Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(19):6468-6478. doi:10.7150/jca.101636 This issue Cite
Research Paper
Identifying CEACAM1 as a potential prognostic biomarker for basal-like breast cancer by bioinformatics analysis and in vitro experiments
1. Department of Clinical Laboratory Center, The First Affiliated Hospital of Anhui University of Chinese Medicine, Hefei, China.
2. Department of Cancer center, The First Dongguan Affiliated Hospital, Guangdong Medical University, Dongguan, China.
3. Department of Medical Oncology, The First Affiliated Hospital of Hainan Medical University, Haikou, China.
4. Department of Clinical Laboratory, Anhui No.2 Provincial People's Hospital, Hefei, China.
5. Department of Medical Oncology, People's Hospital of Wanning, Wanning, Hainan Province, China.
* Both are co-first authors.
Received 2024-7-30; Accepted 2024-9-30; Published 2024-10-21
Abstract
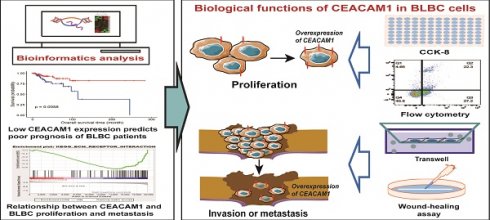
Background: Carcinoembryonic antigen related cell adhesion molecule-1 (CEACAM1) is a very important intercellular adhesion molecule, and its prognostic relevance to breast cancer (BC), especially basal-like breast cancer (BLBC), remains poorly understood.
Methods: CEACAM1 mRNA expression data for BC were sourced from the Cancer Genome Atlas (TCGA) database. Kaplan-Meier survival analysis and Cox regression analysis were used to evaluate the prognostic relationship between CEACAM1 expression and BC. Signaling pathways associated with CEACAM1 were analysed using Gene Set Enrichment Analysis (GSEA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Moreover, cell counting kit-8 (CCK-8), flow cytometry, transwell and wound-healing assays were employed to identify the biological functions of CEACAM1 in BLBC.
Results: CEACAM1 was correlated with overall survival (OS) of BLBC patients. Compared with the subgroup with better prognosis, the levels of CEACAM1 mRNA expression were significantly lower in the subgroup of BLBC with poorer prognosis. Both univariate and multivariate Cox regression analysis suggested that down-regulation of CEACAM1 expression may be an independent factor for poor prognosis in BLBC patients. GSEA and KEGG analysis revealed that CEACAM1 was negatively related with signaling pathways including extracellular matrix (ECM) receptor interaction, focal adhesion, and cell adhesion. The results of in vitro experiments indicated that CEACAM1 not only induced apoptosis of BLBC cells, but also inhibited the invasive and metastatic ability of cancer cells.
Conclusions: CEACAM1 may contribute to improving the OS of BLBC patients due to its ability to inhibit the proliferation and metastasis of cancer cells. Therefore, CEACAM1 could be used as a potential prognostic biomarker and therapeutic target in BLBC.
Keywords: Breast cancer, Basal-like breast cancer (BLBC), CEACAM1, Prognosis, Biomarker
Introduction
As a common malignant tumor all over the world, breast cancer (BC) is one of the leading causes of death in women[1]. Despite the fact that much progress accomplished in the treatment of breast cancer in recent years, some patients still died from cancer recurrence, metastasis and drug resistance[2, 3]. Basal-like breast cancer (BLBC) is a specific category of invasive BC, and most BLBC are triple-negative breast cancers (TNBC), which are negative for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2). In comparison to other types of BC, TNBC is more prevalent in younger women and has a more unfavourable prognosis due to its highly invasive, metastatic ability and high recurrence rate[4, 5]. Early diagnosis and treatment are keys to improving breast cancer survival rate, therefore, seeking and identifying specific biomarkers will help in targeted therapy, early diagnosis and prognosis of BC[6-8].
As a member of the carcinoembryonic antigen (CEA) family, CEA related cell adhesion molecule-1 (CEACAM1) is a single-chain transmembrane glycoprotein belonging to the superfamily of immunoglobulin[9]. In addition to mainly mediating cell-to-cell adhesion, CEACAM1 has a variety of biological functions and participates in various physiological and pathological processes, such as cell proliferation, apoptosis, differentiation, lymphatic vessel neogenesis and angiogenesis[10-12]. Currently, studies have confirmed the strong relationship between CEACAM1 and the progression of tumor, and the CEACAM1 expression levels are generally dysregulated in most tumors[13-16]. It has been found that CEACAM1 expression is significantly up-regulated in cancers such as pancreatic[17] and thyroid cancers[18], which suggests that CEACAM1 may be a cancer-promoting molecule. However, other studies have also confirmed that CEACAM1 expression is significantly decreased in cancers such as hepatocellular carcinoma[19], bladder cancer[20, 21], kidney cancer[22], etc., thereby indicating that CEACAM1 may function as a potential tumour suppressor. The reason for this contradiction may be related to the expression characteristics of CEACAM1 in different tumor tissues (cell membrane-type expression or cytoplasmic expression), but the exact reason is still not very clear. In any case, abnormal expression of CEACAM1 is of great significance in the diagnosis of tumors, the assessment of disease progression, and prognostic judgments[16, 23].
According to different shearing modes, CEACAM1 has 12 isoform structures, of which CEACAM1-3L, 3S, 4L, and 4S are the major isoforms[24]. Different CEACAM1 isoforms play distinct regulatory roles in various tumors. It was found that both CEACAM1-4S and 4L overexpression could promote the invasive and metastatic ability of colon cancer cell line HT29[25, 26], while the invasive and metastatic ability of hepatocellular carcinoma cell line HLF was impaired after overexpression of CEACAM1-4S and 4L[27]. Additionally, overexpression of both CEACAM1-4S and 4L were also reported to attenuate the invasive capability of gastric cancer cells NUGC3[28]. However, CEACAM1-4L, 3L and 4S were capable of promoting the ability of melanoma cells metastasis and invasion, while CEACAM1-3S significantly inhibited the invasion and metastasis of melanoma[29]. Previously, it has been reported that CEACAM1 expression was obviously deregulated in BC and might serve as a diagnostic biomarker in BC[30, 31], but until now there are no reports on the relationship between CEACAM1 and prognosis of patients with BC.
Here the prognostic correlation between CEACAM1 and BLBC and its related potential mechanisms were firstly analysed by bioinformatics in the present study, and then we explored the regulatory roles of the major isoforms CEACAM1-3L and 4L on proliferation, invasion and metastasis of BC.
Materials and methods
Basal-like breast cancer cell lines
MDA-MB-231 (Procell, CL-0150, Wuhan, China), Hs-578T (Procell, CL-0114, Wuhan, China) and normal mammary epithelial cells MCF-10A (Procell, CL-0525, Wuhan, China) were all obtained from Pricella Biotechnology Co., Ltd.(Wuhan, China). Dulbecco's modified eagle's medium (DMEM) (Vivacell, C3113, Shanghai, China) were employed to cultivate MDA-MB-231 and Hs-578T cells. MCF-10A was incubated using specific epithelial culture medium (Procell, CL-0525, Wuhan, China). All cell culture media contained 10% fetal bovine serum (FBS) (Vivacell, C04001-500, Shanghai, China). Cells were all cultured in a humidified environment at 37 ℃, 5% CO2.
Data acquisition
In the present study, gene expression data about CEACAM1 in breast cancer was sourced from the Cancer Genome Atlas (TCGA) database, but was downloaded directly from the database of UCSC Cancer Browser (https://xenabrowser.net/) and cbioportal (http://www.cbioportal.org/). The corresponding clinical information on subtypes, prognosis, and other information about BC samples were also downloaded from UCSC Cancer Browse.
Survival analysis and Cox regression analysis
To investigate the association between CEACAM1 gene expression and survival of breast cancer patients with different subtypes, we collected data of BC patients from the TCGA database. For BC patients who were lost follow-up before death, the time of last follow-up was usually calculated as the time of death. Finally, 927 BC patients in the TCGA database were included in this study. Depending on the median level of CEACAM1 expression as the threshold, all subjects were divided into CEACAM1 high expression group and low expression group. The research used Kaplan-Meier survival analysis and compared the differences of the survival between two groups using the log-rank test.
To further appraise the potential of CEACAM1 as an independent prognostic factor for BLBC, cox regression analysis was carried out on the CEACAM1 data in the database of TCGA-BLBC, and hazard ratios (HRs) and 95% confidence intervals (CI) were calculated. Furthermore, alignment diagram was created to predict one-, three-, and five-year survival probabilities for BLBC patients. A total score was calculated based on the score corresponding to each factor, and the possibility of survival in BLBC was predicted by the total score.
Gene Set Enrichment Analysis (GSEA)
The software of GSEA (http://www.broadinstitute.org/gsea) was used for the purpose of analysing signaling pathways associated with high and low CEACAM1 expression. CEACAM1 gene expression data of BLBC were analysed through the genome database (c2.cp.kegg.v7.4.symbols.gmt) in the software of GSEA. Results such as normalized enrichment scores (NES), FDR corrected pathways and p-values were acquired through the GSEA software. In general, the set of pathway genes with |NES| > 1 and FDR q-values < 0.05 were regarded as statistically significant.
Differentially expressed genes (DEG)
The "limma" package in R software was utilized to examine DEG associated with high or low CEACAM1 expression. According to the Benjamini-Hochberg method, |log2(foldchange)| > 1 and p < 0.05 were chosen as the threshold for screening significantly DEG.
Analysis of protein interaction networks
The online tools (https:/string-db.org/ and http://genemania.org) were used to screen genes whose corresponding proteins interact with CEACAM1.
The pathway analysis of Kyoto Encyclopedia of Gene and Genome (KEGG)
Based on the CEACAM1-related DEG, KEGG pathway analysis were conducted using the package "clusterProfiler" in the software of R. p < 0.05 was deemed to be significantly enriched term. We used the ggplot2 package for R to present the top-ranked pathways in terms of the number of enriched mRNAs in a bubble plot.
RT-qPCR
Cells were collected and RNA was extracted, and cDNA was synthesised using a reverse transcription kit (YEASEN, 11121ES60, Shanghai, China), followed by RT-qPCR using SYBR Green qPCR reagent (Biosharp, BL698A, Beijing, China). Moreover, RT-PCR for this study was performed using primers specific for human CEACAM1-4L (NM001712) were: F: 5'-AAACCAGAGTCTCCCGTCCT-3'; R: 5'-TTGTGCTCTCTGTGAGATCACGC-3'), and specific primers for human CEACAM1-3L (X14831) were: F: 5'-TCACTGATAATATGCTCTACCACAAGA-3'; R: 5'-TTGTGCTCTCTGTGAGATCACGC-3'. The primer for human GAPDH were: F: 5'-GCCATCACGTATCGTGGAAGG-3', R: 5'-GCCATCACGCCACAGTTTC-3'.
Cell transfection
The empty vector of plasmids (Miaoling Biology, P40122, Wuhan, China) served as a control. Plasmids contained CEACAM1-3L and CEACAM1-4L genes were constructed. BLBC cells (MDA-MB-231 or Hs-578T) were seeded into 6-well plates (3 × 105 cells/well) and 1.25μg of plasmid DNA was transfected using lipofectamine2000 reagent (Invitrogen, 11668-019, Carlsbad, USA). After 6 h of transfection, cells were rinsed and continued to be cultured in complete growth medium for 48 h, and then lysed for PCR analysis or other experiments.
Cell counting kit-8 (CCK-8)
The suspension of BLBC cells (100 μL/well, 4500 cells/well) were seeded into 96-well plates and 5 μg of plasmid DNA was transfect using lipofectamine 2000. After culturing the cells for 48 h, 10 μL solution reagent of CCK-8 (Biosharp, BS350C, Beijing, China) was pipetted into each well. 2 h incubation later, the absorbance was detected at 450 nm and the change ratio in cellular activity was calculated.
Analysis of apoptosis by flow cytometry
BLBC cells suspension in 6-well plate (100 μL/well, 105~106 cells/well) were pre-cultured for 24h, transfected 5 μg of plasmid DNA and continued to culture for 48h. The cells were then collected by trypsin digestion without EDTA, and washed twice with PBS and added 5 uL of Annexin V and 5 uL of Propidium Iodide (PI) (Biosharp, BL107B, Beijing, China). After reaction 15min at room temperature avoiding light, the proportion of apoptotic cells were analysed by flow cytometry.
Transwell invasion assay
After overexpression transfection, MDA-MB-231 or Hs-578T cells (1000 per chamber) were seeded into a transwell chamber (LABSELECT, 14361, Shanghai, China) of a 24-well plate and incubated for 48 h. The cancer cells were fixed in the transwell chamber by methanol and stained with 0.1% crystal violet. Finally, the cancer cells in the lower layer of the chamber were photographed after rinsing twice with PBS.
Wound-healing assay
MDA-MB-231 or Hs-578T cells in 6-well plates (1x105 cells/well) were inoculated and cultured to achieve 100% fusion. The cultures were then scraped to form a wound line. After 24h, the wound area was observed with a microscope (magnification ×200; COIC microscope, Chongqing Shiguang, China) and the rate of cell migration was calculated.
Statistical analysis
The statistical software of SPSS 20.0 (IBM SPSS, USA) and R software were used to analyse data of this study. Normally distributed data were described as mean ± standard deviation, and differences between groups were analysed by Student's t-test or one-way ANOVA. Kaplan-Meier survival analysis, cox regression analysis, alignment diagram, DEG analysis, KEGG, and Vann diagram were carried out by the software of R. p < 0.05 was deemed to be statistically significant.
Results
Data preprocessing and survival analysis
The mRNA expression data for breast cancer in TCGA downloaded from the UCSC Cancer Browser was log2(FPKM + 1), which was subsequently converted to log2(TPM + 1). The time frame of the 927 breast cancer samples sourced from the TCGA database ranged from May 2001 to October 2012. Based on molecular classification of the tumors, all samples included 490 Lum A subtypes (52.86%), 192 Lum B subtypes (20.71%), 77 Her2 subtypes (8.31%), and 168 Basal subtypes (18.12%) (Table 1). In 168 BLBC samples of our study, 59 were younger than 50 years and 109 were older than or equal to 50 years (Table 1). In addition, TNM stage in BLBC and the groups according to survival analysis parameters were also expressed as categorical variables (Table 1). However, there was a missing number of cases in the BLBC subgroup due to incomplete clinical data for some patients (Table 1).
CEACAM1 expression levels in different subgroups of BLBC according to TCGA data
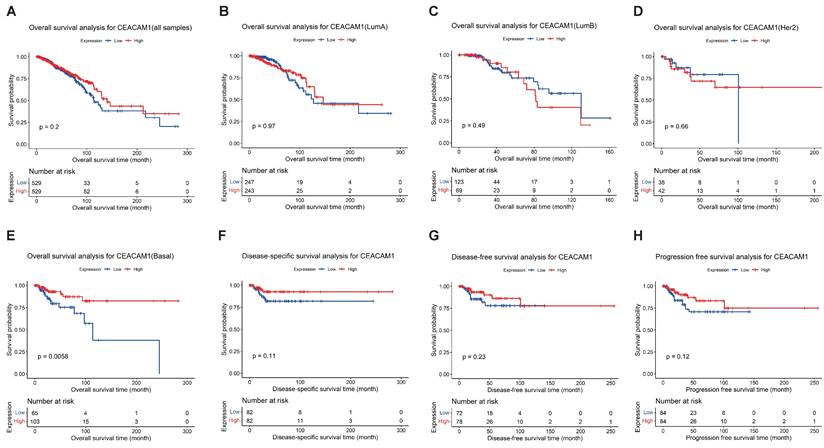
Depending on the data of TCGA-BLBC, we compared the levels of CEACAM1 mRNA in various clinical subgroups of BLBC and noticed that CEACAM1 expression were down-regulated in the subgroups with poorer prognosis (Table 2). Furthermore, in the four clinical variables including stage, OS, DSS, and PFS, the levels of CEACAM1 expression were significantly decreased in the poorer prognostic subgroup by comparison to the better prognostic subgroup (p = 0.0428, 0.0094, 0.0176, 0.0105, respectively) (Table 2 and Suppl. Figure S1). However, as some patients' clinical data were incomplete, this may have affected the comparison of CEACAM1 expression in subgroups of BLBC.
Survival analysis of CEACAM1 in BC on the basis of TCGA data. Overall survival analysis of CEACAM1 in BC and its subgroups (A-E), and DSS, DFS, and PFS analysis of CEACAM1 in BLBC (F-H).
Clinicopathological characteristics of the BC patients from TCGA database.
| Group | Variable | Number | (%) |
|---|---|---|---|
| Subtypes | LumA | 490 | 52.86 |
| LumB | 192 | 20.71 | |
| Her2 | 77 | 8.31 | |
| Basal | 168 | 18.12 | |
| Basal group | |||
| Age | < 50 | 59 | 35.12 |
| ≥ 50 | 109 | 64.88 | |
| Stage | I | 21 | 12.65 |
| II-VI | 145 | 87.35 | |
| T classification | T1 | 31 | 18.56 |
| T2-T4 | 136 | 81.44 | |
| M classification | M0 | 147 | 98.00 |
| M1 | 3 | 2.00 | |
| N classification | N1-N2 | 151 | 89.88 |
| N3 | 17 | 10.12 | |
| Tumor status | Tumor free | 136 | 91.89 |
| With tumor | 12 | 8.11 | |
| OS times (months) | Alive | 146 | 86.90 |
| Dead | 22 | 13.10 | |
| DSS times (months) | Alive or dead tumor free | 150 | 91.46 |
| Dead with tumor | 14 | 8.54 | |
| PFS times (months) | Censored | 143 | 85.12 |
| Progression | 25 | 14.88 | |
| DFS times (months) | DiseaseFree | 133 | 88.67 |
| Recurred/Progressed | 17 | 10.12 |
We performed survival analyses of CEACAM1 expression in all breast cancer samples and observed no correlation between the levels of CEACAM1 expression and overall survival (OS) (Figure 1A-D). The survival analysis of CEACAM1 was also explored in different subtypes of BC patients, and we found that CEACAM1 was significantly correlated with OS in BLBC patients, and high CEACAM1 expression predicted a better prognosis (p = 0.0056)(Figure 1E). In addition, by examining the relationship between CEACAM1 expression and disease-specific survival (DSS), disease-free survival (DFS) and progression-free survival (PFS), it was also suggested that patients with high CEACAM1 expression had a better prognosis, however, the difference of statistics was not significant (p > 0.05)(Figure 1F-H).
Regression analysis suggests that CEACAM1 may be an prognostic biomarker in BLBC
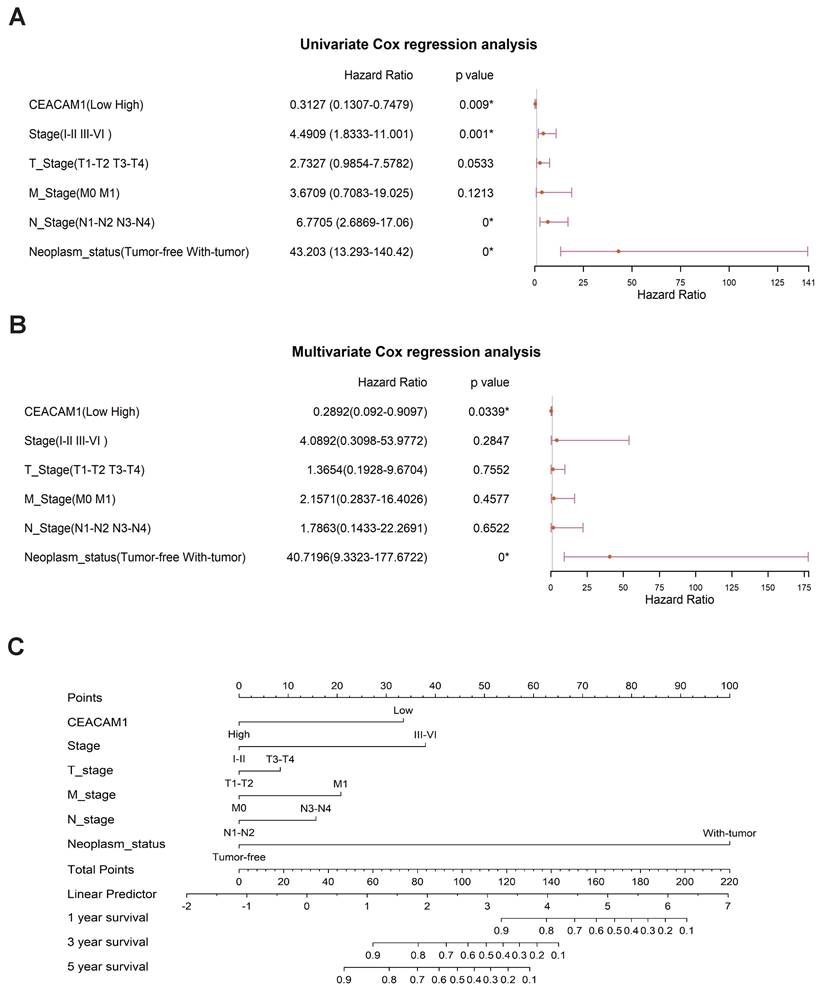
Significant correlations between CEACAM1 expression and BLBC stage (I-II vs III-IV), N-stage (N1-N2 vs N3-N4), and tumor status (tumor absence vs presence) (p = 0.001, 0.000, < 0.0001, respectively) (Figure 2A) were obtained by univariate cox regression analysis. Multivariate cox regression analysis also showed a significant relationship between CEACAM1 expression and the status of tumor presence or absence in BLBC (p < 0.0001) (Figure 2B). Furthermore, we investigated the relationship between CEACAM1 expression and the 1, 3, 5-year survival of BLBC patients by plotting an alignment diagram, and the results showed a higher probability of longer survival in BLBC patients with high CEACAM1 expression (Figure 2C). The aforementioned results collectively indicate that CEACAM1 may be a potential prognostic biomarker in BLBC.
GSEA analysis of CEACAM1-related signaling pathways
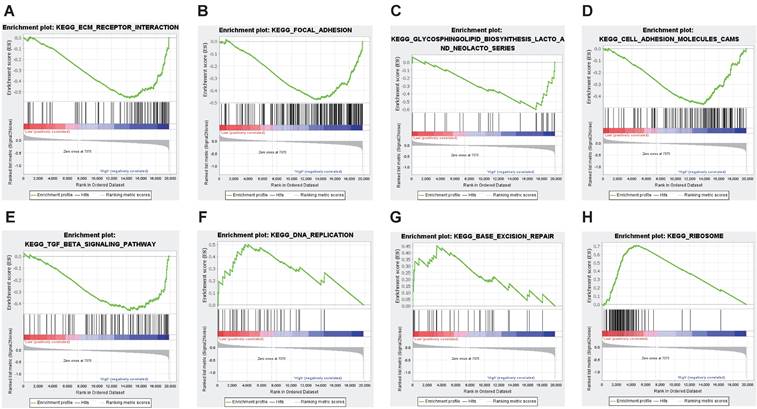
The discrepancy of prognosis between BLBC patients with high and low CEACAM1 expression may be related to some important signaling pathways. Therefore, we conducted GSEA analysis and depending on the set threshold, we obtained a total of 10 pathways enriched in CEACAM1 low expression set and 5 pathways enriched in CEACAM1 high expression set. These findings suggested that CEACAM1 was negatively related to signal pathways such as focal adhesion, extracellular matrix (ECM) receptor interaction, and cell adhesion, etc., while CEACAM1 was positively correlated with signaling pathways such as ribosome, DNA replication, base excision repair signaling pathways (Table 2 and Figure 3).
CEACAM1 expression levels in different clinical subgroups of BLBC patients from TCGA database.
| Variable | Group | Number | Mean ± SD | p |
|---|---|---|---|---|
| Age | < 50 | 59 | 4.6266 ± 1.4976 | 0.2984 |
| ≥ 50 | 109 | 4.3490 ± 1.7075 | ||
| Stage | Ⅰ | 21 | 5.1329 ± 1.5789 | 0.0428* |
| Ⅱ-Ⅳ | 145 | 4.3543 ± 1.6296 | ||
| T classification | T1 | 31 | 4.4785 ± 1.6046 | 0.4923 |
| T2-T4 | 136 | 4.2071 ± 1.9137 | ||
| M classification | M0 | 147 | 4.5544 ± 1.6204 | 0.6056 |
| M1 | 3 | 4.0652 ± 0.9752 | ||
| N classification | N1-N2 | 151 | 4.5046 ± 1.6687 | 0.1734 |
| N3 | 17 | 3.9300 ± 1.2734 | ||
| Tumor status | Tumor free | 136 | 4.4002 ± 1.6343 | 0.2448 |
| With tumor | 12 | 3.8292 ± 1.3415 | ||
| OS | Alive | 146 | 4.5739 ± 1.6085 | 0.0094* |
| Dead | 22 | 3.6011 ± 1.6130 | ||
| DSS | Alive or dead tumor free | 150 | 4.5200 ± 1.6224 | 0.0176* |
| Dead with tumor | 14 | 3.4400 ± 1.3555 | ||
| PFS | Censored | 143 | 4.5817 ± 1.6318 | 0.0105* |
| Progression | 25 | 3.6732 ± 1.4796 | ||
| DFS | Disease Free | 133 | 4.5955 ± 1.6447 | 0.0617 |
| Recurred/Progressed | 17 | 3.8061 ± 1.3804 |
To further explore the function of CEACAM1, we firstly obtained 33 significantly differentially expressed genes that interacted with CEACAM1 in BLBC (Figure 4A-B). Among these genes, only one demonstrated an increase in expression, while 32 exhibited a decrease. And 10 and 20 reciprocal genes were also obtained by String and GeneMANIA, respectively (Figure 4C-D). These CEACAM1-related genes (Totally 55 genes, Figure 4E) were subjected to KEGG pathway analysis, and 23 significantly enriched pathways were found, and we also found that CEACAM1 was correlated with signaling pathways, such as focal adhesion, ECM-receptor interaction, etc (Figure 4F).
Cox regression analysis of CEACAM1 as a prognostic marker for BLBC. Forest plots of cox regression analysis displayed the association between CEACAM1 and clinicopathological parameters (stage, T-stage, N-stage, M-stage, tumor absence or presence) in patients with BLBC (A-B). Alignment diagram of the relationship between CEACAM1 and prognosis of BLBC patients (C).
CEACAM1 regulates proliferation and apoptosis in BLBC cells
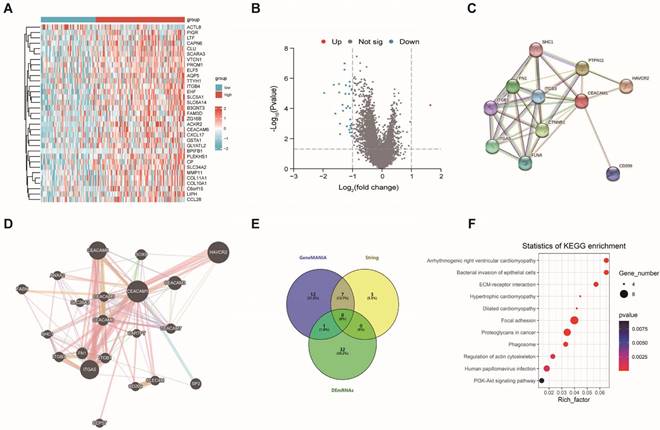
The relative mRNA expression of CEACAM1-3L and 4L in BLBC cells (MDA-MB-231 and Hs-578T) was obviously lower than that in normal cells (MCF-10A) by PCR experiments (Figure 5A-B). Based on the bioinformatics analysis, CEACAM1 was found to have the potential to regulate apoptosis in breast cancer, and overexpression of CEACAM1-3L and 4L in BLBC cells was identified to significantly suppress the proliferative activity of cancer cells by CCK-8 assay (Suppl. Figure S2 and Figure 5C). In addition, flow cytometric analysis of BLBC cells apoptosis after overexpression of CEACAM1-3L and 4L also showed that CEACAM1 remarkably promotes apoptosis of cancer cells (Figure 5D).
GSEA analysis of potential signaling pathways associated with CEACAM1. CEACAM1 was negatively related to pathways such as ECM receptor (A), focal adhesion (B), glycosphingolipid biosynthesis lacto and neolacto series(C), cell adhesion molecules CAMS (D), TGF-beta signaling (E). CEACAM1 was positively correlated with signaling pathways such as ribosome (F), DNA replication (G), base excision repair signaling pathways (H).
CEACAM1-related genes and its KEGG analysis. DEG between BLBC with high and low CEACAM1 expression (A-B). Genes that interacted with CEACAM1 were obtained by String and GeneMANIA (C-D). DEG and related genes obtained from String and GeneMANIA database were performed to produce a Vann diagram to find 55 CEACAM1-related genes (E). KEGG pathway analysis of signaling pathways with CEACAM1-related genes (F).
CEACAM1 regulates invasion and migration of basal-like breast cancer cells
Since bioinformatics analysis suggested that CEACAM1 was significantly negatively related with prognosis of BLBC patients, and the main reason affecting the prognosis of BC patients might be invasion and metastasis. In the current study, the findings through wound-healing experiment also demonstrated that the migration rates of BC cells (MDA-MB-231 and Hs-578T) overexpressing CEACAM1-3L and 4L were significantly less than that in the groups of blank and negative control (NC) (Figure 6A). Moreover, in the transwell experiment, we similarly found that the number of invasive cancer cells in BC cells overexpressing CEACAM1-3L and 4L was also significantly less than that in the blank and NC groups (Figure 6B).
Discussion
As one of the most common malignancies in female, the prognosis of breast cancer, especially BLBC, remains disappointing[1, 3]. Currently, breast cancers are often classified into four major subtypes based on molecular typing: Luminal A and Luminal B, basal-like, and HER2 overexpression. In particular, approximately 75% of BLBCs belong to TNBC, and the lack of ER, PR, and HER receptors limits the use of endocrine and targeted therapies. The prognosis of BLBC is usually related to tumor volume, grade and early recurrence. In addition, BLBC is highly invasive and metastatic, usually spreading to organs such as the brain, bone, and lung, which in turn leads to an unfavourable prognosis. Therefore, development of new biomarkers to elucidate the determinants of invasive and metastatic disease and thus improve the prognosis of this disease is imminent.
As an important molecule mediating cell-cell adhesion, CEACAM1 is also able to regulate cell proliferation, apoptosis, and lymphatic and vascular neogenesis. However, the expression of CEACAM1 in different tumors is contradictory. CEACAM1 is significantly up-regulated in gastric[32], pancreatic[17], and thyroid cancers[18], but decreased in hepatocellular[19], bladder[21], and renal cancers[22], for which the reasons were not completely clarified. Our study found that CEACAM1 expression was reduced in BLBC cells (MDA-MB-231 and Hs-578T) compared to normal breast cells (MCF-10A). Despite the paradoxical trends of CEACAM1 expression in different tumors, CEACAM1 has been verified to play a pivotal role during tumor development. Therefore, the biofunction and clinical values of CEACAM1 in BC remain to be further ascertained. In the present study, we conducted an investigation of the expression level, prognostic significance, and biological function of CEACAM1 in BLBC for the first time.
CEACAM1 expression in BLBC cells and its regulation of cell viability. mRNA expression of CEACAM1-3L and 4L in MCF-10A, MDA-MB-231, and Hs-578T (A-B). The proliferative activity of BLBC cells overexpressed CEACAM1-3L and 4L was measured by CCK-8 assays (C). Flow cytometric analysis of apoptosis in BLBC cells after overexpression of CEACAM1-3L and 4L (D). *** p < 0.001, *** p < 0.0001.
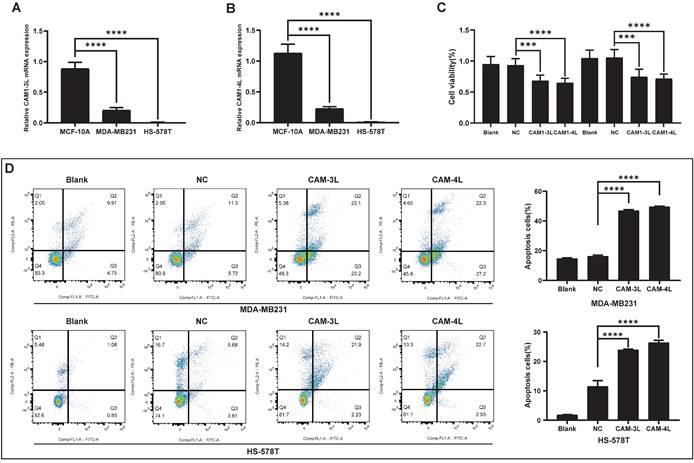
CEACAM1 inhibits BLBC cells migratory and invasive capacity. The migration rate (A) and number of invading cells (B) of MDA-MB-231 and Hs-578T overexpressing CEACAM1-3L and 4L were assessed by the experiments of wound-healing and transwell. *p < 0.05, ** p < 0.01, *** p < 0.001, *** p < 0.0001.
There is a close relationship between abnormal alterations in CEACAM1 expression and tumor progression and prognosis. Previously it was found that the survival of CEACAM1-negative lung adenocarcinoma patients was significantly longer than that of CEACAM1-positive patients[33]. Since there was a significant relationship observed between CEACAM1 and microvessel density (MVD), patients with esophageal squamous carcinoma with high CEACAM1 expression had poorer survival[34]. However, by immunohistochemical analysis of 235 gastric cancer patients, the results revealed that patients with high CEACAM1 expression had significantly longer survival compared with those with low CEACAM1 expression[28]. Furthermore, immunohistochemical analysis of tumor tissue microarrays from 17,747 patients with prostate cancer confirmed that absence of CEACAM1 expression predicted a poor prognosis in prostate cancer[35]. In the present study, by analysing the bioinformatics data, our findings demonstrated that CEACAM1 expression levels were related with survival of BLBC patients, and patients with high CEACAM1 expression exhibited a favourable prognosis. Furthermore, the expression of CEACAM1 was markedly lower in the subgroup with poorer prognosis than in the subgroup with better prognosis for four clinical parameters: stage, OS, DSS, and PFS. Subsequently, cox regression analysis demonstrated a significant relationship between CEACAM1 and the stage of BLBC, which further suggests that CEACAM1 is an potential prognostic marker for BLBC patients.
To further reveal the relationship between CEACAM1 and BLBC prognosis, the biofunctions of CEACAM1 in the pathogenesis of BLBC need to be ascertained. We firstly performed GSEA analysis, which revealed that CEACAM1 was negatively correlated with focal adhesion, ECM receptor interaction, and cell adhesion, but positively correlated with Ribosome, DNA replication, Base excision repair and other pathways. Then, KEGG pathway analysis was performed on DEG between the CEACAM1 low and high expression patients. These differentially expressed genes identified in our study were engaged in the development of breast cancer, including focal adhesion, the interaction of ECM receptor, and PI3K-Akt pathway. The relatively consistent findings were confirmed in oral cancer, where CEACAM1 was associated with these signaling pathways, and low expression of CEACAM1 in oral cancer led to worse prognosis[36]. The modulation between ECM and cells through focal adhesion, ECM receptors and actin cytoskeleton influences the morphology, adhesion and migration status of cells, which is important for tumor invasion and metastasis[37-39]. In addition, PI3K-Akt pathway is also involved in breast cancer proliferation and invasion[40]. Thus, CEACAM1 may modulate these pathways to influence BLBC occurrence and metastasis. Recently, it has been confirmed that decreasing CEACAM1 expression in liver cancer cells (Mahlavu and SK-Hep-1) may inhibit invasion and migration of tumor cells[41]. To further validate the suppressive properties of the CEACAM1 molecule on BC cells in in vitro experiments, we chose the main protein isoforms of CEACAM1-3L and 4L for the study. However, our results revealed that overexpression of CEACAM1-3L and 4L not only induced apoptosis and inhibited the viability of BLBC cells, but also inhibited the invasive and metastatic potential of cancer cells. Therefore, CEACAM1 may contribute to the OS of BLBC patients by inhibiting the proliferation and metastasis of cancer cells.
In our study, we sought to ascertain the correlation between CEACAM1 and prognostic value in BLBC, as well as gained preliminary insight into the biological functions of CEACAM1-3L and 4L in BLBC. However, it is undeniable that there are still potential limitations of this study that deserve further consideration. First, although bioinformatics analysis indicated that low expression of CEACAM1 predicted an unfavorable prognosis for BLBC patients, a large number of clinical trials are required to be conducted to confirm our results. Second, we predicted signaling pathways associated with CEACAM1 based on online databases, but absence of relevant experimental evidence.
Conclusion
This study revealed that low CEACAM1 expression was strongly correlated with poor prognosis in BLBC by bioinformatics. The outcomes of GSEA and KEGG pathway analysis also suggested that CEACAM1 was engaged in regulating the proliferation and metastasis of BLBC. Moreover, we demonstrated the main isoforms of CEACAM1 suppressed the proliferation, invasion and metastasis of BLBC by cytological experiments. Therefore, CEACAM1 may improve patients' OS by inhibiting the proliferative ability and metastasis of BLBC. In the future, more attention should be paid to CEACAM1 and its different isoforms in the study of BLBC, the related molecular mechanisms should be further explored through more experiments, and the clinical significance of CEACAM1-3L or CEACAM1-4L in BLBC patients will be investigated.
Abbreviations
BC: breast cancer; BLBC: basal-like breast cancer; TNBC: triple-negative breast cancer; ER: estrogen receptor; PR: progesterone receptor; HER-2: human epidermal growth factor receptor 2; CEA: carcinoembryonic antigen; CEACAM1: CEA related cell adhesion molecule-1; TCGA: The Cancer Genome Atlas; GSEA: Gene Enrichment Analysis; KEGG: Kyoto Encyclopedia of Gene and Genome; HRs: hazard ratios; CI: confidence intervals; OS: overall survival; ECM: extracellular matrix; DSS: Disease-specific Survival; DFS: Disease-Free Survival; PFS: Progression-Free Survival; RT-qPCR: Real-time fluorescence quantitative polymerase chain reaction; CCK-8: Cell Counting Kit-8
Supplementary Material
Supplementary figures.
Acknowledgements
Funding
This project was supported by Anhui Provincial Health Research Project Fund (AHWJ2023BAa20124, AHWJ2023A20505), the Hainan Province Clinical Medical Center (No. (2021)75 and No. (2021)276), Hainan Provincial Natural Science Foundation of China (No. 820RC765 and 823RC587) and the National Natural Science Foundation of China (No. 82260582).
Author contributions
Boke Zhang, Haixia Huang and Changcheng Yang collected and organized the data; Boke Zhang and Chuanzhu Wang analysed and interpreted the data. Boke Zhang wrote this article and Liu Ran checked it. Changcheng Yang and Ran Liu supervised this manuscript; Final approval of manuscript: All authors.
Availability of data and materials
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kamangar F, Dores G, Anderson W. Patterns of Cancer Incidence, Mortality, and Prevalence Across Five Continents: Defining Priorities to Reduce Cancer Disparities in Different Geographic Regions of the World. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2023;41:5209-24
2. Henriques B, Mendes F, Martins D. Immunotherapy in Breast Cancer: When, How, and What Challenges? Biomedicines. 2021;9:1687
3. Pandey S, Mondal S, Kajal K, Kurmi B, Verma S, Patel P. Current progress in the targeted therapy of breast cancer: Structure-activity correlation and docking studies (2015-2021). Archiv der Pharmazie. 2023;356:e2200602
4. Trillo P, Sandoval J, Trapani D, Nicolò E, Zagami P, Giugliano F. et al. Evolution of biological features of invasive lobular breast cancer: Comparison between primary tumour and metastases. European journal of cancer (Oxford, England: 1990). 2023;185:119-30
5. Grinda T, Antoine A, Jacot W, Cottu P, de la Motte Rouge T, Frenel J. et al. Real-world clinical and survival outcomes of patients with early relapsed triple-negative breast cancer from the ESME national cohort. European journal of cancer (Oxford, England: 1990). 2023;189:112935
6. Subhan M, Parveen F, Shah H, Yalamarty S, Ataide J, Torchilin V. Recent Advances with Precision Medicine Treatment for Breast Cancer including Triple-Negative Sub-Type. Cancers. 2023;15:2204
7. Chaudhuri A, Kumar D, Dehari D, Patil R, Singh S, Kumar D. et al. Endorsement of TNBC Biomarkers in Precision Therapy by Nanotechnology. Cancers. 2023;15:2661
8. Jagannathan G, White M, Xian R, Emens L, Cimino-Mathews A. A New Landscape of Testing and Therapeutics in Metastatic Breast Cancer. Clinics in laboratory medicine. 2023;43:299-321
9. Chen T, Zimmermann W, Parker J, Chen I, Maeda A, Bolland S. Biliary glycoprotein (BGPa, CD66a, CEACAM1) mediates inhibitory signals. Journal of leukocyte biology. 2001;70:335-40
10. LeBlanc S, Arabzadeh A, Benlolo S, Breton V, Turbide C, Beauchemin N. et al. CEACAM1 deficiency delays important wound healing processes. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2011;19:745-52
11. Ergün S, Kilik N, Ziegeler G, Hansen A, Nollau P, Götze J. et al. CEA-related cell adhesion molecule 1: a potent angiogenic factor and a major effector of vascular endothelial growth factor. Molecular cell. 2000;5:311-20
12. Memaj P, Ouzerara Z, Jornayvaz F. Role of Oxidative Stress and Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 in Nonalcoholic Fatty Liver Disease. International journal of molecular sciences. 2023;24:11271
13. Jeon S, Kang M, Jeon M, Chung Y, Kim A, Lee Y. et al. CEACAM1 Marks Highly Suppressive Intratumoral Regulatory T Cells for Targeted Depletion Therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2023;29:1794-806
14. Liu J, Muturi H, Khuder S, Helal R, Ghadieh H, Ramakrishnan S. et al. Loss of Ceacam1 promotes prostate cancer progression in Pten haploinsufficient male mice. Metabolism: clinical and experimental. 2020;107:154215
15. Zhou M, Jin Z, Liu Y, He Y, Du Y, Yang C. et al. Up-regulation of carcinoembryonic antigen-related cell adhesion molecule 1 in gastrointestinal cancer and its clinical relevance. Acta biochimica et biophysica Sinica. 2017;49:737-43
16. Li J, Liu X, Duan Y, Wang H, Su W, Wang Y. et al. Abnormal expression of circulating and tumor-infiltrating carcinoembryonic antigen-related cell adhesion molecule 1 in patients with glioma. Oncology letters. 2018;15:3496-503
17. Zińczuk J, Zaręba K, Romaniuk W, Kamińska D, Nizioł M, Baszun M. et al. Expression of Chosen Carcinoembryonic-Related Cell Adhesion Molecules in Pancreatic Intraepithelial Neoplasia (PanIN) Associated with Chronic Pancreatitis and Pancreatic Ductal Adenocarcinoma (PDAC). International journal of medical sciences. 2019;16:583-92
18. Liu W, Wei W, Winer D, Bamberger A, Bamberger C, Wagener C. et al. CEACAM1 impedes thyroid cancer growth but promotes invasiveness: a putative mechanism for early metastases. Oncogene. 2007;26:2747-58
19. Mao C, Yin H, Ning H, Peng Z, Li K, Ding G. Levels of HBx, VEGF, and CEACAM1 in HBV-related hepatocellular carcinoma and their correlation with cancer prognosis. European review for medical and pharmacological sciences. 2017;21:3827-33
20. Ella-Tongwiis P, Lamb R, Makanga A, Shergill I, Hughes S. The role of antibody expression and their association with bladder cancer recurrence: a single-centre prospective clinical-pilot study in 35 patients. BMC urology. 2020;20:187
21. Igami K, Uchiumi T, Shiota M, Ueda S, Tsukahara S, Akimoto M. et al. Extracellular vesicles expressing CEACAM proteins in the urine of bladder cancer patients. Cancer science. 2022;113:3120-33
22. Kang Z, Wang L, Liu J, Ouyang H, Xie S, Tan Y. Expression of CEACAM1 and CD105 in Renal Cell Carcinoma and Its Correlation with Microvessel Density. Critical reviews in eukaryotic gene expression. 2021;31:1-9
23. Yang F, Zeng Z, Li J, Ren X, Wei F. TIM-3 and CEACAM1 are Prognostic Factors in Head and Neck Squamous Cell Carcinoma. Frontiers in molecular biosciences. 2021;8:619765
24. Kim W, Huang Y, Gandhi A, Blumberg R. CEACAM1 structure and function in immunity and its therapeutic implications. Seminars in immunology. 2019;42:101296
25. Nittka S, Böhm C, Zentgraf H, Neumaier M. The CEACAM1-mediated apoptosis pathway is activated by CEA and triggers dual cleavage of CEACAM1. Oncogene. 2008;27:3721-8
26. Yamaguchi S, Yokoyama S, Ueno M, Hayami S, Mitani Y, Takeuchi A. et al. CEACAM1 is associated with recurrence after hepatectomy for colorectal liver metastasis. The Journal of surgical research. 2017;220:353-62
27. Kiriyama S, Yokoyama S, Ueno M, Hayami S, Ieda J, Yamamoto N. et al. CEACAM1 long cytoplasmic domain isoform is associated with invasion and recurrence of hepatocellular carcinoma. Annals of surgical oncology. 2014;21:505-14
28. Takeuchi A, Yokoyama S, Nakamori M, Nakamura M, Ojima T, Yamaguchi S. et al. Loss of CEACAM1 is associated with poor prognosis and peritoneal dissemination of patients with gastric cancer. Scientific reports. 2019;9:12702
29. Ullrich N, Heinemann A, Nilewski E, Scheffrahn I, Klode J, Scherag A. et al. CEACAM1-3S Drives Melanoma Cells into NK Cell-Mediated Cytolysis and Enhances Patient Survival. Cancer research. 2015;75:1897-907
30. Yang C, Cao M, Liu Y, He Y, Yang C, Du Y. et al. Inhibition of cell invasion and migration by CEACAM1-4S in breast cancer. Oncology letters. 2017;14:4758-66
31. Yang C, He P, Liu Y, He Y, Yang C, Du Y. et al. Assay of serum CEACAM1 as a potential biomarker for breast cancer. Clinica chimica acta; international journal of clinical chemistry. 2015;450:277-81
32. Shi J, Xu S, He P, Xi Z. Expression of carcinoembryonic antigen-related cell adhesion molecule 1(CEACAM1) and its correlation with angiogenesis in gastric cancer. Pathology, research and practice. 2014;210:473-6
33. Laack E, Nikbakht H, Peters A, Kugler C, Jasiewicz Y, Edler L. et al. Expression of CEACAM1 in adenocarcinoma of the lung: a factor of independent prognostic significance. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:4279-84
34. Qian W, Huang P, Liang X, Chen Y, Guan B. High expression of carcinoembryonic antigen-associated cell adhesion molecule 1 is associated with microangiogenesis in esophageal squamous cell carcinoma. Translational cancer research. 2020;9:4762-9
35. Luebke A, Ricken W, Kluth M, Hube-Magg C, Schroeder C, Büscheck F. et al. Loss of the adhesion molecule CEACAM1 is associated with early biochemical recurrence in TMPRSS2:ERG fusion-positive prostate cancers. International journal of cancer. 2020;147:575-83
36. Sai M, Zhonghua W, Chao L, Zhenli L, Xuan Z, Liheng L. et al. CEACAM1 as a molecular target in oral cancer. Aging (Albany NY). 2023;15:8137-8154
37. Chellini L, Caprara V, Spadaro F, Sestito R, Bagnato A, Rosanò L. Regulation of extracellular matrix degradation and metastatic spread by IQGAP1 through endothelin-1 receptor signalling in ovarian cancer. Matrix Biology. 2019;81:17-33
38. Sun L, Liu L, Liu X, Wang Y, Li M, Yao L. et al. MGr1-Ag/37LRP induces cell adhesion-mediated drug resistance through FAK/PI3K and MAPK pathway in gastric cancer. Cancer Science. 2014;105:651-9
39. Hu H-H, Wang S-Q, Shang H-L, Lv H-F, Chen B-B, Gao S-G. et al. Roles and inhibitors of FAK in cancer: current advances and future directions. Frontiers in Pharmacology. 2024;15:1274209
40. Tian Y, Zhao L, Gui Z, Liu S, Liu C, Yu T. et al. PI3K/AKT signaling activates HIF1α to modulate the biological effects of invasive breast cancer with microcalcification. npj Breast Cancer. 2023;9:93
41. Chueh-Tan C, Chian-Feng C, Tung-Yi L, Wei-Jyun H, Kate H, Ching-Yao T. et al. Traditional Chinese medicine Kuan-Sin-Yin decoction inhibits cell mobility via downregulation of CCL2, CEACAM1 and PIK3R3 in hepatocellular carcinoma cells. J Ethnopharmacol. 2023;317:116834
Author contact
![]() Corresponding author: Changcheng Yang, yanggreatwalledu.cn.
Corresponding author: Changcheng Yang, yanggreatwalledu.cn.

 Global reach, higher impact
Global reach, higher impact