Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(19):6232-6238. doi:10.7150/jca.101385 This issue Cite
Research Paper
METTL14 Induced N6-Methyladenosine Modification of FOXP4 mRNA in HBV-HCC
1. Department of Endoscopy, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, 200438, China.
2. Department of General Surgery, The Second Affiliated Hospital of Nanchang University, Nanchang University School of Medicine, Nanchang, Jiangxi Province, China.
#Co-auther contribute equal to this article.
Received 2024-7-24; Accepted 2024-9-28; Published 2024-10-14
Abstract
Chronic hepatitis B virus infections are a significant cause of liver cirrhosis and cancer. Our research reveals that HBV infection leads to a marked increase in m6A modification of Foxp4 mRNA, resulting in enhanced stability of the mRNA and a subsequent increase in Foxp4 mRNA levels. Analysis of biopsy samples from chronic HBV patients demonstrated consistent upregulation of m6A-modified Foxp4 mRNA levels alongside increased Foxp4 mRNA levels. Functionally, Foxp4 was found to promote proliferation, migration, and invasion of hepatocellular carcinoma (HCC) cells in laboratory settings. Additionally, HBV gene expression was shown to activate the PI3K/AKT pathway by modulating Foxp4 mRNA stability in HCC cells. This study provides valuable insights into the underlying mechanisms of HBV infection and its potential implications for cancer development.
Keywords: epitranscriptomic modification, HBV, Foxp4, METTL14, HCC
Introduction
Hepatitis B virus (HBV) infection is a serious concern as it can progress to chronic hepatitis and increase the risk of developing HCC (1-3). Research suggests that HBV can manipulate the host's gene expression to facilitate viral replication and ensure its survival (4). This manipulation ultimately contributes to the progression of chronic hepatitis B to liver cirrhosis and potentially HCC (5). However, the specific mechanisms or triggers that lead to HBV-related HCC are not yet fully understood.
Viruses have the ability to manipulate host gene expression profiles, facilitating their own replication and establishment within the host (6-9). A common epigenetic alteration employed by viruses is the modification of RNA, specifically the N6-methyladenosine (m6A) modification of mRNAs (10-13). Foxp4 plays a critical role in tumorigenesis and cancer progression (14-17). In this study, we delve into the HBV's interaction with the host gene Foxp4, a key player in tumorigenesis and cancer progression. We reveal that HBV specifically targets Foxp4 mRNA for m6A modification, resulting in a stabilization of Foxp4 mRNA levels. The identified mechanism suggests a novel avenue for HBV-associated hepatocellular carcinoma (HCC) development.
Materials and Methods
Cell culture and transfection
Hepatocellular carcinoma (HCC) cells from ATCC were cultured as outlined previously. Primary human hepatocytes were grown following the manufacturer's instructions. Lipofectamine RNAiMAX was used to transfect shRNAs as per the protocol (18). The OE-NC, OE-Foxp4, sh-NC, sh-Foxp4, sh-METTL3, sh-METTL14, sh-FTO, sh-KIAA1429, sh-ALKBH5, and HBV 1.3-mer, FLAG-YTHDF1, 2, and 3 plasmids were acquired from Shanghai Yuan Zhi Biotechnology Co., LTD. The effects of stable knockdown and overexpression effect are shown in Supplementary Figure 1. shRNA sequences used for genes knockdown was listed in Supplementary table 1.
qRT-PCR
RNA isolation was performed with Trizol reagent, and qPCR was conducted using SYBR Green supermix following the manufacturer's instructions.
Cell proliferation assays
We employed the YF 555 Click-iT EdU kit (C6016L, US Everbright® Inc., China) to conduct the EdU assay following the manufacturer's instructions.
Cell migration & invasion assays
To assess cell migration and invasion was performed following the manufacturer's instructions. Finally, the migrated/invaded cells were counted using an inverted microscope.
Statistical analysis
All presented results were replicated in three independent experiments. Statistical significance was determined using a one-tailed unpaired Student's t-test.
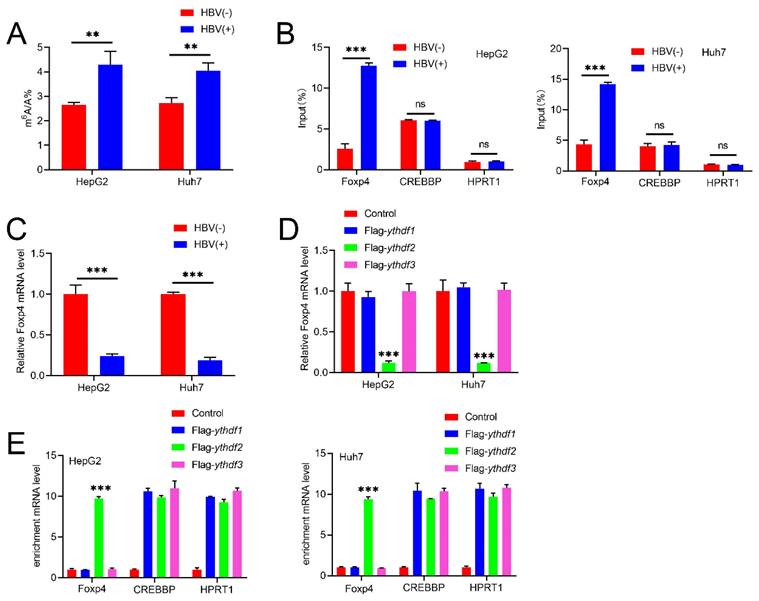
HBV modulates m6A modification on Foxp4. (A) m6A level in indicated HCC cells. (B)MeRIP-PCR was performed on m6A modified HBV transcripts and Foxp4 mRNA. (C)The mRNA level of Foxp4 in HCC cells was determined using qRT-PCR. (D) Enriched Foxp4, CREBBP, and HPRT1 mRNA was quantified by qRT-PCR. (E) HCC cells were transfected with FLAG-YTHDF2 and Foxp4 mRNA quantified. Ns P > 0.05; **P <0.01; ***P < 0.001.
Results
HBV modulates m6A modification on Foxp4
LC-MS/MS assay revealed a significant increase in m6A abundance in HBV-infected HCC cell lines (Figure.1A). To further investigate the mechanism, we conducted MeRIP assays, which showed that HBV transfection elevates m6A modification levels of Foxp4 mRNA. To validate our findings, we used CREBBP (a cellular RNA known to be modified by m6A) as a positive control and HPRT1 (a cellular RNA that lacks m6A modification) as a negative control (19). The results indicated that HBV-induced m6A modification of Foxp4 mRNA positively regulates its expression levels (Figure 1C). The stability and translation of m6A-modified RNAs are governed by "reader proteins" such as YTHDFs (20). FLAG-YTHDF1 immunoprecipitated samples demonstrated an enrichment of Foxp4 mRNAs compared to control samples and those immunoprecipitated with FLAG-YTHDF2 or FLAG-YTHDF3. CREBBP and HPRT1 RNAs served as positive and negative controls, respectively (Figure 1D). Transient expression of YTHDF1 in HCC cells resulted in elevated Foxp4 mRNA levels (Figure 1E). Collectively, these results establish that Foxp4 mRNA undergoes m6A modification, which is induced by HBV infection.
HBV enhances FOXP4 mRNA stability via modulation of m6A modification
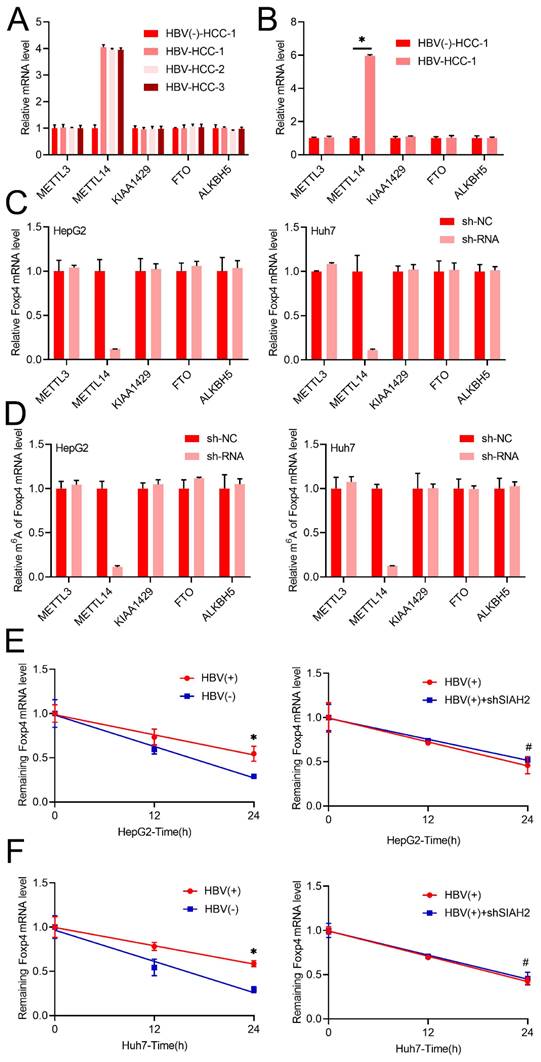
Following we noted that METTL14 mRNA levels increased significantly in both chronic HBV patient samples and primary human hepatocytes infected with HBV, other m6A regulators such as METTL3, KIAA1429, ALKBH5, and FTO remained unchanged (Figure 2A and B). Interestingly, only knock down METTL14, while not other m6A regulators such as METTL3, KIAA1429, ALKBH5, and FTO decreased m6A and mRNA level of Foxp4 in HCC cells (Figure 2C and D). Additionally, transient HBV expression was shown to increase the half-life of Foxp4 mRNA, while the absence of METTL14 prevented HBV-induced changes in Foxp4 mRNA levels (Figure 2E and F).
FoxP4 potentiates the malignant phenotype of HCC cells
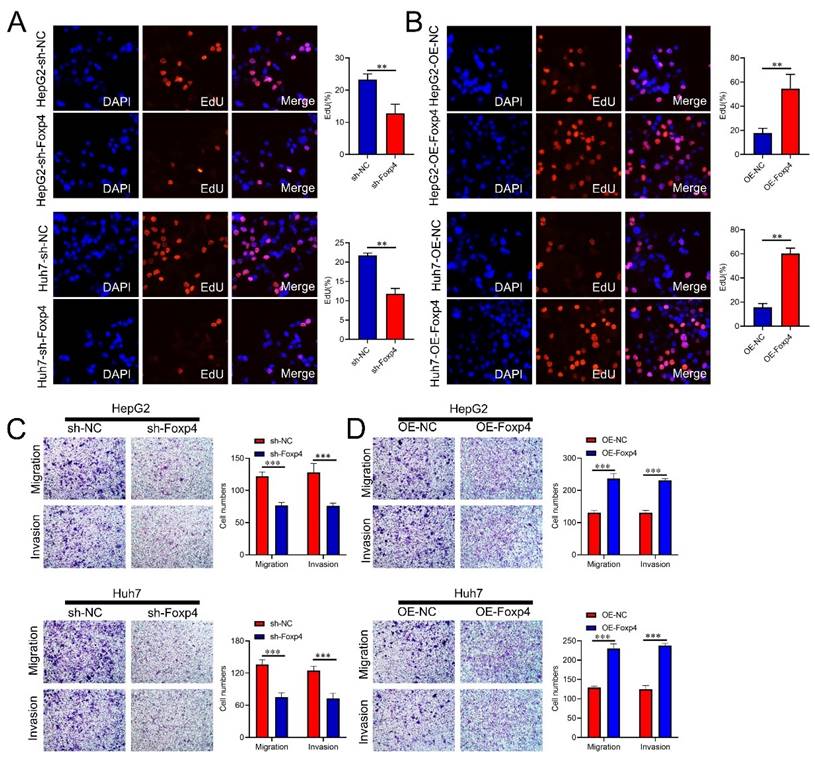
The Foxp4 knockdown significantly reduced the proportion of EdU-positive cells in HCC cells compared to the control group (Figure 3A), while Foxp4 overexpression had the opposite effect (Figure 3B). Transwell assays demonstrated that downregulation of Foxp4 significantly inhibited cell migration and invasion (Figure 3C), while overexpression of Foxp4 enhanced these abilities (Figure 3D).
HBV stimulates the PI3K/AKT pathway by enhancing m6A modification of Foxp4 mRNA
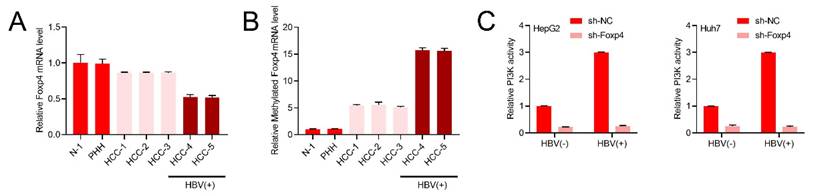
Our research revealed a significant decrease in Foxp4 mRNA expression levels in both HBV-positive and negative HCC patients (Figure 4A). m6A methylation of Foxp4 mRNA was found to be elevated in all HCC samples, with HBV-positive patients exhibiting higher levels compared to HBV-negative cases (Figure 4B). Further investigation into the PI3K activity in HBV expressing cells supported this notion (Figure 4C).
Discussion
Hepatitis B virus has been identified as playing a crucial role in promoting tumor development by affecting the epigenetic regulation of host cells. However, the extent to which newly defined epitranscriptomics regulation is involved in HBV-induced epigenetic instability remains largely unexplored (21-25). The inhibition of Foxp4 by HBV may serve as one of the mechanisms that accelerates tumor development in cases of chronic virus infection. It has been observed that HBV transfection leads to alterations in cellular m6A profiling, prompting further investigation into how HBV specifically impacts m6A modification of cellular RNAs. Interestingly, research has found that HBV infection results in an increase in METTL14 expression levels in chronic HBV patients, HBV-positive hepatocellular carcinoma (HCC) patients, and primary human hepatocytes infected with HBV. This raises the compelling possibility that the upregulation of METTL14 expression by HBV is linked to the development of liver tumors. This hypothesis was supported by analyses showing changes in METTL14 expression levels in HBV-negative HCC patient samples and HepG2 cells transfected with HBV. Furthermore, studies demonstrate that m6A modification plays a crucial role in regulating mRNA stability, splicing, nuclear export, and translation of target mRNAs. Specifically, the increase in Foxp4 mRNA levels induced by METTL14 was found to be primarily due to the slowed degradation of Foxp4 mRNA, orchestrated by the m6A reader protein YTHDF1 (26), resulting in elevated Foxp4 protein levels. These findings strongly suggest that HBV-induced upregulation of METTL14 in normal liver cells promotes m6A modification of Foxp4 mRNA, leading to increased RNA stability and potentially contributing to the development of HCC.
Interferon-alpha is a powerful tool in the treatment of HBV infection (27-31). Adjuvant interferon therapy following hepatectomy has been proven to reduce the recurrence of HBV-related HCC and enhance overall survival rates in patients (30,31). Our research has discovered increased expression of Foxp4 during HBV infection, whether it potentially inhibits p-IRF-3 nuclear import and disrupts the IFN signaling pathway has not been studied and is worth exploring in the future.
The fork head box (FOX) protein, specifically FOXP4, is a crucial player in various biological processes, particularly in HCC (14-17). This research sheds light on the critical role of FOXP4 in the HBV-HCC progression, providing valuable insights for potential clinic-therapy interventions. This study marks a significant contribution to understanding the complex functions of FOXP4 in cancer development.
HBV enhances FOXP4 mRNA stability via modulation of m6A modification. (A and B) The mRNA levels of key enzymes involved in m6A modification were measured in the liver biopsies and PHH infected with HBV. (C) qRT-PCR quantification of Foxp4 mRNA levels in HCC cells with shMETTL3/METTL14/KIAA1429/FTO/ALKBH5. (D). MeRIP-qRT-PCR analysis was m6A level of Foxp4 mRNA in HCC cells with shMETTL3/METTL14/KIAA1429/FTO/ALKBH5. (E) qRT-PCR analysis of remaining Foxp4 mRNA. (F) The HBV transfected HCC cells were depleted for remaining Foxp4 mRNA. *P < 0.05.
FoxP4 potentiates the malignant phenotype of HCC cells. (A-B) Cell viability of HCC cells was examined by EdU assays, magnification x 400. (C and D) The transwell assays were performed in indicated HCC cells. Magnification x 400. ** P < 0.01; *** P < 0.001.
HBV stimulates the PI3K/AKT pathway by enhancing m6A modification of Foxp4 mRNA. (A) qRT-PCR quantification of Foxp4 mRNA levels. (B) MeRIP-qRT-PCR analysis of samples in (A). (C) PI3K activity in Indicated HCC cells.
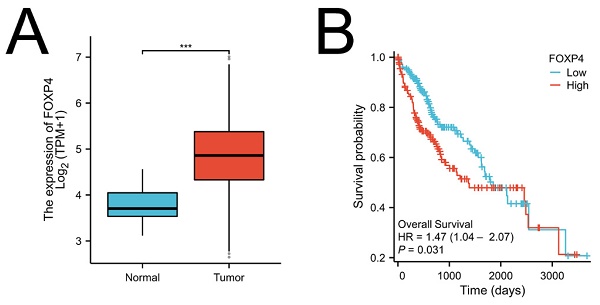
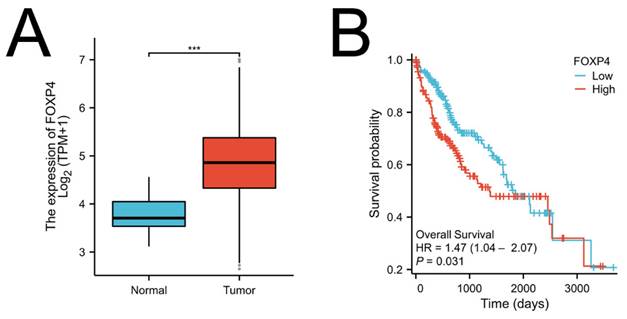
Foxp4 levels linked to poor survival in HCC. (A) Foxp4 mRNA expression in HCC tissues and normal tissues. (B) Kaplan-Meier curve of Foxp4 mRNA expression for overall survival (OS) in HCC. The high/low Foxp4 group was cut based on the mean expression in HCC.The high Foxp4 group=185, the low Foxp4 group=184. *** P < 0.001.
Shortcomings of the paper, we are unable to obtain sufficient clinical samples for validation the correlation of expression profiles of METTL14, N6-Methyladenosine, and FOXP4 with any of clinic parameters. In summary, our findings shed light on how HBV manipulates host gene expression to facilitate long-term infection by influencing m6A modification processes. Specifically, the virus-induced modification of Foxp4 through m6A regulation may play a role in the development of liver cancer linked to HBV infection. These insights reveal new pathways for potential therapeutic interventions against HBV-related hepatocarcinogenesis.
Supplementary Material
Supplementary figure and table.
Acknowledgements
Funding
This study was supported by The Natural Science Foundation of Jiangxi Province (20212BCJ23024).
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
All authors performed data analysis and interpretation of results, and participated in planning and performing the experiments. Ting-ting Fan and Xin Ye conceived and planned the experiments. All authors contributed to the article and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314
2. Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68
3. Hu J, Protzer U, Siddiqui A. Revisiting Hepatitis B Virus: Challenges of Curative Therapies. J Virol. 2019;93:e01032-19
4. An P, Xu J, Yu Y, Winkler CA. Host and Viral Genetic Variation in HBV-Related Hepatocellular Carcinoma. Front Genet. 2018;9:261
5. Chen CY, Chen J, He L, Stiles BL. Foxp4: Tumor Suppressor and Metabolic Regulator. Front Endocrinol (Lausanne). 2018;9:338
6. Carnero A, Paramio JM. The Foxp4/PI3K/AKT Pathway in vivo, Cancer Mouse Models. Front Oncol. 2014;4:252
7. Li S, Zhu MZ, Pan RG, Fang T, Cao YY, Chen SL, Zhao XL. et al. The tumor suppressor Foxp4 has a critical role in antiviral innate immunity. Nature Immunology. 2016;17:241-249
8. Lamontagne J, Mell JC, Bouchard MJ. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog. 2016;12:e1005438
9. Kim GW, Lee SH, Cho H, Kim M, Shin EC, Oh JW. Hepatitis C Virus Core Protein Promotes miR-122 Destabilization by Inhibiting GLD-2. PLoS Pathog. 2016;12:e1005714
10. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343-1355
11. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-328
12. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE. et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1:16011
13. Gonzales-van Horn SR, Sarnow P. Making the Mark: The Role of Adenosine Modifications in the Life Cycle of RNA Viruses. Cell Host Microbe. 2017;21:661-669
14. Gu T, Pan H, Zhang F, Wang L, Cheng Z. The FOXP4-AS1/miR-3130-3p/SP4 feedback loop is associated with prostate cancer. Cell Mol Biol (Noisy-le-grand). 2022Sep30;68(10):161-166
15. Luo SW, Tang L, Zhou D, Bo H, Fan LQ. FOXP4 promotes proliferation of human spermatogonial stem cells. Asian J Androl. 2023;25(3):322-330
16. Li X, Zou S, Tu X, Hao S, Jiang T, Chen JG. Inhibition of Foxp4 Disrupts Cadherin-based Adhesion of Radial Glial Cells, Leading to Abnormal Differentiation and Migration of Cortical Neurons in Mice. Neurosci Bull. 2023Jul;39(7):1131-1145
17. Zhang G, Zhang G. Upregulation of FoxP4 in HCC promotes migration and invasion through regulation of EMT. Oncol Lett. 2019Apr;17(4):3944-3951
18. Kim G-W. et al. HBV-Induced Increased N6 Methyladenosine Modification of Foxp4 RNA Affects Innate Immunity and Contributes to HCC. Hepatology. 2021;73:533-547
19. Zhen H. et al. Decreased Expression of Programmed Death Ligand-L1 by Seven in Absentia Homolog 2 in Cholangiocarcinoma Enhances T-Cell-Mediated Antitumor Activity. Front Immunol. 2022;13:834193
20. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-120
21. Kisan A, Chhabra R. Modulation of gene expression by YTH domain family (YTHDF) proteins in human physiology and pathology. J Cell Physiol. 2023;238:5-31
22. Wang Y, Wang Y, Gu J, Su T, Gu X, Feng Y. The role of RNA m6A methylation in lipid metabolism. Front Endocrinol (Lausanne). 2022Sep8;13:866116
23. Rizzo GEM, Cabibbo G, Craxì A. Hepatitis B Virus-Associated Hepatocellular Carcinoma. Viruses. 2022May7;14(5):986
24. Premkumar M, Anand AC. Overview of Complications in Cirrhosis. J Clin Exp Hepatol. 2022;12(4):1150-1174
25. Fang Q, Chen H. The significance of m6A RNA methylation regulators in predicting the prognosis and clinical course of HBV-related hepatocellular carcinoma. Mol Med. 2020Jun17;26(1):60
26. Yen YP, Chen JA. The m6A epitranscriptome on neural development and degeneration. J Biomed Sci. 2021May27;28(1):40
27. Ye J, Chen J. Interferon and Hepatitis B: Current and Future Perspectives. Front Immunol. 2021Sep7;12:733364
28. Zhang F, Yuan Y, Ma F. Function and Regulation of Nuclear DNA Sensors During Viral Infection and Tumorigenesis. Front Immunol. 2021Jan11;11:624556
29. Sun J, Wu G, Pastor F, Rahman N, Wang WH, Zhang Z, Merle P, Hui L, Salvetti A, Durantel D, Yang D, Andrisani O. RNA helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis B virus replicating hepatocytes. Gut. 2022May;71(5):991-1005
30. Zhao LN, Yuan HF, Wang YF, Yun HL, Zheng W, Yuan Y, Geng Y, Zhao M, Feng LY, Zhang XD. IFN-α inhibits HBV transcription and replication by promoting HDAC3-mediated de-2-hydroxyisobutyrylation of histone H4K8 on HBV cccDNA minichromosome in liver. Acta Pharmacol Sin. 2022Jun;43(6):1484-1494
31. Xiong J, Jiang Y, Zhang J, Chen Y, Hu Y. CK1α upregulates the IFNAR1 expression to prompt the anti-HBV effect of type I IFN in hepatoma carcinoma cells. Virol Sin. 2022Dec;37(6):894-903
Author contact
![]() Corresponding authors: Xin Ye. Department of Endoscopy, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, 200438, China. Email: charliebraunedu.cn. Ting-ting Fan. Department of Endoscopy, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, 200438, China. Email: funtingtingcom.
Corresponding authors: Xin Ye. Department of Endoscopy, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, 200438, China. Email: charliebraunedu.cn. Ting-ting Fan. Department of Endoscopy, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, 200438, China. Email: funtingtingcom.

 Global reach, higher impact
Global reach, higher impact