Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(20):6658-6667. doi:10.7150/jca.98769 This issue Cite
Research Paper
Mendelian Randomization Analysis of Non-small Cell Lung Cancer and Five Exposure Factors Based on Pathway Enrichment Analysis
1. Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang, China, 330006.
2. Department of Gastroenterology, Traditional Chinese Medicine Hospital of Wannian, Wannian, China, 335599.
3. Department of Cardiovascular Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang, China, 330006.
† These authors contributed to the work equally and should be regarded as co-first authors.
Received 2024-5-22; Accepted 2024-10-18; Published 2024-10-28
Abstract
Background: Increasing knowledge has made it crucial to identify and minimize potential risk factors in order to prevent non-small cell lung cancer (NSCLC). This initiative aims to utilize Mendelian randomization analysis to identify exposure factors that could be causally linked to NSCLC. The results will help create new ways of controlling and preventing NSCLC.
Methods: The GEO database's NSCLC data were used to find differentially expressed genes, which were further analyzed for GO and KEGG pathway enrichment. Use pathway enrichment analysis as a guide to screen exposure factors. The exposure variables that are causally associated with NSCLC were screened using a two-sample Mendelian randomization technique. Heterogeneity and pleiotropy analyze were employed to assess the validity of the study's findings.
Results: Coronary atherosclerosis, cell adhesion molecule 3 (a molecule that maintains the normal structure and function of the lungs), dipeptidase 1 (one of the major adhesion receptors for neutrophils), thimet oligopeptidase (involved in hydrolyzing a variety of vasoactive signal peptides), and dipeptidyl peptidase 2 (an intracellular protease involved in the cell differentiation process and preventing cell death). The above five exposure factors were discovered to have an inverse relationship with NSCLC. Essentially, this implies that higher levels of these components can decrease the likelihood of developing lung cancer. No heterogeneity or pleiotropy was detected, and the study results were reliable.
Conclusion: The study identified five potential exposure variables for NSCLC, laying the groundwork for treatment and prevention strategies and suggesting a new path for future research.
Keywords: Mendelian randomization analysis, Non-small cell lung cancer, Exposure factors, Pathway enrichment analysis
Introduction
Non-small cell lung cancer (NSCLC) is the predominant form of lung cancer, accounting for around 80% to 85% of all occurrences. Its increasing morbidity and mortality have brought a certain burden to the world [1]. With growing awareness, it is crucial to pinpoint potential risk factors for lung cancer and implement enhanced strategies for early prevention of the illness. For example, smoking is a prominent risk factor for the development and occurrence of lung cancer. Avoiding smoking or stopping smoking at the earliest opportunity can greatly reduce the occurrence of lung cancer and enhance the outlook for individuals diagnosed with lung cancer [2, 3].
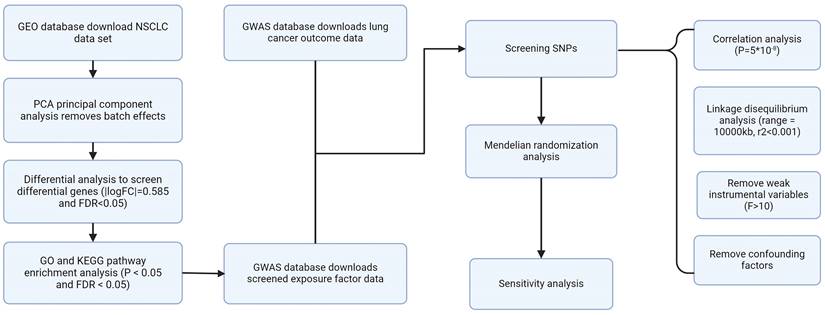
Flow chart.
Pathway enrichment analysis, as a bioinformatics tool, is a powerful method used to uncover the mechanisms behind the occurrence and progression of diseases. This method analyzes sets of genes in genomic data linked to certain biological pathways to get insights into the molecular processes of disease [4].
Mendelian randomization (MR) is a novel epidemiological approach that efficiently reduces bias and provides advantages in terms of time and cost. It can accurately assess the cause-and-effect relationship between exposure variables and lung cancer outcomes while preventing any bias induced by reverse causality or confounding factors [5].
The study aims to utilize pathway enrichment analysis to thoroughly investigate the biological pathways associated with NSCLC in order to discover relevant exposure factors. Mendelian randomization was used to confirm the causal association between the screened exposure factors and NSCLC.
Data and Methods
Acquisition and processing of data related to lung cancer
The GEO website obtained four groups of NSCLC data, including GSE19188, GSE44077, GSE75037, and GSE116959. Principal Component Analysis (PCA) was used to perform batch correction on the four acquired data sets to eliminate batch effects. Subsequently, differential analysis was employed to identify genes that were expressed differently in NSCLC (Table S1). The screening conditions were |logFC|= 0.585 and FDR<0.05. The screening process was completed through the "limma" R code package. Figure 1 shows our research process.
Performing GO and KEGG pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway enrichment analysis was conducted utilizing genes that were expressed differently. Pathway analysis was conducted utilizing the "ClusterProfiler" R tools (P < 0.05 and FDR < 0.05) [6].
Mendelian randomization data sources
The lung cancer outcome data required for Mendelian randomization (MR) and the exposure factor data screened for pathway enrichment analysis originate from genome-wide association research [7, 8]. Table S2 fully presents the exposure data information used in the study.
Mendelian randomization analysis
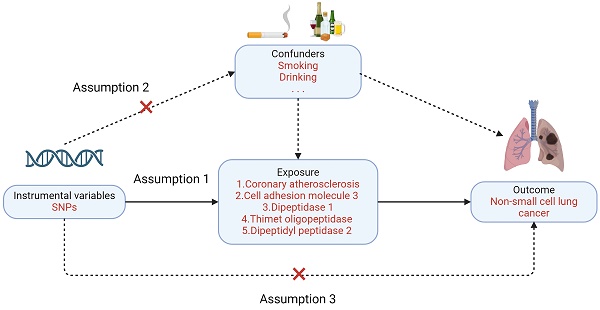
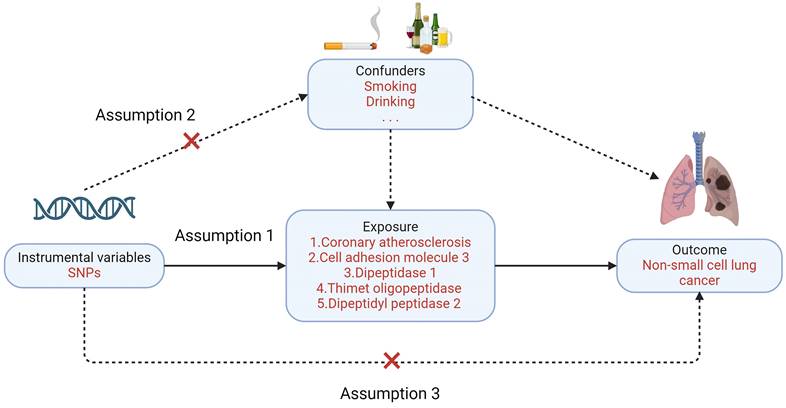
Mendelian randomization must satisfy three primary assumptions: 1. Exposure factors and instrumental variables have a significant association. 2. Confounding variables and instrumental variables are unrelated to one another; 3. Instrumental variables do not directly affect the results but affect the results through correlation with the degree of exposure. In this study, NSCLC was the outcome variable (Figure 2). Enrichment analysis is used to guide the screening of lung cancer-related exposure factors, and single nucleotide polymorphisms (SNPs) related to exposure factors are instrumental variables (IV) [9].
In the association analysis, we initially used a significance threshold of P<5×10-8 to identify SNPs that had a stronger correlation with the exposure component. Subsequently, to ensure the independence of the instrumental variables, we performed a linkage disequilibrium (LD) analysis according to the criteria of a 10,000 kb range and a r2 value below 0.001 [10]. Afterwards, the F test was used to exclude factor variables that were considered to be weak, using a standard threshold of F >10 [11]. Five approaches were employed to establish the causative association between the exposure variables and NSCLC outcomes. These methods include the inverse variance weighted (IVW) method, simple mode, weighted median, MR Egger, and weighted mode [12]. The IVW method is widely acknowledged as the primary approach for establishing causal linkages because of its strong analytical results and efficient handling of measurement mistakes [13]. To ensure the rigor of the study, we calculated the IVW and MR Egger's Cochran Q statistics to assess the heterogeneity among different SNPs, and the MR-Egger intercept test tested the pleiotropy of the study results. The test standards for heterogeneity and pleiotropy were P>0.05 [14]. Ultimately, a leave-one-out technique was used to evaluate the influence of individual SNPs on the causal relationship between exposure variables and NSCLC outcomes. The experiments were conducted using R software (version 4.2.2). The code package used for analysis is the "TwoSampleMR" package. The screening criteria used for all methodologies are shown in Table S3.
Results
Data processing and pathway enrichment analysis
We conduct PCA (principal component analysis) on the data to remove batch effects across various data sets. The visualization graphic is shown in Figure S1. Next, using differential analysis, we screened genes differentially expressed in lung cancer for further analysis. (logFC = 0.585, FDR<0.05) (Figure S2).
Analysis of pathway enrichment
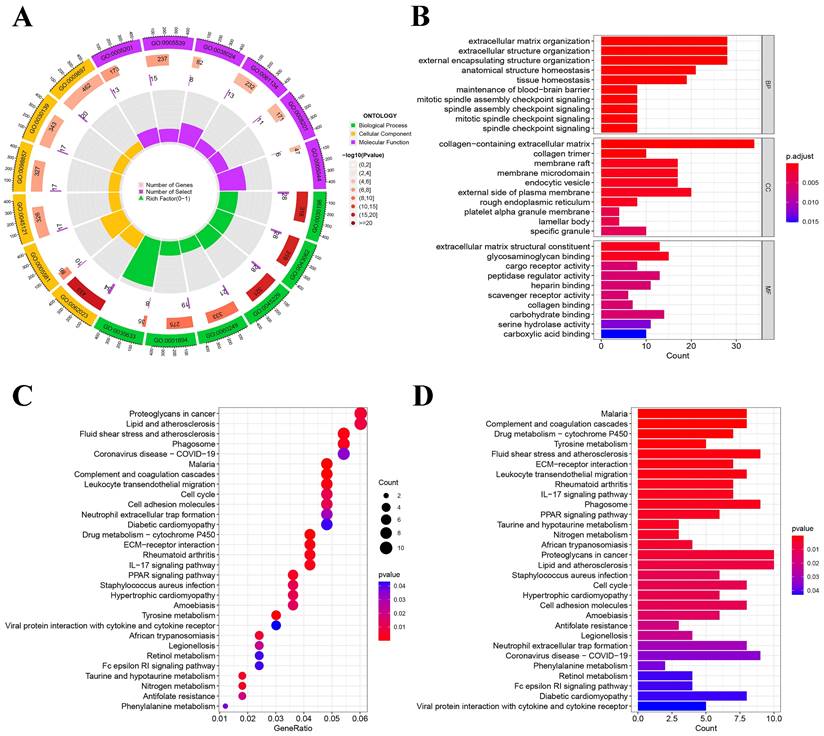
We conducted KEGG and GO pathway enrichment analyses on NSCLC using genes that were differentially expressed. Highly enriched pathways in GO enrichment analyses include GO:0062023 collagen-containing extracellular matrix, GO:0030198 extracellular matrix organization, and GO:0005539 glycosaminoglycan binding (Figure 3A-B). hsa05205: Proteoglycans in cancer, hsa05417: lipid, and atherosclerosis were highly enriched in KEGG enrichment analysis (Figure 3C-D). After screening and validating the entire enriched pathway, we identified several enriched pathways to guide the screening of exposure factors. For example: hsa05418: fluid shear stress and atherosclerosis; hsa05417: lipid and atherosclerosis; GO:0052547: regulation of peptidase activity; GO:0010466: negative regulation of peptidase activity; GO:0010811: positive regulation of cell-substrate adhesion, etc. We present all pathways obtained from our analysis in Table S4 and Table S5.
Three major assumptions of Mendelian randomization.
Non-small cell lung cancer pathway enrichment analysis. (A) Circle chart of GO enrichment analysis; (B) Histogram of GO enrichment analysis; (C) Bubble chart of KEGG enrichment analysis; (D) Histogram of KEGG enrichment analysis.
Acquisition of exposure factors
After screening After screening guided by pathway enrichment analysis, five exposure factors were included in the study, including coronary atherosclerosis, cell adhesion molecule 3, dipeptidase 1, thimet oligopeptidase (THOP1), and dipeptidyl peptidase 2. After quality evaluation of exposure factors, the number of SNPs screened out was 77, 17, 14, 14, and 9, respectively. We provide the SNP information for valid IVs in Table S6. All single nucleotide polymorphisms (SNPs) had F values exceeding 10, suggesting the effectiveness of all five instrumental variables (IVs).
Two-sample Mendelian randomization between five exposure factors and NSCLC
We employed five methodologies to evaluate the presence of a causal connection between the screened exposure factors and the risk of NSCLC (including MR Egger, IVW, weighted median, simple mode, and weighted mode). The IVW technique is the primary approach used in our study assessment. The findings indicate that there is an inverse causal association between the five exposure factors we examined and the prediction of NSCLC using the IVW technique (Figure 4). Among them, coronary atherosclerosis (OR = 0.85, 95% CI: 0.73-0.98); P = 0.02), cell adhesion molecule 3 (OR = 0.84, 95% CI: 0.73-0.97); P = 0.02), dipeptidase 1 (OR = 0.85, 95% CI: 0.74-0.99); P = 0.03), THOP1 (OR = 0.92, 95% CI: 0.86-0.98); P = 0.01), and dipeptidyl peptidase 2 (OR = 0.92, 95% CI: 0.85-1.00); P = 0.04). Consequently, elevations in these variables can diminish the likelihood of developing lung cancer. We display the specific values analyzed in the table and visualize them in Table 1.
Validation of Mendelian randomization findings
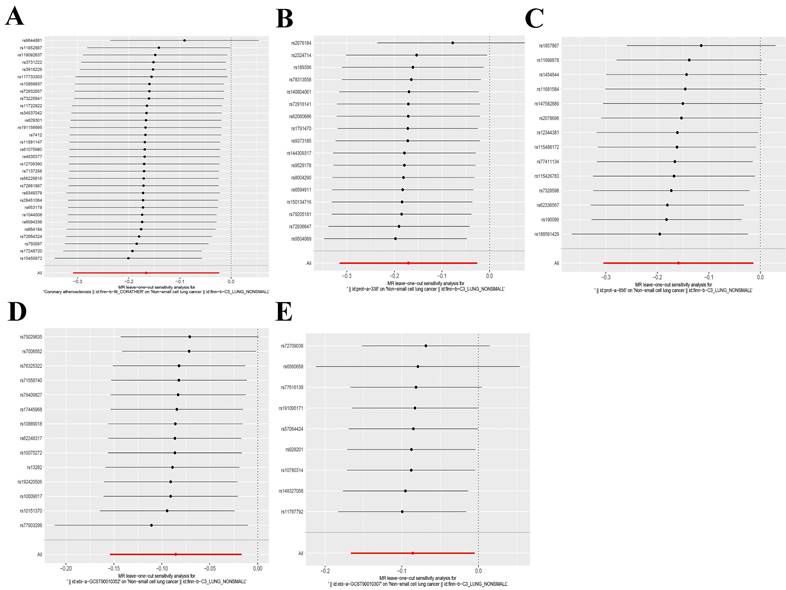
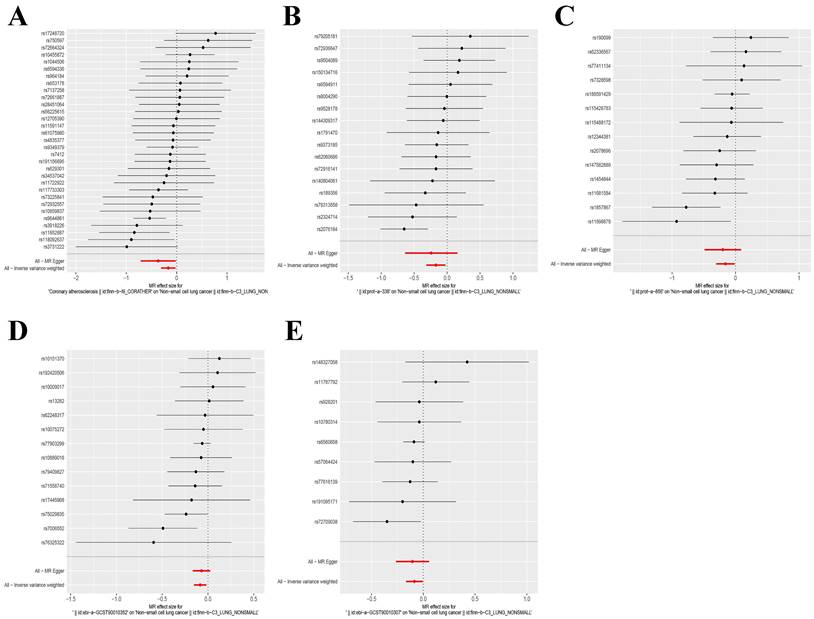
The IVW and MR-Egger approaches demonstrated the absence of heterogeneity in the five examined exposure factors. (P>0.05). In addition, the MR-Egger regression analysis demonstrated that there was no horizontal pleiotropy among the SNPs of all exposure factors. Table 2 displays the findings from the analyses of heterogeneity and pleiotropy. Leave-one-out sensitivity analysis revealed no SNPs that significantly affected causality (Figure 5). Figure 6 displays the forest plot of SNPs in the MR study., and the funnel plot can be seen in Figure S3. The above results show that the results obtained by IVW are reliable.
Verification of results
To verify the analytic results of coronary atherosclerosis, we utilised distinct exposure data (GWAS ID: ukb-d-I9_CORATHER). The IVW analysis approach confirms a significant inverse causal association between coronary atherosclerosis and NSCLC (odds ratio = 0.01, 95% confidence interval: 0.001-0.43) (Table S7-S8). Unfortunately, the validity of the remaining four exposure factors could not be confirmed due to the unavailability of relevant external data. The visualisation of the verification results is depicted in Figure S4.
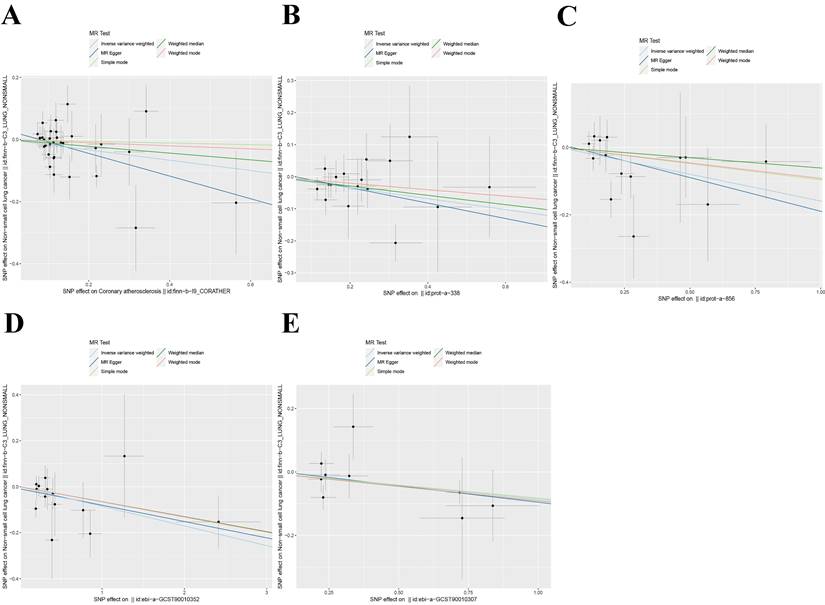
Scatter plot of Mendelian randomization analysis of five exposure factors. (A) Coronary atherosclerosis; (B) Cell adhesion molecule 3; (C) Dipeptidase 1; (D) Thimet oligopeptidase; (E) Dipeptidyl peptidase 2.
Leave-one-out analysis of five exposure factors. (A) Coronary atherosclerosis; (B) Cell adhesion molecule 3; (C) Dipeptidase 1; (D) Thimet oligopeptidase; (E) Dipeptidyl peptidase 2.
Mendelian randomization-evaluated exposure factors for the presence of non-small cell lung cancer.
| Exposure | Method | Beta | OR (95%CI) | P value |
|---|---|---|---|---|
| Coronary atherosclerosis | MR Egger | -0.37 | 0.69(0.49-0.98) | 0.05 |
| Weighted median | -0.11 | 0.90(0.73-1.11) | 0.32 | |
| Inverse variance weighted | -0.17 | 0.85(0.73-0.98) | 0.02 | |
| Simple mode | -0.03 | 0.97(0.65-1.46) | 0.90 | |
| Weighted mode | -0.05 | 0.95(0.63-1.43) | 0.81 | |
| Cell adhesion molecule 3 | MR Egger | -0.24 | 0.79(0.53-1.17) | 0.25 |
| Weighted median | -0.14 | 0.87(0.70-1.06) | 0.17 | |
| Inverse variance weighted | -0.17 | 0.84(0.73-0.97) | 0.02 | |
| Simple mode | -0.10 | 0.90(0.63-1.29) | 0.59 | |
| Weighted mode | -0.10 | 0.90(0.64-1.28) | 0.58 | |
| Dipeptidase 1 | MR Egger | -0.20 | 0.82(0.61-1.09) | 0.20 |
| Weighted median | -0.06 | 0.94(0.77-1.15) | 0.55 | |
| Inverse variance weighted | -0.16 | 0.85(0.74-0.99) | 0.03 | |
| Simple mode | -0.10 | 0.91(0.68-1.22) | 0.53 | |
| Weighted mode | -0.09 | 0.91(0.72-1.15) | 0.45 | |
| Thimet oligopeptidase | MR Egger | -0.07 | 0.93(0.85-1.03) | 0.17 |
| Weighted median | -0.07 | 0.94(0.85-1.03) | 0.19 | |
| Inverse variance weighted | -0.09 | 0.92(0.86-0.98) | 0.01 | |
| Simple mode | -0.07 | 0.94(0.79-1.10) | 0.45 | |
| Weighted mode | -0.07 | 0.94(0.85-1.03) | 0.20 | |
| Dipeptidyl peptidase 2 | MR Egger | -0.10 | 0.90(0.77-1.06) | 0.24 |
| Weighted median | -0.09 | 0.91(0.83-1.01) | 0.07 | |
| Inverse variance weighted | -0.09 | 0.92(0.85-1.00) | 0.04 | |
| Simple mode | -0.08 | 0.92(0.79-1.08) | 0.34 | |
| Weighted mode | -0.09 | 0.91(0.83-1.00) | 0.09 |
Abbreviation: P value: Probability; OR: Odds ratio; 95%CI: 95% Confidence Interval.
Heterogeneity analysis and MR-Egger regression of 5 exposure factors associated with lung cancer.
| Exposure | Heterogeneity P | MR-Egger regression | |||
|---|---|---|---|---|---|
| MR Egger | IVW | Intercept | Intercept P | ||
| Coronary atherosclerosis | 0.2628 | 0.2379 | 0.0287 | 0.2269 | |
| Cell adhesion molecule 3 | 0.4751 | 0.5383 | 0.0142 | 0.7147 | |
| Dipeptidase 1 | 0.2973 | 0.3614 | 0.0105 | 0.7493 | |
| Thimet oligopeptidase | 0.5252 | 0.5926 | -0.0093 | 0.6835 | |
| Dipeptidyl peptidase 2 | 0.4097 | 0.5089 | 0.0089 | 0.7967 | |
Abbreviation: Heterogeneity P: Heterogeneity P-value; IVW: Inverse variance weighted; Intercept P: Intercept P-value.
Forest plot of SNPs for five exposure factors. (A) Coronary atherosclerosis; (B) Cell adhesion molecule 3; (C) Dipeptidase 1; (D) Thimet oligopeptidase; (E) Dipeptidyl peptidase 2.
Discussion
During the previous few years, with the continuous advancement of science and technology, the examination and treatment methods for lung cancer have been continuously improved [15, 16]. The perception of lung cancer danger among the general public is gradually rising. Identifying potential risk factors linked to the onset of lung cancer and taking steps to avoid or minimize exposure are crucial methods for preventing and managing lung cancer in the present day. Our work employed pathway enrichment analysis to direct the screening of exposure variables in order to identify additional risk factors that are strongly linked to lung cancer. A total of 5 exposure factors were screened out, including coronary atherosclerosis, cell adhesion molecule 3, dipeptidase 1, THOP1, and dipeptidyl peptidase 2. The results of Mendelian randomization revealed a causal association in the opposite direction between these five exposure factors and the occurrence of NSCLC. More precisely, these five characteristics of exposure were linked to a reduced likelihood of getting non-small cell lung cancer.
Our research discovered a correlation between coronary atherosclerosis and a decreased likelihood of developing NSCLC. This conclusion was corroborated by the verification of various exposure data. A study examining the correlation between coronary artery calcium (CAC) and different forms of cancer revealed that male patients with a CAC level of 400 or above had a heightened susceptibility to lung cancer [17]. Chen et al.'s study also discovered that those with coronary artery disease (CAD) have a heightened susceptibility to lung cancer [18]. This is contrary to our findings. Although the aforementioned investigations have partially elucidated the correlation between coronary atherosclerosis and lung cancer, they have not fully accounted for the impact of smoking and other confounding variables. This is the key reason that the above studies are contrary to ours. Researchers conducted a study on tumor necrosis factor (TNF) and discovered that higher levels of TNF were linked to an increased risk of coronary artery disease, while lower levels of TNF were connected with a decreased risk of lung cancer [19]. While there is ongoing debate over the impact of TNF on lung cancer, it is generally accepted that TNF has a role in promoting the onset and progression of lung cancer [20, 21]. Tumor necrosis factor (TNF) is a probable crucial element in the connection between coronary artery disease (CAD) and lung cancer. Cell adhesion molecules have been shown to be involved in the induction and maintenance of tissue differentiation, and their loss or downregulation is associated with the formation of various tumors [22]. Our research suggests a correlation between cell adhesion molecule 3 and a reduced risk of developing NSCLC. A study on cell adhesion molecule 4 (CADM4) showed that CADM4 can impede the proliferation and spread of non-small cell lung cancer (NSCLC) cells by blocking the Akt signaling pathway [23]. CADM3 may also decrease the likelihood of developing lung cancer in the same manner. The expression of CADM4 in gallbladder cancer has been observed to have a negative correlation with the poor prognosis of individuals with this type of cancer [24]. However, there are still few studies on cell adhesion molecule 3. In our study, there was a reverse causal relationship between dipeptidase-1 (DPEP-1) and NSCLC. Dipeptidase-1 is a zinc-dependent metalloprotease shown to be a major adhesion receptor on the liver and lung endothelium, and its reduction reduces neutrophil recruitment to the lungs and liver [25]. Dysregulation of DPEP-1 in a variety of cancers can have an impact on cancer progression. For example, in hepatoblastoma (HB), poor patient prognosis is associated with upregulation of DPEDP-1, while in breast cancer, DPEP-1 target genes may be a tumor suppressor gene [26, 27]. Neutrophil recruitment is a crucial cellular characteristic of lung cancer, and DPEP-1 is believed to have a causal connection with lung cancer by influencing this cellular characteristic [28]. THOP1 also has a reverse causal relationship with NSCLC. THOP1 is linked to a higher susceptibility to NSCLC. Studies have shown that the expression of THOP1 in NSCLC is less than that found in healthy lung tissue. Additionally, a drop in THOP1 expression is linked to a reduction in the 5-year disease-free survival and overall survival of individuals with NSCLC [29]. THOP1 has the ability to break down a wide range of active peptides, with a particular emphasis on bradykinin (BK) [30]. Bradykinin is a significant signaling molecule that plays a crucial role in tumor-associated blood vessel formation and the progression of tumors [31]. THOP1, acting as an antagonist of BK, will likely reduce the likelihood of developing lung cancer by decreasing BK. Finally, our investigation confirmed a reverse causal connection between DPP2 and NSCLC. DPP2 is an enzyme that belongs to the serine protease family and has a crucial function in preserving cellular quiescence. It has been shown to be an important predictive factor in chronic lymphocytic leukemia (CLL). Inhibition of DPP2 can induce apoptosis in CLL cells [32, 33]. However, there are currently few studies on its use in cancer, and it still has certain research potential.
We performed GO and KEGG pathway enrichment analyses to discover the pathways enriched in NSCLC. We subsequently identified and screened exposure variables associated with these pathways using the provided information. Mendelian randomization was used to investigate the causal link between exposure factors and the outcomes of NSCLC. And finally, we screened out five exposure factors, including coronary atherosclerosis, cell adhesion molecule 3, dipeptidase 1, THOP1, and dipeptidyl peptidase 2. Mendelian randomization provides evidence that these five exposure factors have a causal association in the opposite direction with NSCLC. This method effectively eliminates the impact of confounding factors, hence enhancing the accuracy of assessing causality. It provides new research directions and a basis for the prevention and treatment of NSCLC. However, our study also has certain shortcomings. First, our research relies on public databases, and the relevant results still need further experimental verification. Secondly, some of our research methods failed to produce the same results as the IVW method, but the SNPs included in the study all met the valid IV hypothesis, and the results obtained by the study did not show heterogeneity and pleiotropy, so our results are reliable.
Conclusion
Based on GO and KEGG pathway enrichment analyses of NSCLC, we screened exposure factors related to lung cancer. Through Mendelian randomization analysis, five exposure factors were screened out, including coronary atherosclerosis, cell adhesion molecule 3, dipeptidase 1, THOP1, and dipeptidyl peptidase 2. The IVW method proves that these five exposure factors have a reverse causal relationship with NSCLC and may reduce the risk of NSCLC. Heterogeneity and pleiotropic effects analysis proved the accuracy of the study results. The specific mechanism of the interaction between exposure factors and NSCLC still requires further experimental verification.
Abbreviations
BK: bradykinin; CAC: coronary artery calcium; CAD: coronary artery disease; CADM4: cell adhesion molecule 4; CLL: chronic lymphocytic leukemia; DPEP-1: Dipeptidase-1; DPP2: dipeptidyl peptidase 2; GO: Gene Ontology; HB: hepatoblastoma; IV: instrumental variables; IVW: inverse variance weighting; KEGG: Kyoto Encyclopedia of Genes; LD: linkage disequilibrium; MR: Mendelian randomization; NSCLC: Non-small cell lung cancer; PCA: Principal Component Analysis; SNPs: single nucleotide polymorphisms; THOP1: Thimet oligopeptidase; TNF: tumor necrosis factor.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The authors appreciate all the public health workers who participated in the TCGA database and R language developers.
Funding
This study was supported by Natural Science Foundation of Jiangxi Province (Grant number: 20224BAB206071). Role of the Funding: The funding had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Availability of data and material
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
Wenxiong Zhang had full access to all the data in the manuscript and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Concept and design: All authors.
Acquisition, analysis, or interpretation of data: All authors.
Statistical analysis: Zishun Guo, Jieshu Zhang, Zhuozheng Hu, and Weijun Zhou.
Drafting of the manuscript: Zishun Guo, Jieshu Zhang, and Wenxiong Zhang.
Critical revision of the manuscript for important intellectual content: Zishun Guo, Jieshu Zhang, Wenxiong Zhang, and Shuqiang Zhu.
Supervision: Zishun Guo, Wenxiong Zhang, and Shuqiang Zhu.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Venkatesan P. European Lung Cancer Congress 2023. Lancet Respir Med. 2023;11(6):e59-e60
2. Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20(9):624-639
3. Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48(3):889-902
4. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A. et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14(2):482-517
5. Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods. 2019;10(4):486-496
6. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2(3):100141
7. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018:7:e34408.
8. Hemani G, Tilling K, Davey Smith G. Correction: Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(12):e1007149
9. Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc Nephrol. 2016;27(11):3253-3265
10. Pan S, Zhang Z, Pang W. The causal relationship between bacterial pneumonia and diabetes: a two-sample mendelian randomization study. Islets. 2024;16(1):2291885
11. Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ. et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21(3):223-42
12. Zhou H, Zhang Y, Liu J, Yang Y, Fang W, Hong S. et al. Education and lung cancer: a Mendelian randomization study. Int J Epidemiol. 2019;48(3):743-750
13. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880-906
14. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389
15. Miller M, Hanna N. Advances in systemic therapy for non-small cell lung cancer. Bmj. 2021;375:n2363
16. Chaft JE, Rimner A, Weder W, Azzoli CG, Kris MG, Cascone T. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat Rev Clin Oncol. 2021;18(9):547-557
17. Dzaye O, Berning P, Dardari ZA, Mortensen MB, Marshall CH, Nasir K. et al. Coronary artery calcium is associated with increased risk for lung and colorectal cancer in men and women: the Multi-Ethnic Study of Atherosclerosis (MESA). Eur Heart J Cardiovasc Imaging. 2022;23(5):708-716
18. Chen HH, Lo YC, Pan WS, Liu SJ, Yeh TL, Liu LY. Association between coronary artery disease and incident cancer risk: a systematic review and meta-analysis of cohort studies. PeerJ. 2023;11:e14922
19. Yuan S, Carter P, Bruzelius M, Vithayathil M, Kar S, Mason AM. et al. Effects of tumour necrosis factor on cardiovascular disease and cancer: A two-sample Mendelian randomization study. EBioMedicine. 2020;59:102956
20. Gong K, Guo G, Gerber DE, Gao B, Peyton M, Huang C. et al. TNF-driven adaptive response mediates resistance to EGFR inhibition in lung cancer. J Clin Invest. 2018;128(6):2500-2518
21. Gong K, Guo G, Beckley N, Zhang Y, Yang X, Sharma M. et al. Tumor necrosis factor in lung cancer: Complex roles in biology and resistance to treatment. Neoplasia. 2021;23(2):189-196
22. Duraivelan K, Samanta D. Emerging roles of the nectin family of cell adhesion molecules in tumour-associated pathways. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188589
23. Luo F, Zhao Y, Liu J. Cell adhesion molecule 4 suppresses cell growth and metastasis by inhibiting the Akt signaling pathway in non-small cell lung cancer. Int J Biochem Cell Biol. 2020;123:105750
24. Bang S, Jee S, Son H, Cha H, Song K, Park H. et al. Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role. Int J Mol Sci. 2023;24(8):6898
25. Choudhury SR, Babes L, Rahn JJ, Ahn BY, Goring KR, King JC. et al. Dipeptidase-1 Is an Adhesion Receptor for Neutrophil Recruitment in Lungs and Liver. Cell. 2019;178(5):1205-1221.e1217
26. Cui X, Liu X, Han Q, Zhu J, Li J, Ren Z. et al. DPEP1 is a direct target of miR-193a-5p and promotes hepatoblastoma progression by PI3K/Akt/mTOR pathway. Cell Death Dis. 2019;10(10):701
27. Green AR, Krivinskas S, Young P, Rakha EA, Paish EC, Powe DG. et al. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res Treat. 2009;113(1):59-66
28. Aloe C, Wang H, Vlahos R, Irving L, Steinfort D, Bozinovski S. Emerging and multifaceted role of neutrophils in lung cancer. Transl Lung Cancer Res. 2021;10(6):2806-2818
29. Qi L, Li SH, Si LB, Lu M, Tian H. Expression of THOP1 and its relationship to prognosis in non-small cell lung cancer. PLoS One. 2014;9(9):e106665
30. Shrimpton CN, Smith AI, Lew RA. Soluble metalloendopeptidases and neuroendocrine signaling. Endocr Rev. 2002;23(5):647-64
31. Ikeda Y, Hayashi I, Kamoshita E, Yamazaki A, Endo H, Ishihara K. et al. Host stromal bradykinin B2 receptor signaling facilitates tumor-associated angiogenesis and tumor growth. Cancer Res. 2004;64(15):5178-85
32. Danilov AV, Klein AK, Lee HJ, Baez DV, Huber BT. Differential control of G0 programme in chronic lymphocytic leukaemia: a novel prognostic factor. Br J Haematol. 2005;128(4):472-481
33. Danilov AV, Danilova OV, Brown JR, Rabinowitz A, Klein AK, Huber BT. Dipeptidyl peptidase 2 apoptosis assay determines the B-cell activation stage and predicts prognosis in chronic lymphocytic leukemia. Exp Hematol. 2010;38(12):1167-1177
Author contact
![]() Corresponding authors: Wenxiong Zhang, MD, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, 1 Minde Road, Nanchang, 330006, China. Email: Ndefy01261edu.cn. Shuqiang Zhu, MD, Department of Cardiovascular Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, 1 Minde Road, Nanchang, 330006, China. Email: 276446377com.
Corresponding authors: Wenxiong Zhang, MD, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, 1 Minde Road, Nanchang, 330006, China. Email: Ndefy01261edu.cn. Shuqiang Zhu, MD, Department of Cardiovascular Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, 1 Minde Road, Nanchang, 330006, China. Email: 276446377com.

 Global reach, higher impact
Global reach, higher impact