiRNA-1-33-Gly-GCC-2-M3 | - | Fresh tumor tissues | [75] |
| tiRNA-1-34-Lys-CTT-1-M2 | - | Fresh tumor tissues | [75] | |
| Diffuse glioma | ts-26, tRFdb-3012a, and tRFdb-3012b | tRF-1 and tRF-3 | tRF explorer and tRFdb databases | [76] |
| Osteosarcoma | tRF-33-6SXMSL73VL4YDN, tRF-32-6SXMSL73VL4YK, tRF-32- M1M3WD8S746D2, tRF-35-RPM830MMUKLY5Z, tRF-33-K768WP9N1EWJDW and tRF-32-MIF91SS2P46I3 | tRFs | Blood samples | [77] |
| Chronic lymphocytic leukemia (CLL) | ts-46 and ts-47 | - | Blood samples | [78] |
| ts-3676 and ts-4521 | - | Blood samples | [78] | |
| ts-42, ts-70 and ts-36 | - | Blood samples | [79] | |
| ts-43 and ts-44 | tRF-5 | Blood samples | [79] | |
| Acute myeloid leukemia (AML) | tsRNA20 and tsRNA66 | - | Blood samples | [80] |
| Nasopharyngeal carcinoma (NPC) | tRF-1:28-Val-CAC-2 and tRF-1:24-Ser-CGA-1-M3 | tRF-5c and tRF-5b | NPC tissues | [81] |
| tRF-55:76-Arg-ACG-1-M2 | tRF-3b | NPC tissues | [81] | |
| Oral submucous fibrosis (OSF) | tiRNA-Val-CAC-002 | 5'tiRNA | buccal OSF tissues | [82] |
| tRF-Gly-TCC-016 | tRF-5c | buccal OSF tissues | [82] | |
| Papillary thyroid cancer (PTC) | tRF-39-0VL8K87SIRMM12E2 | tRF-3 | PTC tissues | [83] |
| Laryngeal squamous cell carcinoma (LSCC) | tRF-33-Q1Q89P9L842205 | 5'tiRNA | LSCC tumor tissues | [84] |
| Esophageal squamous cell carcinoma (ESCC) | tRNA-GlyGCC-5 | - | Salivary exosomes | [85] |
tsRNAs potentially used for tumor early-state screening and diagnosis
In the clinic, a large number of tumors are discovered and diagnosed at advanced stage, and have already missed the best opportunity for treatment. Therefore, improving the early screening rate and diagnosis rate has become the focus of current cancer treatment research. Recently, the discovery of tsRNA and its abnormal expression in tumors can effectively distinguish tumor patients from healthy people. And tsRNA may be a new diagnostic biomarker for tumors, and may be used for tumor early-state screening in theory. Many studies have found a significant rise of tsRNA expression in tumor tissue, cell line, or serum (Table 2). Such as, the expression of tRF-Leu-CAG was significantly upregulated in NSCLC tissues, cell lines and serum and was positively correlated with tumor stage, which proved that tRF-Leu-CAG may be used as a diagnostic marker in stage NSCLC IV cases [44]. Meanwhile, some studies found that tRF-21-RK9P4P9L0 and tRF-16-PSQP4PE were highly elevated in LUAD tissues compared with normal tissues. These findings indicated that tsRNA can be utilized as a diagnostic biomarker for LUAD, but a single tsRNA is less efficient than a combination of multiple tsRNAs [46]. The expression levels of tRF-Arg-CCT-017 in HER-2 subtypes, and expression levels of tRF-Gly-CCC-001 and tiRNA-Phe-GAA-003 differ between luminal BC and TNBC, reflecting obvious heterogeneity, suggesting that tRF-Arg-CCT-017, tRF-Gly-CCC-001 and tiRNA-Phe-GAA-003 can serve as novel diagnostic biomarkers for BC (Figure 2E) [49]. Moreover, tRF-1:28-Val-CAC-2 has a relatively good ability to distinguish between primary NPC and healthy control (Figure 2D) [81]. tRF-39-0VL8K87SIRMM12E2 was verified by qPCR to be significantly upregulated in PTC cell lines and tissue samples [83]. In the tsRNAs used in diagnosis, ts-N102 was significantly upregulated in HCC tissues, while the highly related hsa-mir-215 was a tumor suppressor in CRC and multiple myeloma [60]. As an important part of the biobank, blood sample has rich biomolecules that are used for disease diagnosis, stage identification, and prognosis prediction.
Compared with tissue sample, blood sample has advantages of easy access, continuous sampling, and high patient acceptance. Therefore, simultaneous analysis of biomarkers found that tsRNAs in blood sample may improve an accuracy and convenience of cancer detection (Table 2). Some reports suggested that half of 5′-tRNA-Arg-CCT, 5'-tRNA-Glu-CTC, and 5'-tRNA-Lys-TTT were downregulated in clear cell RCC patient serum and tissues, possibly as a non-invasive biomarker [69]. RNAs in exosomes are relatively more stable, more resistant to physical degradation, and exosomes are easily accessible and carry a variety of molecules associated with specific diseases. The four tsRNA (tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5, and tRNA-GluCTC-5) were identified to be highly expressed in exosomes derived from the plasma of HCC patients [62]. The saliva-derived exosomal tsRNA (tRNA-GlyGCC-5) was found to be could distinguish in the patients with ESCC, and it was served as a non-invasive, convenient and reliable diagnostic biomarker. tRNA-GlyGCC-5 was supposed to be a preoperative biomarker to select patients who benefit from adjuvant therapy (Figure 2B) [85]. In EC patients, the exosomal tRF-20-S998LO9D could potentially be used as a non-invasive biomarker [72].
tsRNA is a novel type of regulatory non-coding RNA that has attracted a great attention across multiple subfields of biology, particular in recent studies to its participation in a variety of biological processes under diverse pathological and physiological conditions (Table 2). Yue Huang et al. used bioinformatics analysis to predict that tDR-7816-mediated xenobiotic metabolic processes supporting tumorigenesis in BC, and identified tDR-7816 as a potential biomarker and intervention target for non-TNBC [51]. tsRNA-26576 may act as an oncoprotein by inhibiting the expression of SPEN and FAT4. Normally, tsRNA remains stable and detectable in the blood, and its dysregulation has been identified to be associated with BC development or progression, suggesting that tsRNA-26576 may be a valid marker for BC diagnosis [47]. In OSF formation, tRF-Gly-TCC-016 may promote OSF formation and progression through cytokine-cytokine receptor interaction and cAMP signaling pathway, so it was documented to have an important significance in early screening and diagnosis of OSF, and may serve as a potential diagnostic marker [82]. In addition, androgen-dependent tsRNAs (5'-tRNA-Glu-CUC) may be used as a biomarker to monitor and predict progression in PC [70].
Moreover, tsRNA expression is tissue specific, particularly spatiotemporal, and many studies have emerged on tsRNA as a clinical marker (Table 2). Some studies found that combining 2-tsRNAs features have some disease specificity in PC, suggesting that serum tRFPro-AGG-004 and tRF-Leu-CAG-002 can serve as a new promising biomarker, even in early-stage screening (Figure 2C) [68]. In addition, some specific tsRNAs (tsRNA-ValTAC-41 and tsRNA-MetCAT-37) may be highly sensitive, non-invasive, and effective biomarkers for PDAC [67]. Bing Xu et al. identified four tRNA fragments from tRNA-Leu-CAA in sncRNA-seq dataset of glioma samples, and found that three tsRNAs (ts-26, tRFdb-3012a and tRFdb-3012b) were significantly downregulated in glioma, indicating that tRNA-leu-caa-derived tsRNA may serve as a diagnostic and prognostic biomarke for diffuse glioma [76]. Hongxia Deng et al. identified a 5′-tiRNA, tRF-33-Q1Q89P9L842205, which is specifically cleaved by angiogenin in the anticodon of mature tRNA-Gly-CC and closely associated with LSCC, while tRF-33-Q1Q89P9L842205 has promising applications as a biomarker of LSCC [84]. tRF-Gln-TTG-006 is significantly better diagnostic than AFP in the early stage of HCC [61]. tRF-22-WB86Q3P92, tRF-22-WE8SPOX52, tRF-22-WE8S68L52, and tRF-18-8R1546D2 were found to have some diagnostic and prognostic potential for in CRC (Figure 2A) [63].
tsRNA-targeted therapy in tumors
Some tsRNAs have been described as functional molecules to promote cancer progression, rendering them as promising therapeutic targets or agents (Table 3). For example, in NSCLC, tRF-Leu-CAG could suppress the proliferation of NSCLC cells and inhibit G0/G1 cell-cycle progression through targeting AURKA (Figure 3A) [44]. The 5'tiRNA-His-GTG can inhibit the expressions of pro-proliferation and anti-apoptosis related genes through hippo signaling pathway by targeting LATS2 [66]. tRF-03357 has predictive targeting, while inhibition of tRF-03357 expression can directly upregulate HMBOX1 in HGSOC to inhibit cell proliferation, migration and invasion (Figure 3E) [73]. These studies indicate that tsRNAs have great potential as therapeutic drugs. However, there is still a long way to go before clinical transformation application.
tsRNAs for tumor early-stage screening and diagnosis
| Tumour type | tsRNA | Expression | Screening or diagnosis | Refs |
|---|---|---|---|---|
| NSCLC | tRF-Leu-CAG | High | Diagnosis | [44] |
| LUAD | tRF-21-RK9P4P9L0 and tRF-16-PSQP4PE | High | Screening | [46] |
| BC | tsRNA-26576 | High | Screening | [47] |
| tRF-Arg-CCT-017, tRF-Gly-CCC-001 and tiRNA-Phe-GAA-003 | High | Diagnosis | [49] | |
| Non-TNBC | tDR-7816 | Low | Screening | [51] |
| HCC | ts-N102 | High | Diagnosis | [60] |
| tRF-Gln-TTG-006 | High | Diagnosis | [61] | |
| Liver cancer | tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5 | High | Screening | [62] |
| CRC | tRF-22-WB86Q3P92, tRF-22-WE8SPOX52, tRF- 22-WE8S68L52 and tRF-18-8R1546D2 | Low | Diagnosis | [63] |
| PDAC | tsRNA-ValTAC-41 and tsRNA-MetCAT-37 | High | Diagnosis | [67] |
| PC | tRF-Pro-AGG-004 and tRF-Leu-CAG-002 | High | Diagnosis | [68] |
| ccRCC | 5'-tRNA-Arg-CCT, 5'-tRNA-Glu-CTC, and 5'-tRNA-Lys-TTT | Low | Diagnosis | [69] |
| CaP | 5'-tRNA-Glu-CUC-half | High | Diagnosis | [70] |
| EC | tRF-20-S998LO9D | Low | Screening | [72] |
| Diffuse glioma | ts-26, tRFdb-3012a, and tRFdb-3012b | Low | Screening | [76] |
| NPC | tRF-1:28-Val-CAC-2 | High | Screening | [81] |
| OSF | tRF-Gly-TCC-016 | High | Screening | [82] |
| PTC | tRF-39-0VL8K87SIRMM12E2 | High | Diagnosis | [83] |
| LSCC | tRF-33-Q1Q89P9L842205 | Low | Diagnosis | [84] |
| ESCC | tRNA-GlyGCC-5 | High | Diagnosis | [85] |
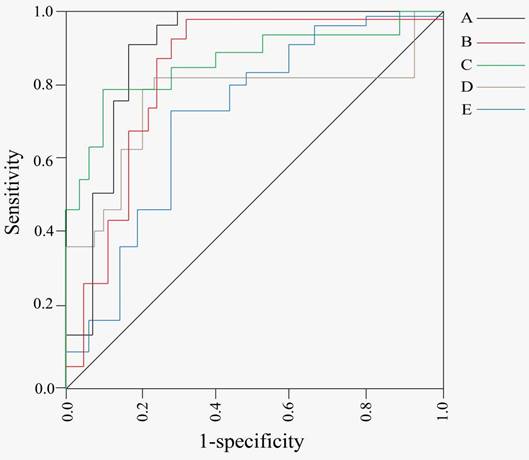
ROC curve of tsRNAs of tumor early-stage screening and diagnosis. A, According to ROC curve analysis, tRF-22-WB86Q3P92, tRF-22-WE8SPOX52, tRF-22-WE8S68L52, and tRF-18-8R1546D2 may have some diagnostic and prognostic potential in CRC (Reproduced with permission from Front Oncol publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [63]. B, According to ROC curve analysis, tRNA-GlyGCC-5 has a value as a preoperative biomarker for ESCC diagnosis (Reproduced with permission from Mol Cancer publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [85]. C, According to ROC curve analysis, tRFPro-AGG-004 and tRF-Leu-CAG-002 has useful as a potential diagnostic biomarker for PC (Reproduced with permission from Mol Cancer publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [68]. D, According to the ROC curve, tRF-1:28-Val-CAC-2 may be a potential diagnostic marker in NPC cases (Reproduced with permission from Front Mol Biosci publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [81]. E, According to ROC curve, tRF-Arg-CCT-017, tRF-Gly-CCC-001 and tiRNA-Phe-GAA-003 can be used as a diagnostic index for BC (Reproduced with permission from NPJ Breast Cancer publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [49].
tsRNAs used for tumor targeted therapy and mechanism. A, Inhibition of tRF-Leu-CAG expression in NSCLC can inhibit cell proliferation and hinder cell cycle by direct downregulation of AURKA expression or inhibition of AURKA expression by regulating other genes involved in cancer process [44]; B, tiRNA-Val-CAC-001 downregulates protein level of LRP 6 and β-catenin, while upregulates p-β-catenin, inhibiting cell migration and proliferation and promoting apoptosis of GC cells [53]; C, tRF-Val-CAC-016 regulates MAPK pathway by targeting CACNA1d, thereby inhibiting GC cell proliferation [54]; D, Overexpression of tRF-20-S998LO9D upregulates SESN2 expression, thus inhibiting the proliferation, migration and invasion of EC cells, and promoting cell apoptosis [72]; (E) Inhibition of tRF-03357 expression directly downregulate HMBOX1 in HGSOC to inhibit cell proliferation, migration and invasion [73]; F, Overexpression of tRFdb-3003a/b directly binds to VAV 2 and downregulates VAV 2 expression in gliomas, thus regulating tumor progression [74]; (G) tRF-33-Q1Q89P9L842205 inhibits cell growth, proliferation, migration, invasion and induced apoptosis of LSCC cells by directly silencing PIK3CD [84].
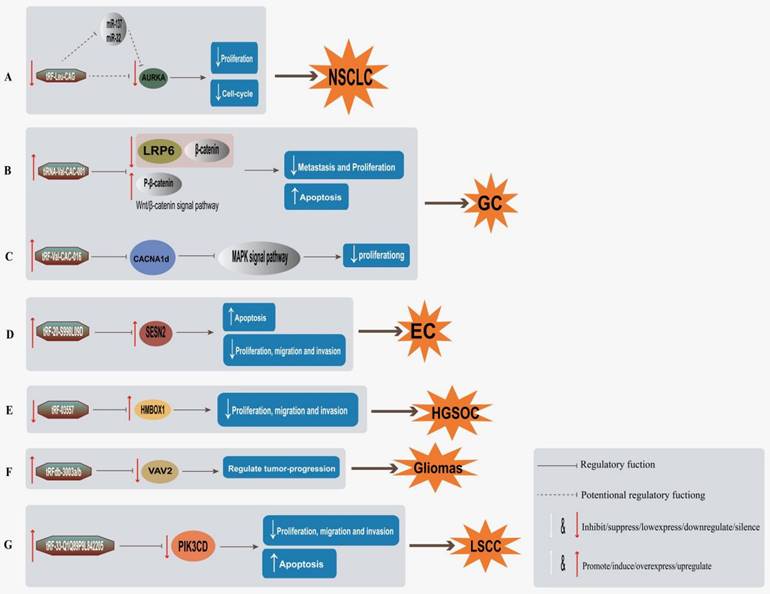
Some tsRNAs can not only promote cell proliferation as a carcinogen, but also inhibit the development of cancer as an inhibitor. tsRNA could realize its targets through regulating a certain target gene protein by some tumor signaling pathways, and then achieve an targeted therapeutic effect (Table 3). For example, tiRNA-Val-CAC-001 works as a cancer suppressor in GC by targeting LRP6 via Wnt/β-catenin signaling pathway, and upregulating tiRNA-Val-CAC-001 inhibited metastasis and proliferation but promoting apoptosis (Figure 3B) [53]. Meanwhile, tRF-Val-CAC-016 in GC was found to modulate CACNA1d-mediated transduction of MAPK signaling pathway, thus inhibiting the proliferation of GC (Figure 3C) [54]. tsRNA may also directly upregulate or downregulate the related target genes to inhibit tumor progression (Table 3). For example, tRF-20-S998LO9D is an EC repressor, inhibits migration, proliferation and invasion, and promotes apoptosis by upregulating SESN2 (Figure 3D) [72]. The tRF-33-Q1Q89P9L842205 induces apoptosis in LSCC cells and inhibits cell growth, migration and invasion via directly downregulating PIK3CD expression in LSCC (Figure 3G) [84]. Furthermore, Jian Ren et al. found that overexpression of tRFdb-3003a/b may play a key role in tumor progression of gliomas, tRFdb-3003a/b may inhibit tumor proliferation and growth by directly binding to VAV2 to regulate VAV2 expression in gliomas (Figure 3F) [74]. Meanwhile some investigators have also found that downregulated tRFdb-3012a/b may directly target the 3'untranslated region of RBM 43, and ts-26 may directly target the 3'untranslated region of HOXA13 to play a tumor suppressor role in glioma progression through specific signaling pathways [76]. Clinically, there are also some specific targets and pathways of tsRNA that have not yet been discovered, but they play an important role in cancer therapy through some specific mechanisms of action (Table 3). For example, in the transformation of OSF to oral squamous cell carcinoma (OSCC), tiRNA-Val-CAC-002 acts as a suppressor of oncogenic cytokines, which affects the course of OSCC by regulating tumor cell proliferation and invasion and mesenchymal-epithelial transformation (MET) [82]. Some reports found that ts-46 and ts-47 strongly downregulated in invasive CLL [78], suggesting that these tsRNAs are potential tumor suppressors, meanwhile, ts-42 was inactivated in CLL mainly through promoter methylation, thus acting as tumor suppression [79]. tRF-0009 (tDR-7336) may play a key regulatory role in hypoxia-induced chemoresistance in TNBC [50]. These studies suggest that, in tumor therapeutic strategy, tsRNAs may inhibit tumor onset or progress mainly through suppressing tumor cell proliferation, growth, migration, and invasion, and promoting cell apoptosis, to achieve therapeutic effects.
tsRNAs used for prognosis of the patients with tumor
Increasing number of reports indicate that tsRNA has a great potential as biomarker for tumor prognosis. Some tsRNAs are downregulated in tumors, they are associated with higher survival in cancer (Table 4). Zuo et al., using multivariate COX survival analysis, raised that one tsRNAs (ts-N22) was poorly expressed and was associated with a lower overall survival rate in patients with HCC [60]. Xu et al. found that the tumor patients with three tsRNAs (ts-26, tRFdb-3012a, and tRFdb-3012b) had significantly lower clinical survival than those with high expression through Kaplan-Meier curve analysis and log-rank comparison, indicating that downregulated tsRNAs (ts-26 and tRFdb-3012a/b) were associated with poor survival prognosis in glioma patients (Figure 4C) [76].
tsRNAs used for tumor targeted therapy and application
| Tumor type | tsRNA | Targeted gene | Mechanism | Therapeutic efficacy | Refs |
|---|---|---|---|---|---|
| NSCLC | tRF-Leu-CAG | AURKA | Decreases proliferation and cause G0/G1 cell-cycle progression via signaling pathways by targeting AURKA. | Well | [44] |
| GC | tiRNA-Val-CAC-001 | LRP6 | Decreases metastasis and proliferation and promote apoptosis via Wnt/β-catenin pathway by targeting LRP6. | High | [53] |
| tRF-Val-CAC-016 | CACNA1d | Suppress proliferation via MAPK pathway by targeting CACNA1d. | High | [54] | |
| EC | tRF-20-S998LO9D | SESN2 | Decreases migration, proliferation, and invasion and promote apoptosis by upregulating SESN2. | High | [72] |
| HGSOC | tRF-03357 | HMBOX1 | Decreases proliferation, migration, and invasion partly through downregulating HMBOX1. | Well | [73] |
| Gliomas | tRFdb-3003a/b | VAV2 | Inhibit cell proliferation and tumor growth by directly binding to VAV2 to regulate its expression. | High | [74] |
| tRFdb-3003a/b and ts-26 | RBM43 and HOXA13 | Play a tumor suppressor role in progression by directly targeting 3'untranslated region of RBM43 and HOXA13. | Medium | [76] | |
| LSCC | tRF-33-Q1Q89P9L842205 | PIK3CD | Induces apoptosis and inhibit cell growth, migration and invasion via directly downregulating PIK3CD. | High | [84] |
| CRC | 5'tiRNA-His-GTG | LATS2 | Decreases proliferation and anti-apoptosis via “turning off” hippo signaling pathway by targeting LATS2. | Well | [66] |
| TNBC | tDR-0009 (tDR-7336) | STAT3 | Inhibits activation of STAT3 phosphorylation. | High | [50] |
| CLL | ts-46 and ts-47 | - | - | Medium | [78] |
| ts-42 | - | Inactivates mostly by promoter methylation. | Medium | [79] | |
| OSF | tiRNA-Val-CAC-002 | - | Inhibits proliferation and invasion of cells and regulates MET. | Medium | [82] |
tsRNAs in the prognosis of tumors
| Tumor type | tsRNA | Expression | Prognosis | Refs |
|---|---|---|---|---|
| LUAD | tsRNA-5001a | High | Poor | [45] |
| tRF-21-RK9P4P9L0 | [46] | |||
| BC | tRF-Arg-CCT- 017 and tiRNA-Phe-GAA-003 | High | Poor | [49] |
| HCC | ts-N22 | Low | Well | [60] |
| CRC | 5'tiRNA-Pro-TGG | High | Poor | [64] |
| PDAC | tsRNA-ValTAC-41 | High | Poor | [67] |
| PC | tRF-Pro-AGG-004 and tRF-Leu-CAG-002 | High | Poor | [68] |
| Diffuse glioma | ts-26, tRFdb-3012a and tRFdb-3012b | Low | Well | [76] |
| Osteosarcoma | tRF-33-6SXMSL73VL4YDN, tRF-32-6SXMSL73VL4YK, tRF-32-M1M3WD8S746D2, tRF-35-RPM830MMUKLY5Z, tRF-33-K768WP9N1EWJDW, and tRF-32-MIF91SS2P46I3 | High | Poor | [77] |
| CLL | ts-3676 and ts-4521 | High | Poor | [78] |
| AML | tsRNA20 and tsRNA66 | Low | Well | [80] |
Kaplan-Meier curve analysis of tsRNAs expression for OS. A, 5'tiRNA-Pro-TGG is negatively associated with CRC prognosis (Reproduced with permission from Cancer Cell Int publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [64]. B, tRF-Pro-AGG-004 and tRF-Leu-CAG-002 are negatively associated with PC prognosis (Reproduced with permission from Mol Cancer publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [68]. C, ts-26 and tRFdb-3012a/b are positively associated with Diffuse glioma prognosis (Reproduced with permission from Cancer Manag Res publisher, the link of the Creative Commons licence: https://www.dovepress.com/terms.php & http://creativecommons.org/licenses/by-nc/3.0/.) [76]. D, tRF-33-6SXMSL73VL4YDN, tRF-32-6SXMSL73VL4YK, tRF-32-M1M3WD8S746D2, tRF-35-RPM830MMUKLY5Z, tRF-33-K768WP9N1EWJDW, and tRF-32-MIF91SS2P46I3 are negatively associated with Osteosarcoma prognosis (Reproduced with permission from Front Oncol publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [77]. E, tRF-21-RK9P4P9L0 is negatively associated with LUAD prognosis (Reproduced with permission from Cancer Cell Int publisher, the link of the Creative Commons licence: http://creativecommons.org/licenses/by/4.0/.) [46].
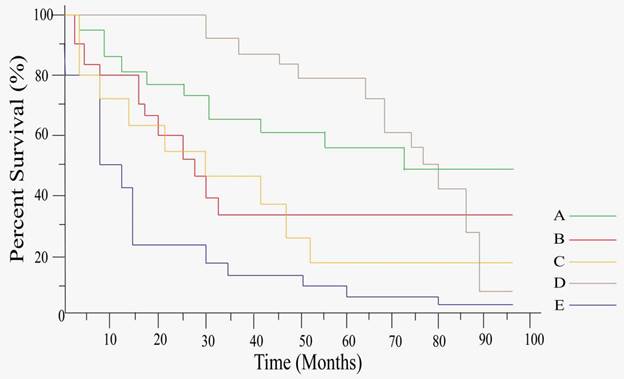
Moreover, Bill M et al. found that only KD of tsRNA20 and tsRNA66 reduced the proliferation capacity of THP-1 and OCI-AML 3 cells, indicating that upregulated tsRNAs (tsRNA20 and tsRNA66) improved the overall survival of CN-AML [80]. Some tsRNAs highly expressed in the tumors, and they were also associated with the lower survival of the tumor patients (Table 4). Hu et al. reported that tsRNA-5001a was significantly upregulated in LUAD tissues, and function assay showed that overexpression of tsRNA-5001a significantly increased the risk of postoperative recurrence in LUAD patients and was associated with poor prognosis [45]. Meanwhile, Wang et al. showed that, in LUAD tissue, only tRF-21-RKP4P9L0 was highly elevated and significantly associated with a poor prognosis, in mechanism, overexpression of tRF-21-RK9P4P9L0 promotes the proliferation, migration and invasion of LUAD (A549 and H1299) cells (Figure 4E) [46]. Wang et al. showed that the expression of tiRNA-1:33-Pro-TGG-1 (5′tiRNA-Pro-TGG) was higher in SSLs than that in healthy population, while 5′tiRNA-Pro-TGG and heparanase 2 (HPSE2) were found a significant negative correlation between their expression levels, another analysis of survival outcomes in CRC patients demonstrated that the lower level of HPSE2 was associated with poorer prognosis (Figure 4A) [64]. Wang et al. found that the breast cancer patients with higher levels of tRF-Arg-CCT-017 or tiRNA-Phe-GAA-003 were associated with worse disease-free survival rate (DFS) and overall survival rate (OS) through statistical Kaplan-Meier curves to analyze DFS and OS [49]. Xue et al. found the patients with low serum tsRNA-ValTAC-41 level had a significantly longer OS than those with high level [67]. Jin et al. found that tRF-Pro-AGG-004 and tRF-LeuCAG-002 played a cancer-promoting role in PC, with which the patients have a poor prognosis (Figure 4B) [68]. Tang et al. identified six high tsRNAs including tRF-33-6 SXMSL-6VL4YDN, SXMSL73VL4YK, tRF-32-M1M-MWD8S746D2, tRF-35-RPM830-MMUKLY5Z, tRF-33-K768WP9N1EWJDW, and tRF-32-MIF91SS2P46I3 in osteosarcoma, they were associated with poor survival in osteosarcoma patients (Figure 4D) [77]. Furthermore, Balatti et al. found that ts-3676 and ts-4521 had a prognostic value for specific cell subgroups of CLL, with ts-3676 and ts-4521 almost twice the patients showing both aggressive clinical course and markers in patients with poor prognostic markers but indolent disease [78].
Conclusions and prospects
tsRNA has now attracted significant interest, establishing diverse tsRNA types and abundance of functions. According to tsRNA corresponding positions in the parental tRNA transcripts, tsRNAs are classified into tRF-1s, tRF-3s, tRF-5s, tiRNA, and tRF-2s/i-tRFs. Each particular type of tsRNA has a specific structure and production process. tsRNA is expected to be used as a marker for early-stage screening, diagnosis and prognosis of tumors, and as a target for treatment, with broad application prospects. However, the exact underlying mechanisms of action and the corresponding functions of each tsRNA type have not been fully elucidated. Therefore, further researches are needed to elucidate tsRNA mechanism in tumorigenesis and estimate tsRNA's value in clinical application. Meanwhile, with the development of high-throughput sequencing technology and the emergence of even more advanced research methods, increasingly more tsRNA will be discovered, with a high diversity of types and functions, which would extend their potential beneficial applications.
Abbreviations
AML: Acute Myeloid Leukem-ia; ANG: Angiogenin; BC: Breast cancer; CaP: prostate cancer; ccRCC: clear cell renal cell carcinoma; CLL: Chronic lymphocytic leukemia; CRC: Colorectal cancer; EC: Endometrial carcinoma; ESCC: Esophageal squamous cell carcinoma; GC: Gastric cancer; GBMs: Glioblastomas; HCC: Hepatocellular arcinoma; HGSOC: High-grade serous ovarian cancer; LSCC: laryngeal squamous cell carcinoma; LUAD: Lung adenocarcinoma; miRNA: microRNA; MIBC: Muscle-invasive bladder cancer; NPC: Nasopharyngeal carcinoma; NSCLC: Non-small cell lung cancer; nts: nucleotides; OSF: Oral submucous fibrosis; PC: Pancreatic cancer; PDAC: Pancreatic ductal adenocarcinoma; piRNA: PIWI-interacting RNA; pre-tRNAs: precursor tRNAs; PTC: Papillary thyroid cancer; RNase Z/ELAC2: endonuclease Z; siRNA: small interfering RNA; sncRNAs: small non-coding RNAs; snRNA: small nuclear RNA; snoRNA: small nucleolar RNA; tiRNAs(tRHs): tRNA halves; TNBC: Triple-negative breast cancer; Non-TNBC: Notriple-negative breast cancer; tRFs: tRNA-derived fragments; tsRNA: tRNA-derived small RNA.
Acknowledgements
We thank the members in Hunan Key Laboratory of Oncotarget Gene for providing us with programming services.
Funding
This work was supported in part by Major Scientific and Technological Innovation Project of Hunan Province (2021SK1020-4), Natural Science Foundation of Hunan Province (2018JJ6131, 2019JJ40175, and 2021JJ70098), Changsha Science and Technology Project (kg1801107 and kq2004136), and Research Projects of Hunan Health Commission (B2019084).
Author contributions
L.C., Q.T., and Y. W. performed data analyses. L.C. prepared all the figures and wrote the manuscript. F.T. supervised the project. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary data.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-Derived Small RNAs in Cancer: New Potential Biomarkers and Target for Therapy. Adv Cancer Res. 2017;135:173-187
2. Balatti V, Nigita G, Veneziano D. et al. tsRNA signatures in cancer. Proc Natl Acad Sci U.S.A. 2017;114:8071-8076
3. Goodarzi H, Liu X, Nguyen Hoang CB. et al. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790-802
4. Haussecker D, Huang Y, Lau A. et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. Rna. 2010;16:673-695
5. Xu D, Qiao D, Lei Y. et al. Transfer RNA-derived small RNAs (tsRNAs): Versatile regulators in cancer. Cancer Lett. 2022;546:215842
6. Peng EY, Shu Y, Wu Y. et al. Presence and diagnostic value of circulating tsncRNA for ovarian tumor. Mol Cancer. 2018;17:163
7. Kim HK, Fuchs G, Wang S. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57-62
8. Chen Q, Yan M, Cao Z. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397-400
9. Hide G. Trypanosomiasis and leishmaniasis: novel biology versus practical disease control. Trends Parasitol. 2002;18:477-478
10. Zhang Y, Zhang Y, Shi J. et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172-174
11. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2017;19:45-58
12. Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2014;10:553-563
13. Li Z, Ender C, Meister G. et al. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787-6799
14. Cole C, Sobala A, Lu C. et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. Rna. 2009;15:2147-2160
15. Kawaji H, Nakamura M, Takahashi Y. et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157
16. Rounge TB, Furu K, Skotheim RI. et al. Profiling of the small RNA populations in human testicular germ cell tumors shows global loss of piRNAs. Mol Cancer. 2015;14:153
17. Keam SP, Young PE, McCorkindale AL. et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42:8984-8995
18. Xu S-y, Liao J-Y, Ma L-M. et al. Deep Sequencing of Human Nuclear and Cytoplasmic Small RNAs Reveals an Unexpectedly Complex Subcellular Distribution of miRNAs and tRNA 3′ Trailers. PLoS ONE. 2010;5:e10563
19. Telonis AG, Loher P, Honda S. et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6:24797-24822
20. Cuciniello R, Filosa S, Crispi S. Novel approaches in cancer treatment: preclinical and clinical development of small non-coding RNA therapeutics. J Exp Clin Cancer Res. 2021;40:383
21. Xiao L, Wang J, Ju S. et al. Disorders and roles of tsRNA, snoRNA, snRNA and piRNA in cancer. J Med Genet. 2022;59:623-631
22. Yamasaki S, Ivanov P, Hu G-f. et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35-42
23. Lee YS, Shibata Y, Malhotra A. et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639-2649
24. Shen Y, Yu X, Zhu L. et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med. 2018;96:1167-1176
25. Zhu P, Yu J, Zhou P. Role of tRNA-derived fragments in cancer: novel diagnostic and therapeutic targets tRFs in cancer. Am J Cancer Res. 2020;10:393-402
26. Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends in Biochem Sci. 2016;41:679-689
27. Chen H, Xu Z, Liu D. Small non-coding RNA and colorectal cancer. J Cell and Mol Med. 2019;23:3050-3057
28. Li S, Chen Y, Sun D. et al. Angiogenin Prevents Progranulin A9D Mutation-Induced Neuronal-Like Cell Apoptosis Through Cleaving tRNAs into tiRNAs. Mol Neurobiol. 2017;55:1338-1351
29. Wong DTW, Xiao X, Grogan TR. et al. Characterization of Human Salivary Extracellular RNA by Next-generation Sequencing. Clin Chem. 2018;64:1085-1095
30. Zhu L, Ge J, Li T. et al. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett. 2019;452:31-37
31. Jiang P, Yan F. tiRNAs & tRFs Biogenesis and Regulation of Diseases: A Review. Curr Med Chem. 2019;26:5849-5861
32. Chen Q, Zhang X, Shi J. et al. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci. 2021;46:790-804
33. Su Z, Wilson B, Kumar P. et al. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu Rev Genet. 2020;54:47-69
34. Kumar P, Mudunuri SB, Anaya J. et al. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141-D145
35. Kumar P, Anaya J, Mudunuri SB. et al. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
36. Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol. 2012;227:2822-2826
37. Thompson DM, Parker R. Stressing Out over tRNA Cleavage. Cell. 2009;138:215-219
38. Li Y, Luo J, Zhou H. et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048-6055
39. Fu H, Feng J, Liu Q. et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Letters. 2008;583:437-442
40. Lee SR, Collins K. Starvation-induced Cleavage of the tRNA Anticodon Loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744-42749
41. Thompson DM, Lu C, Green PJ. et al. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. Rna. 2008;14:2095-2103
42. Jöchl C, Rederstorff M, Hertel J. et al. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677-2689
43. Donovan J, Rath S, Kolet-Mandrikov D. et al. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. Rna. 2017;23:1660-1671
44. Shao Y, Sun Q, Liu X. et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730-738
45. Hu F, Niu Y, Mao X. et al. tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Trans Lung Cancer Res. 2021;10:3957-3972
46. Wang J, Liu X, Cui W. et al. Plasma tRNA-derived small RNAs signature as a predictive and prognostic biomarker in lung adenocarcinoma. Cancer Cell Int. 2022;22:59
47. Zhou J, Wan F, Wang Y. et al. Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer Manag Res. 2019;11:3945-3956
48. Farina NH, Scalia S, Adams CE. et al. Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer. J Cell Physiol. 2020;235:5318-5327
49. Wang J, Ma G, Ge H. et al. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ Breast Cancer. 2021;7:4
50. Cui Y, Huang Y, Wu X. et al. Hypoxia-induced tRNA-derived fragments, novel regulatory factor for doxorubicin resistance in triple-negative breast cancer. J Cell Physiol. 2018;234:8740-8751
51. Huang Y, Ge H, Zheng M. et al. Serum tRNA-derived fragments (tRFs) as potential candidates for diagnosis of nontriple negative breast cancer. J Cell Physiol. 2019;235:2809-2824
52. Shen Y, Yu X, Ruan Y. et al. Global profile of tRNA-derived small RNAs in gastric cancer patient plasma and identification of tRF-33-P4R8YP9LON4VDP as a new tumor suppressor. Int J Med Sci. 2021;18:1570-1579
53. Zheng J, Li C, Zhu Z. et al. A 5`-tRNA Derived Fragment NamedtiRNA-Val-CAC-001 Works as a Suppressor in Gastric Cancer. Cancer Manag Res. 2022;14:2323-2337
54. Xu W, Zheng J, Wang X. et al. tRF-Val-CAC-016 modulates the transduction of CACNA1d-mediated MAPK signaling pathways to suppress the proliferation of gastric carcinoma. Cell Commun Signal. 2022;20:68
55. Zhang F, Shi J, Wu Z. et al. A 3′-tRNA-derived fragment enhances cell proliferation, migration and invasion in gastric cancer by targeting FBXO47. Arch Biochem Biophys. 2020;690:108467
56. Huang Y, Zhang H, Gu X. et al. Elucidating the Role of Serum tRF-31-U5YKFN8DYDZDD as a Novel Diagnostic Biomarker in Gastric Cancer (GC). Front Oncol. 2021;11:723753
57. Zhu L, Li Z, Yu X. et al. The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem Cell Res Ther. 2021;12:418
58. Shen Y, Xie Y, Yu X. et al. Clinical diagnostic values of transfer RNA-derived fragment tRF-19-3L7L73JD and its effects on the growth of gastric cancer cells. J Cancer. 2021;12:3230-3238
59. Gu X, Ma S, Liang B. et al. Serum hsa_tsr016141 as a Kind of tRNA-Derived Fragments Is a Novel Biomarker in Gastric Cancer. Front Oncol. 2021;11:679366
60. Zuo Y, Chen S, Yan L. et al. Development of a tRNA-derived small RNA diagnostic and prognostic signature in liver cancer. Genes Dis. 2022;9:393-400
61. Zhan S, Yang P, Zhou S. et al. Serum mitochondrial tsRNA serves as a novel biomarker for hepatocarcinoma diagnosis. Front Med. 2022;16:216-226
62. Zhu L, Li J, Gong Y. et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18:74
63. Zhu Y, Chen S, Ling Z. et al. Comprehensive Analysis of a tRNA-Derived Small RNA in Colorectal Cancer. Front Oncol. 2021;11:701440
64. Wang X-Y, Zhou Y-J, Chen H-Y. et al. 5'tiRNA-Pro-TGG, a novel tRNA halve, promotes oncogenesis in sessile serrated lesions and serrated pathway of colorectal cancer. World J of Gastrointest Oncol. 2023;15:1005-1018
65. Li S, Shi X, Chen M. et al. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer. 2019;145:1395-1407
66. Tao E-W, Wang H-L, Cheng WY. et al. A specific tRNA half, 5'tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J Exp Clin Cancer Res. 2021;40:67
67. Xue M, Shi M, Xie J. et al. Serum tRNA-derived small RNAs as potential novel diagnostic biomarkers for pancreatic ductal adenocarcinoma. Am J Cancer Res. 2021;11:837-848
68. Jin F, Yang L, Wang W. et al. A novel class of tsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer. Mol Cancer. 2021;20:95
69. Zhao C, Tolkach Y, Schmidt D. et al. 5′-tRNA Halves are Dysregulated in Clear Cell Renal Cell Carcinoma. J Urol. 2018;199:378-383
70. Zhao C, Tolkach Y, Schmidt D. et al. tRNA-halves are prognostic biomarkers for patients with prostate cancer. Urol Oncol. 2018;36:503.e501-503.e507
71. Qin C, Chen Z-H, Cao R. et al. Differential Expression Profiles and Bioinformatics Analysis of tRNA-Derived Small RNAs in Muscle-Invasive Bladder Cancer in a Chinese Population. Genes. 2022;13:601
72. Qian T, Yu X, Xu A. et al. tRF-20-S998LO9D inhibits endometrial carcinoma by upregulating SESN2. Epigenomics. 2022;14:1563-1577
73. Zhang M, Li F, Wang J. et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. OncoTargets Ther. 2019;12:6371-6383
74. Ren J, Wu X, Shang F-F. et al. The tRNA-Cys-GCA Derived tsRNAs Suppress Tumor Progression of Gliomas via Regulating VAV2. Dis Markers. 2022;2022:1-16
75. Tu M, Zuo Z, Chen C. et al. Transfer RNA-derived small RNAs (tsRNAs) sequencing revealed a differential expression landscape of tsRNAs between glioblastoma and low-grade glioma. Gene. 2023;855:147114
76. Xu B, Liang J, Zou H. et al. Identification of Novel tRNA-Leu-CAA-Derived tsRNAs for the Diagnosis and Prognosis of Diffuse Gliomas. Cancer Manag Res. 2022;14:2609-2623
77. Tang Z, Zhang S, Ling Z. Development of a tRNA-Derived Small RNA Prognostic Panel and Their Potential Functions in Osteosarcoma. Front Oncol. 2021;11:652040
78. Balatti V, Palamarchuk A, Rizzotto L. et al. Role of Ts-RNAs in CLL. Blood. 2016;128:2016-2016
79. Veneziano D, Tomasello L, Balatti V. et al. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2019;116:24252-24258
80. Bill M, Veneziano D, Kohlschmidt J. et al. Prognostic and Biologic Significance of Transfer RNA-Derived Small RNAs (tsRNAs) Expression in Younger Adult Patients (Pts) with Cytogenetically Normal Acute Myeloid Leukemia (CN-AML). Blood. 2018;132:89-89
81. Lu Z, Su K, Wang X. et al. Expression Profiles of tRNA-Derived Small RNAs and Their Potential Roles in Primary Nasopharyngeal Carcinoma. Front Mol Biosci. 2021;8:780621
82. Zeng L, Peng H, Yu H. et al. Expression profiles of tRNA-derived small RNA and their potential roles in oral submucous fibrosis. J Oral Pathol Med. 2021;50:1057-1066
83. Shan S, Wang Y, Zhu C. A comprehensive expression profile of tRNA-derived fragments in papillary thyroid cancer. J Clini Lab Anal. 2020;35:e23664
84. Deng H, Wang J, Ye D. et al. A 5'-tiRNA fragment that inhibits proliferation and migration of laryngeal squamous cell carcinoma by targeting PIK3CD. Genomics. 2022;114:110392
85. Li K, Lin Y, Luo Y. et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer. 2022;21:21
Author contact
![]() Corresponding author: Dr. Faqing Tang, Hunan Key Laboratory of Oncotarget Gene, Clinical Laboratory of Hunan Cancer Hospital, the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 410013, Changsha, China. E-mail: tangfqorg.cn; Tel.: 86-731-89762688; Fax: 86-731-89762688.
Corresponding author: Dr. Faqing Tang, Hunan Key Laboratory of Oncotarget Gene, Clinical Laboratory of Hunan Cancer Hospital, the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 410013, Changsha, China. E-mail: tangfqorg.cn; Tel.: 86-731-89762688; Fax: 86-731-89762688.

 Global reach, higher impact
Global reach, higher impact