Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(4):1137-1148. doi:10.7150/jca.103182 This issue Cite
Review
The regulatory role and therapeutic potential of long non-coding RNA in non-small cell lung cancer
1. Donghai County People's Hospital affiliated to Kangda College of Nanjing Medical University, Lianyungang 222300, Jiangsu, China.
2. Department of General Surgery, Donghai County People's Hospital, Lianyungang 222300, Jiangsu, China.
3. School of Medicine, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu, China.
4. Department of Basic Medicine, Kangda College of Nanjing Medical University, Lianyungang 222000, Jiangsu, China.
5. Department of Oncology, The First Affiliated Hospital of Soochow University, Suzhou 215006, Jiangsu, China.
Received 2024-9-3; Accepted 2024-12-24; Published 2025-1-6
Abstract
Lung cancer remains the leading cause of cancer-related mortality worldwide, with non-small cell lung cancer (NSCLC) being the predominant subtype. Recent advances in transcriptome sequencing have highlighted the critical role of long non-coding RNAs (lncRNAs) in NSCLC, with lncRNAs influencing gene expression through epigenetic, transcriptional, and post-transcriptional mechanisms. Despite the growing understanding of lncRNAs, challenges such as delayed diagnosis and drug resistance continue to complicate NSCLC management. This review explores novel findings in the role of lncRNAs (e.g., MALAT1, HOTAIR, and GAS5) in NSCLC, with a particular focus on their encoded small peptides and N6-methyladenosine (m6A) modifications. We further discuss how the interplay between lncRNAs, their encoded peptides, and m6A modifications can provide new strategies for improving NSCLC diagnosis, treatment, and overcoming drug resistance. This review also highlights emerging research avenues that could lead to innovative clinical interventions in NSCLC.
Keywords: LncRNAs, proliferation, metastasis, targeted therapies, NSCLC
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide, accounting for approximately one in five cancer deaths[1, 2]. Non-small cell lung cancer (NSCLC) constitutes approximately 80-85% of all lung cancer cases[3]. The poor survival rates of patients with lung cancer are primarily due to the lack of effective early diagnostic biomarkers and the development of resistance to chemotherapy or targeted therapies in the advanced stages of the disease[4, 5]. Recent advancements in molecular biology have highlighted the critical role of non-coding RNAs (ncRNAs) in gene expression regulation and their implications in cancer biology, including lung cancer. NcRNAs are categorized based on nucleotide length into long non-coding RNAs (lncRNAs) and small or short non-coding RNAs. LncRNA, typically exceeding 200 nucleotides in length, represent one of the largest and most diverse families of RNAs. Despite their lack of protein-coding ability, lncRNAs make up a substantial portion of the non-coding RNA population[6-8]. Recent studies have demonstrated that the dysregulation, deletion or mutation of lncRNAs is closely associated with various diseases, including lung cancer. LncRNAs regulate gene expression through multiple mechanisms, including epigenetic modifications[9], mRNA splicing[10], interactions with microRNA[11], lncRNA-protein interactions[12], and lncRNA-mRNA interactions[13]. Furthermore, the regulatory mechanisms of lncRNA in tumor cell progression vary depending on their intracellular localization. Nuclear lncRNAs can influence biological functions by modulating chromatin structure and transcription and serving as structural scaffolds to anchor nuclear domains, thereby regulating biological processes[14]. Contrastingly, cytoplasmic lncRNAs often impact cell signaling by regulating post-transcriptional modifications and mRNA translation[15]. Currently, accumulating studies have revealed that certain lncRNAs can encode small open reading frame (sORF)-derived peptides, which are believed to contribute to the sensitivity of cancer treatment[16]. Additionally, RNA modification at the N6 position of internal adenosine (m6A) in ncRNAs by the methyltransferase is essential for malignant maintenance of tumor cells[17]. In summary, the regulatory roles of lncRNAs in NSCLC are multifaceted and profound. Understanding these roles not only provides insight into the molecular underpinnings of NSCLC but also opens up new avenues for diagnostic and therapeutic innovations. This review explores the diverse functions of lncRNAs in NSCLC, emphasizing their mechanisms of action, impact on tumor biology and potential as therapeutic targets.
NSCLC-linked lncRNAs
In lung cancer, lncRNAs play pivotal roles in both oncogenic and tumor-suppressive pathways. They regulate key processes such as cell proliferation[18], apoptosis[19], metastasis[20], and angiogenesis[21], contributing to the complexity of the tumor microenvironment[22, 23]. Dysregulated expression of specific lncRNAs, including MRPL23-AS1, MALAT1 and GAS5, has been associated with tumor progression, poor prognosis and therapy resistance. For instance, MALAT1 promotes cell migration and invasion[24], while MRPL23-AS1 is involved in EMT and metastasis[25]. Conversely, GAS5 functions as a tumor suppressor by inducing apoptosis and inhibiting cell growth[26]. The regulatory mechanisms of lncRNAs are diverse and involve interactions with DNA, RNA and proteins to modulate key signaling pathways. LncRNAs may act as molecular scaffolds, decoys, guides, or enhancers, influencing gene expression at epigenetic, transcriptional and post-transcriptional levels[27-29]. This multifaceted regulatory capability highlights the importance of lncRNAs as central nodes within the intricate networks that drive lung cancer development and progression.
LncRNAs and the hallmarks of NSCLC
LncRNAs are integral to several hallmarks of NSCLC, including cell proliferation, metastasis, apoptosis and angiogenesis. Their regulatory roles in these processes are crucial for understanding lung cancer progression and developing targeted therapies. The regulation of lncRNA expression and function in cancer often mirrors the mechanisms that govern known oncogenes and tumor suppressors, including DNA methylation, gene amplification or deletion or mutations. Furthermore, advances in bioinformatics and biochemical methodologies have revealed that ncRNAs previously considered noncoding may actually encode small biologically active peptides. These coding products play a significant role in the progression of NSCLC. This section will highlight examples of well characterized lncRNAs that exhibit oncogenic or tumor-suppressive roles in NSCLC.
Cell proliferation and cell cycle regulation
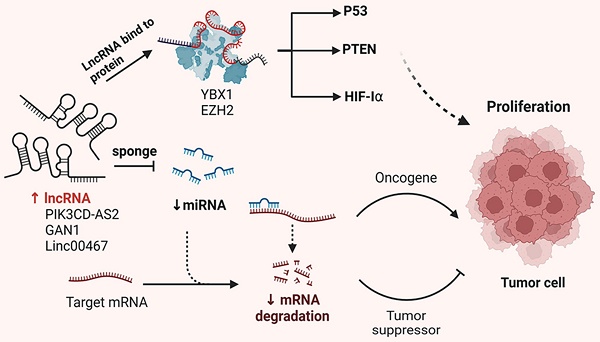
The development and progression of malignant tumors are often marked by dysregulation of the cell cycle and apoptotic signaling pathways. Emerging evidence suggests that lncRNAs play either promotive or inhibitory roles in tumor development and progression. Like protein-coding genes, lncRNAs can be categorized into oncogenic and tumor-suppressive subtypes. This section provides an overview of the molecular mechanisms by which lncRNAs influence tumor proliferation, including their functions as miRNA sponges, protein scaffolds and coding peptides (Figure 1).
LncRNA-XIST is aberrantly expressed in various tumor and promotes malignant biological behavior. Zhou et al. demonstrated that XIST competes with miR-16 through the ceRNA mechanism, thereby reversing its suppression of the downstream target gene CDK8. This interaction leads to cell cycle arrest of NSCLC cells in the G0-G1 phase, ultimately promoting proliferation and tumor progression[30]. Additionally, RIP experiments have shown that XIST and miR-186-5p are enriched in Ago2 immuno-precipitation. XIST binds to miR-186-5p, inhibiting its tumor-suppressive effects and affecting cell proliferation through cell cycle regulation[31]. Chen et al. reported that lncRNA-HOTAIR is upregulated in NSCLC cell lines. Silencing HOTAIR significantly inhibits A549 proliferation by promoting miR-217 expression[32]. Furthermore, Li et al. found that HOTAIR acts as a competitive endogenous RNA for miR-149-5p, thereby promoting the growth and invasion of NSCLC cells[33]. MALAT1, one of the earliest identified dysregulated lncRNAs in NSCLC, is highly expressed in most tumor tissues. MALAT1 enhances cell proliferation by modulating the expression of cell cycle-related genes. It interacts with various transcription and splicing factors to promote the transition from the G1 to S phase, facilitating rapid cell division. Li et al. observed that MALAT1 was significantly overexpressed in various NSCLC cell lines. The knockdown of MALAT1 in A549 and H460 cell lines significantly increased miR-124 levels, which also inhibited MA-LAT1 expression. Experimental results showed that MALAT1 and miR-124 were highly enriched in Ago2, indicating their interaction. Therefore, the MA-LAT1/miR-124/STAT3 pathway promotes NSCLC cell proliferation[34]. Another study found that both MALAT1 and MDM4 were highly expressed in A549 and H460 cell lines, with a positive correlation in NSCLC tissues. MALAT1 enhances the expression of its target gene MDM4 by downregulating miR-185-5p levels, thereby promoting NSCLC cell proliferation[35]. DANCR is significantly overexpressed in NSCLC and enhances the binding of EZH2 to p21, increasing H3K27me3 modification at the p21 promoter region, which inhibits p21 expression and promotes NSCLC cell proliferation[36]. Yu et al. reported that DANCR competes with miR-216a to regulate β-catenin expression, thus promoting cell proliferation via the Wnt/β-Catenin signaling pathway[37]. Meng et al. found that JPX interacts with miR-145-5p and upregulates downstream cyclin D2 expression through the ceRNA mechanism, thus regulating cell cycle progression[38]. Additionally, SNHG15 binds competitively to miR-486, leading to upregulation of CDK14 mRNA expression and affecting NSCLC cell growth and proliferation[39]. XIST has been shown to upregulate the anti-apoptotic factor Bcl-2 by binding to miR-449a, thereby inhibiting cell apoptosis[40]. Conversely, Wu et al. reported that MEG3 binds to miR-7-5p, inhibits Bcl-2 expression and promotes BAX expression, thereby inducing apoptosis in NSCLC cells[41]. Overall, these studies indicate that lncRNAs play a crucial role in regulating NSCLC cell proliferation, cell cycle and apoptosis. A summary of these mechanisms is presented in Table 1.
Mechanisms of lncRNA regulation of NSCLC proliferation. In NSCLC cells, lncRNAs carry specific sequences that can adsorb miRNAs, which act like sponges, thus preventing miRNAs from binding to their targets. Conversely, lncRNAs interact with proteins, affecting their post-translational modifications, protein stability, transcriptional and translational activities, ultimately affecting the proliferation of tumor cells.
Summarizing the roles and mechanisms of lncRNAs in NSCLC cell proliferation, cell cycle, and apoptosis
| LncRNA | Expression | Mechanism | Function | References |
|---|---|---|---|---|
| PIK3CD-AS2 | up | YBX1/p53 | Proliferation, Apoptosis | [42] |
| MALAT1 | up | miR-185-5p/MDM4 | Proliferation, Apoptosis | [43] |
| MALAT1 | up | MALAT1/FOXP3/GINS1 | Proliferation | [44] |
| UPLA1 | up | DSP/Wnt/β-catenin | Proliferation, Cycle | [45] |
| XIST | up | miR-744/RING1/Wnt/β-catenin | Proliferation | [46] |
| UFC1 | up | UFC1/EZH2/PTEN/PI3K/Akt | Proliferation | [47] |
| LNC00525 | up | miR-338-3p/IRS2 | Proliferation | [48] |
| LNC00525 | up | EZH2/RBMS2/p21 | Proliferation, Cycle | [49] |
| AZIN1-AS1 | up | miR4435-2HG/TGF-β1 | Proliferation | [50] |
| AZIN1-AS1 | up | miR-513b-5p/DUSP11 | Proliferation | [51] |
| GAN1 | down | miR-26a-5p/PTEN | Proliferation, Apoptosis | [52] |
| KTN1-AS1 | up | KTN1-AS1/miR-130a-5p/PDPK1 | Proliferation, Apoptosis | [53] |
| KTN1-AS1 | up | miR-23b/DEPDC1 | Proliferation | [54] |
| LNC00467 | up | AKT | Proliferation | [55] |
| LNC00467 | up | Wnt/β-catenin | Proliferation | [56] |
| LNC00467 | up | miR-4779 and miR-7978 | Proliferation, Apoptosis | [57] |
| RMRP | up | TGFBR1/SMAD2/SMAD3 | Proliferation | [58] |
| LNC00301 | up | EZH2/EAF2/pVHL/HIF-1α | Proliferation, Apoptosis | [59] |
Metastasis and invasion
Metastasis and invasion of tumor cells are critical factors contributing to the poor prognosis of patients with NSCLC. Numerous studies have demonstrated that lncRNAs are involved in regulating cell migration and invasion. This section summarizes and discusses lncRNAs associated with tumor cell migration and invasion (Figure 2).
SNHG6 is significantly overexpressed in NSCLC tissues and cells. Knockdown of SNHG6 inhibits NSCLC cell invasion. SNHG6 competitively binds to miR-944 and miR-181d-5p, promoting the expression of the transcription factor ETS1. ETS1, in turn, binds to the WIPF1 promoter region, enhancing its expression and promoting NSCLC cell invasion by regulating RhoA[60]. High SNHG6 expression is also associated with advanced pathological stage and lymph node infiltration in patients with NSCLC and can serve as an independent predictor of tumor recurrence. In A549 cells, SNHG6 binds to miR-101-3p, downregulating its expression and promoting the expression of its downstream target CDYL, which enhances the invasion ability of NSCLC cells[61].
MEG3 and GAS5 are known tumor suppressor lncRNAs [62]. MEG3 is expressed at low levels in NSCLC tissues, and its upregulation inhibits NSCLC cell migration and invasion while promoting apoptosis through p53[63]. Lv et al. found that MEG3 inhibits migration and invasion of NSCLC cells and enhances PTEN expression via the PI3K/AKT signaling pathway, suggesting its potential as a novel therapeutic target for NSCLC[64]. GAS5, another tumor suppressor in NSCLC, exerts effects through both p53-dependent and p53-independent pathways[65]. Dong et al. reported that the downregulation of GAS5 significantly promotes the growth, migration and invasion of NSCLC cells, while the miR-205/PTEN axis can partially counteract these effects. GAS5 upregulation inhibits NSCLC growth, migration and invasion through the miR-205/PTEN axis[66]. Studies have shown that LNC00673 interacts with LSD1 at the NCALD promoter region, promoting H3K4me2 demethylation, which suppresses NCALD transcription and reduces protein expression. This enhances the metastatic capability of NSCLC cells[67]. LSINCT5 interacts with the metastasis-associated transcription factor HMGA2, protecting it from proteasome-mediated degradation and increasing NSCLC cell migration[68]. Furthermore, NKILA was found to be downregulated in NSCLC tissues and correlates with lymph node metastasis and TNM staging. Mechanistically, NKILA transcription is regulated by the TGF-β signaling pathway, which inhibits NF-κB activity by reducing IKBα phosphorylation, thereby suppressing NSCLC cell migration and invasion[69]. Additionally, NKILA can inhibit EMT by modulating the IL-11/STAT3 signaling pathway [70]. Overall, these studies indicate that lncRNAs play significant roles in regulating NSCLC cell EMT, invasion and metastasis. A summary of these mechanisms is presented in Table 2.
The abnormal expression and interaction of lncRNAs influence NSCLC metastasis by regulating various ceRNA networks and post-translational modifications.
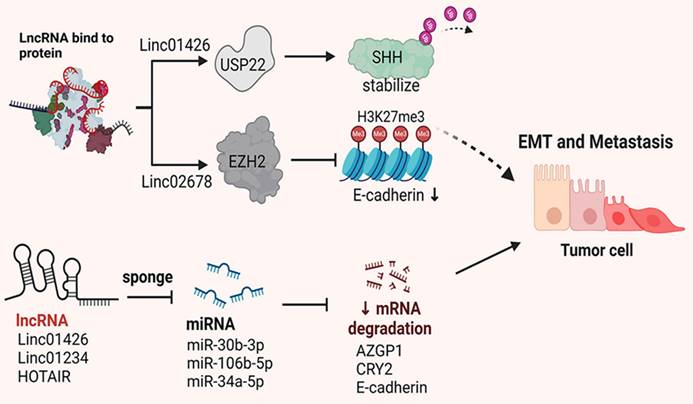
Summarizing the roles and mechanisms of lncRNAs in NSCLC EMT, invasion and metastasis
| LncRNA | Expression | Mechanism | Function | Reference |
|---|---|---|---|---|
| CRYBG3 | up | Bub3 regulation | Migration | [71] |
| LNC01426 | up | Hsa-miR-30b-3p/AZGP1 regulation | Migration, invasion | [72] |
| LNC01426 | up | USP22/SHH regulation | Migration, EMT | [73] |
| LNC02678 | up | EZH2/H3K27me3/CDKN1B regulation | Migration, invasion, EMT | [74] |
| LNC01234 | up | HNRNPA2B1/miR-106b-5p/CRY2/c-Myc | Migration, invasion | [75] |
| LNC01234 | up | EZH2/LSD1/BTG2 regulation | Migration | [76] |
| HOTAIR | up | miR-34a-5p/E-cadherin/vimentin/snail | Migration, invasion, EMT | [77] |
| LNC01123 | up | miR-199a-5p/c-Myc pathway | Invasion, EMT | [78] |
Apoptotic response and cell death
LncRNAs play crucial roles in regulating apoptosis in NSCLC. Apoptosis can be categorized into extrinsic and intrinsic pathways. In the extrinsic pathway, death receptors such as TNFR, Fas/CD95 and TRAIL interact with extracellular ligands like TNF and FasL, leading to the sequential activation of death receptor procaspase-8, caspase-8 and caspase-3. This cascade ultimately regulates DNA fragmentation factors and caspase-activated DNases, culminating in apoptosis[79, 80]. In the intrinsic pathway, cytotoxic stimuli, including carcinogenic stress, chemotherapy drugs and developmental signals selectively activate BH3 family members while suppressing Bcl-2 proteins. This activation triggers pro-apoptotic effectors BAX and BAK, disrupting the outer mitochondrial membrane[81]. The resulting release of cytochrome C and ATF endonuclease from the mitochondria initiates the formation of the apoptosome through the bindings of cytochrome C to APAF1, which subsequently activates caspase-9[82]. Activated caspase-9 then induces caspase-3, caspase-6 and caspase-7, leading to protein cleavage and the execution of apoptosis. Numerous lncRNAs are involved in modulating these apoptotic processes[83]. GAS5, a well-established tumor suppressor lncRNA, induces apoptosis by sequestering the glucocorticoid receptor, thereby inhibiting its anti-apoptotic effects and increasing the sensitivity of cancer cells to apoptotic signals. Conversely, HOTAIR inhibits apoptosis by upregulating anti-apoptotic genes, which contribute to tumor survival and resistance to therapy. Han et al. found that lnc0218 is upregulated in NSCLC and promotes cell proliferation by regulating the miR-4677-3p/SEC61G axis, thereby suppressing apoptosis. Notably, the knockdown of lnc0218 enhances apoptosis in NSCLC cells[84]. Wang et al. discovered that lncRNA-ATB promotes NSCLC cell apoptosis by inhibiting miR-200a expression, which leads to the upregulation of β-catenin expression[85]. Zhang et al. demonstrated that XIST upregulates the anti-apoptotic factor Bcl-2 by binding to miR-449a, thereby inhibiting cell apoptosis[40]. While some lncRNAs inhibit apoptosis, others promote it. For example, Wu et al. showed that MEG3 binds to miR-7-5p, suppressing Bcl-2 expression and promoting BAX expression, thereby inducing apoptosis in NSCLC cells[86]. Pei et al. found that the lncRNA AFAP1-AS1 encodes a small peptide called ATMLP, which localizes to mitochondria and promotes the malignant progression of NSCLC. ATMLP (not the lncRNA itself) is a driver of tumor progression, and the higher the level of ATMLP in a patient's serum, the worse the prognosis. Moreover, its translation process is regulated by m6A methylation. Mechanistically, ATMLP binding to NIPSNAP1 disrupts its translocation within mitochondria and affects the process of tumor cell apoptosis[87].
LncRNA regulates drug resistance in NSCLC cells
Due to the subtle early symptoms of NSCLC, many patients are diagnosed with metastasis at the time of initial presentation, making surgical resection unfeasible. For patients with NSCLC who are ineligible for surgery, chemotherapy and targeted therapy drugs are essential treatment modalities. However, the development of drug resistance and reduced tolerance to treatment significantly diminishes treatment efficacy and overall survival rates[88, 89]. LncRNAs have been implicated in the development of drug resistance in NSCLC. For instance, certain lncRNAs have been shown to influence specific drug pathways, including those targeted by tyrosine kinase inhibitors (TKIs). Notably, the lncRNA HOTAIR has been found to be upregulated in NSCLC cells that have developed resistance to TKIs. HOTAIR is thought to regulate the expression of genes involved in drug metabolism and efflux, thereby contributing to the resistance phenotype. This section summarizes and discusses the roles of lncRNAs in cellular drug resistance (Figure 3).
Mechanisms by which lncRNAs regulate tumor angiogenesis. Certain lncRNAs have sponge-like properties, wherein they absorb miRNAs, inhibit the binding of miRNAs to target genes and reduce degradation of target mRNAs, thereby affecting tumor drug resistance.
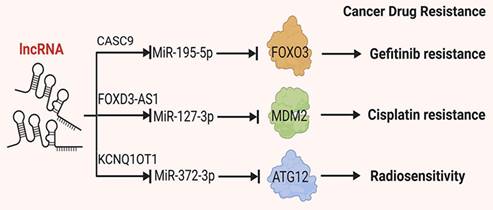
Platinum-based chemotherapy regimens are foundational in the clinical treatment of NSCLC, making it crucial to understand the role of lncRNAs in platinum resistance. Li et al. demonstrated that in Cisplatin-resistant A549 and H460 cells, the expression levels of UCA1, miR-495 RNA and NRF2 protein differed significantly from those in non-resistant cells. Mechanistically, UCA1 directly binds to miR-495, leading to its downregulation. As NRF2 is a direct target of miR-495, its expression is negatively correlated with miR-495 levels. Thus, UCA1 mediates platinum resistance in NSCLC cells through the UCA1/miR-495/NRF2 pathway[90]. Additionally, Wang and Gao showed that LINC01116 is associated with Cisplatin resistance in lung adenocarcinoma[91]. Furthermore, Yao indicated that LINC01116 influences Cisplatin resistance by regulating iron metabolism and AKT signaling in NSCLC[92, 93]. Additionally, lncRNAs also contribute to resistance to other chemotherapeutic agents. Pan et al. found that lnc-ROR is elevated in tumor tissues from docetaxel-resistant patients. In vitro experiments revealed that lnc-ROR affects NSCLC cell sensitivity to docetaxel through the lnc-ROR/miR-145/FSCN1 pathway[94].
Targeted therapies are becoming increasingly important in NSCLC treatment. Resistance to these therapies is often mediated by lncRNAs. For example, over 46% of patients with NSCLC having EGFR mutations respond to EGFR-TKIs such as erlotinib and gefitinib[95, 96]. In EGFR-TKI resistant cells, various lncRNAs are aberrantly expressed and participate in modulating resistance to EGFR-TKIs[97]. In EGFR-TKI-resistant cells, various lncRNAs are aberrantly expressed, modulating resistance to EGFR-TKIs [94]. In gefitinib-resistant NSCLC cells (e.g. SPC-A1 and H1299), researchers identified five dysregulated lncRNAs: three upregulated (UCA1, NEAT1 and CASC9) and two downregulated (EWAST1 and LNC00524). Mechanistically, the co-expression of CASC9 and EWAST1 regulates multiple pathways, including cell growth and apoptosis, thus influencing EGFR-TKI sensitivity[97-99].
Chen et al. found that in gefitinib-resistant PC9 cells, CASC9 (candidate tumor susceptibility gene 9) binds specifically to EZH2, facilitating its binding to the DUSP1 promoter, upregulating H3K27me3 levels and suppressing DUSP1 protein expression. This process enhances NSCLC resistance to gefitinib[100]. Similarly, in extracellular vesicles released from gefitinib-resistant PC-9 cells, UCA1 is upregulated. Xu et al. found that in gefitinib-resistant tumor cells, UCA1 binds directly to EZH2, increasing H3K27me3 levels at the CDKN1A promoter region and reducing downstream CDKN1A expression, thereby mediating gefitinib resistance[101]. These findings highlight the importance of lncRNAs in drug resistance and support potential therapeutic targets. Further studies indicate that KCNQ1OT1 plays a significant role in radiotherapy resistance. Wang et al. demonstrated that KCNQ1OT1 promotes cell proliferation, migration and invasion by modulating the miR-129-5p/JAG1 pathway[102]. Additionally, KCNQ1OT1 influences cell proliferation, autophagy and apoptosis through the miR-204-5p/ATG3 pathway, contributing to radiotherapy resistance in lung adenocarcinoma[103]. Furthermore, He et al. found that KCNQ1OT1 is significantly upregulated in radiotherapy-resistant A549 and H1975 cells. KCNQ1OT1 binds to miR-372-3p, suppressing its expression and promoting the expression of downstream ATG5 and ATG12 mRNA and proteins, which reduced NSCLC cells' sensitivity to radiotherapy[104].
Moreover, TTTY15 inhibits the binding of DNMT3A to the TBX4 promoter, thereby regulating TBX4 expression and affecting NSCLC cells' sensitivity to radiotherapy[105]. Jiao et al. discovered that lncRNA SNHG14 regulates HMGB1 expression by sponging miR-34a. Silencing SNHG14 enhances the sensitivity of NSCLC cells to carboplatin, suggesting SNHG14 is a potential target to overcome drug resistance[106]. Interestingly, lncRNAs can both induce and inhibit drug resistance in tumor cells[107]. For instance, lncRNA FENDRR binds directly to the 3' untranslated region (3'UTR) of the MDR1 gene, preventing RNA-binding proteins from associating with the MDR1 3'UTR, reducing MDR1 expression, and inhibiting drug resistance[108]. Nakano et al. identified LNC00460 as a novel marker associated with poor response and prognosis in EGFR-TKI therapy. In lung cancer cells, LNC00460 enhances EGFR-TKI resistance by acting as a competitive endogenous RNA (ceRNA) for miR-149-5p, promoting interleukin-6 expression and inducing an EMT-like phenotype. Knocking out LNC00460 in gefitinib-resistant NSCLC cells restore their sensitivity to EGFR-TKI. Moreover, high LNC00460 expression correlates with shorter progression-free survival (PFS) and overall survival (OS) after gefitinib treatment[109]. Wang et al. found that LINC01116 is upregulated in gefitinib-resistant NSCLC cells and tissues. Silencing LINC01116 increases IFI44 expression, and IFI44 overexpression reverses gefitinib resistance in PC9/R cells, indicating that LINC01116 modulates gefitinib resistance partly through IFI44[43]. Additionally, Li et al. reported that RHPN1-AS1 is downregulated in gefitinib-resistant patients and NSCLC cell lines, with this downregulation linked to poor prognosis. Overexpression of RHPN1-AS1 restores gefitinib sensitivity in resistant NSCLC cells by interacting directly with miR-299-3p to positively regulate TNFSF12 expression, modulating gefitinib resistance[110]. Additionally, lncRNAs such as SNHG15[111], CASC9[112], LNC554202[113] and BLACAT1[114] also significantly impact resistance to EGFR-TKI therapy. Understanding the interaction mechanisms between lncRNAs and drug resistance can provide new therapeutic strategies to enhance treatment efficacy. A summary of some of the mechanisms is presented in Table 3.
The advantage and challenge of therapeutic targeting of lncRNA
Targeted lncRNA therapy offers a novel and promising strategy for the treatment of NSCLC. The precise modulation of lncRNAs involved in tumor progression and metastasis could offer new avenues for effective cancer therapies[124, 125]. Over recent decades, substantial clinical investments have been directed toward RNA-based therapeutic approaches. Presently, there are several RNA-based therapeutic modalities available, including antisense oligonucleotides (ASOs), RNA interference (RNAi), small molecule inhibitors, microRNA mimics, and CRISPR-Cas9 gene editing. Among these, ASOs and siRNAs are the most extensively utilized[126, 127]. Preclinical studies have shown that synthetic miRNA-based therapeutic molecules, combined with various protective coating techniques, have enabled effective delivery and anti-tumor activity[128, 129]. MALAT1 was the first lncRNA reported to be associated with tumor metastasis. In a mouse model of metastatic NSCLC, subcutaneous injection of MALAT1 ASO significantly reduced the burden of lung nodules, indicating the therapeutic potential of targeting MALAT1 in metastatic lung cancer[130, 131]. Similarly, in patients with metastatic renal cell carcinoma, high expression of the lncRNA ARSR is associated with shorter overall survival. LncARSR promotes resistance to sunitinib by inhibiting AXL and MET expression through the adsorption of miR-34 and miR-449.
Summarizing the roles and mechanisms of lncRNAs in NSCLC drug resistance
| LncRNA | Expression | Mechanism | Function | Reference |
|---|---|---|---|---|
| SNHG15 | up | MiR-451/MDR-1 | Gefitinib resistance | [111] |
| CASC9 | up | EZH2/DUSP1 | Gefitinib resistance | [100] |
| CASC9 | up | MiR-195-5p/FOXO3 | Gefitinib resistance | [112] |
| UCA1 | up | MiR-143/FOSL2 | Gefitinib resistance | [115] |
| UCA1 | up | MiR-495/NRF2 pathway | Cisplatin resistance | [116] |
| LNC01116 | up | EZH2/CDKN1A | Cisplatin resistance | [101] |
| LNC01116 | up | IFI44 | Gefitinib resistance | [43] |
| FGD5-AS1 | up | MiR-140-5p/WEE1 pathway | Cisplatin resistance | [117] |
| BLACAT1 | up | STAT3 | Afatinib resistance | [114] |
| BLACAT1 | up | Cyclin D1 | Cisplatin resistance | [118] |
| FOXD3-AS1 | up | MiR-127-3p/MDM2 pathway | Cisplatin resistance | [119] |
| LNC00665 | up | LNC00665/EZH2/CDKN1C | Cisplatin resistance | [120] |
| SPRY3 | down | IGF2BP3 | Radiosensitivity | [121] |
| KCNQ1OT1 | up | MiR-372-3p/ATG5/ATG12 | Radiosensitivity | [104] |
| KCNQ1OT1 | up | MiR-27b-3p/HSP90AA1 pathway | Radiosensitivity | [122] |
| SNHG15 | up | MiR-451/MDR-1 | Gefitinib resistance | [123] |
In an in vivo xenograft model of renal cell carcinoma, intravenous injection of l ASOs targeting lncARSR effectively restored sensitivity to Sunitinib treatment[132]. In patients with colon cancer, high expression of the lncRNA RAMS11 is associated with poor prognosis. Interestingly, an FDA-approved drug screen revealed that elevated RAMS11 expression confers resistance to fluorodeoxyuridine (FUDR) and topoisomerase inhibitors. Subsequent in vitro experiments demonstrated that CRISPR-mediated knockdown of RAMS11 expression increased cellular sensitivity to FUDR and topoisomerase inhibitors[133, 134]. Furthermore, lncRNAs also play a crucial role in the organ-specific target of tumor cells for metastasis. For instance, CRISPR-mediated knockdown of the lncRNA MAYA in breast cancer cells with bone metastasis significantly inhibited the activation of the YAP pathway, thereby impairing osteoclast differentiation and the bone resorption process[135, 136]. Additionally, research has revealed that m6A modifications on ncRNAs influence tumorigenesis and drug response, highlighting the therapeutic potential of targeting ncRNAs through m6A regulatory factors in cancer therapy[137]. The success of miRNA-based cancer research has spurred further investigations into other ncRNA families that may serve as potential therapeutic targets in cancer.
Despite the promising preclinical results, the translation of lncRNA-targeted therapies to the clinic faces several challenges. When targeting lncRNAs with ASO therapies, it is crucial to minimize off-target effects. Emerging evidence indicates that ASO can also induce premature transcription termination, potentially damaging target RNAs. This possibility should be carefully considered when evaluating the efficacy and safety of ASO therapies. The delivery of lncRNA-targeted therapeutics to tumor cells in the lung remains a significant hurdle due to the anatomical and physiological barriers. The abundance of lncRNAs exhibit overlapping sequences and functional similarities, complicating the design of targeted therapies that can selectively affect a single lncRNA without impacting others. While some lncRNAs have well-defined functions, many remain poorly understood. This lack of knowledge hinders the development of targeted therapies and complicates the prediction of potential off-target effects. Additionally, the potential off-target effects and toxicity of these therapies need to be thoroughly evaluated[138]. In conclusion, although the field of ncRNA-based therapeutics is still in its infancy, we are witnessing its burgeoning potential in cancer therapy. These therapeutics are anticipated to have a wide array of applications in the near future.
Conclusions and future perspectives
Increasingly, studies have shown that lncRNAs play crucial roles in various essential cellular processes, and their roles in cancer, including NSCLC, are becoming increasingly apparent. LncRNAs interact with chromatin, RNA, and proteins, regulating gene expression at multiple stages and influencing important signaling path-ways. Numerous studies have demonstrated that lncRNAs participate in regulating processes such as proliferation, apoptosis, invasion, EMT, migration, and drug resistance in NSCLC cells. While certain lncRNAs have well-characterized mechanisms of action, many others remain to be fully elucidated. Concurrently, the application of multi-omics approaches, including encompassing genomics, transcriptomics, proteomics, and metabolomics, provides a comprehensive framework for investigating the functional roles of lncRNAs in NSCLC. This integrative perspective facilitates the identification of lncRNA interactions within cellular networks and their impact on critical signaling pathways, thereby enhancing our understanding of their contributions to tumor biology and supporting the development of more effective lncRNA-targeted therapies. Several lncRNAs show abnormal expression in NSCLC tissues and the circulation of NSCLC patients, suggesting their potential as diagnostic biomarkers for NSCLC. Unlike miRNAs, the expression levels of lncRNAs can better reflect the disease status, and their tissue-specific expression suggests potential diagnostic applications for specific diseases. LncRNAs are instrumental in modulating sensitivity to radiotherapy and chemotherapy in NSCLC, positioning them as promising targets for novel therapeutic agents that could revolutionize treatment strategies for NSCLC patients. To mitigate the risks associated with potential off-target effects of lncRNA-based therapies and to enhance therapeutic efficacy, advancements in drug delivery systems are critical. By optimizing delivery mechanisms, such as nanoparticle-based carriers and cell-affinity peptides, researchers seek to enhance the bioavailability and specificity of lncRNA-targeted drugs. This approach aims to address existing barriers that limit therapeutic potential, ultimately maximizing patient benefit and improving treatment outcomes.
In summary, lncRNAs play crucial roles in the development and progression of tumors, including NSCLC. Continued in-depth research into lncRNA could yield valuable insights and innovative approaches for the diagnosis and treatment of NSCLC. The development of novel diagnostic biomarkers and clinical therapeutic strategies targeting lncRNAs has the potential to guide future research directions and contribute to more effective cancer treatments.
Acknowledgements
Figures were created with BioRender.com.
Funding
This research was funded by National Natural Science Foundation of China (82203773, 81972174), the Natural Science Foundation of Jiangsu Province (BK20231233), Lianyungang Key R&D Program (Social Development) (SF2207), Suzhou Science and Technology Program-Applied and Fundamental Medical Research-Key Clinical Technology Research (SKY2023009), Scientific Research and Development Foundation of Kangda College of Nanjing Medical University (KD2022KYJJZD158).
Author contributions
All authors contributed to the study's conception and design. S.X. and X.L. performed the investigation and wrote the first draft of the manuscript. W.W., X.P. and J.C. illustrated the figures. S.W. and Z.W. reviewed and edited the draft. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33
2. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299-311
3. Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R. et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med. 2017;377:1919-1929
4. Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720-731
5. Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: a National Cancer Database survey. J Thorac Oncol. 2010;5:29-33
6. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-1927
7. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42-54
8. Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications. Genet Res Int. 2012;2012:737416
9. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300-307
10. Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169-2184
11. Guo T, Li J, Zhang L, Hou W, Wang R, Zhang J, Gao P. Multidimensional communication of microRNAs and long non-coding RNAs in lung cancer. J Cancer Res Clin Oncol. 2019;145:31-48
12. Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56:876-885
13. Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang AX. et al. Comprehensive Analysis of lncRNA and mRNA Expression Profiles in Lung Cancer. Clin Lab. 2017;63:313-320
14. Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018;34:142-157
15. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471
16. Zhang Y. LncRNA-encoded peptides in cancer. J Hematol Oncol. 2024;17:66
17. Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M. et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/beta-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol Cancer. 2022;21:168
18. Yang Q, Wang M, Xu J, Yu D, Li Y, Chen Y. et al. LINC02159 promotes non-small cell lung cancer progression via ALYREF/YAP1 signaling. Mol Cancer. 2023;22:122
19. Iwai M, Kajino T, Nakatochi M, Yanagisawa K, Hosono Y, Isomura H. et al. Long non-coding RNA TILR constitutively represses TP53 and apoptosis in lung cancer. Oncogene. 2023;42:364-373
20. Fan C, Wang Q, Kuipers TB, Cats D, Iyengar PV, Hagenaars SC. et al. LncRNA LITATS1 suppresses TGF-beta-induced EMT and cancer cell plasticity by potentiating TbetaRI degradation. EMBO J. 2023;42:e112806
21. He Z, Zhong Y, Regmi P, Lv T, Ma W, Wang J. et al. Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway. Mol Cancer. 2024;23:65
22. Ao YQ, Gao J, Jiang JH, Wang HK, Wang S, Ding JY. Comprehensive landscape and future perspective of long noncoding RNAs in non-small cell lung cancer: it takes a village. Mol Ther. 2023;31:3389-3413
23. Karger A, Mansouri S, Leisegang MS, Weigert A, Gunther S, Kuenne C. et al. ADPGK-AS1 long noncoding RNA switches macrophage metabolic and phenotypic state to promote lung cancer growth. EMBO J. 2023;42:e111620
24. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X. et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2021;14:32
25. Chen CW, Fu M, Du ZH, Zhao F, Yang WW, Xu LH. et al. Long Noncoding RNA MRPL23-AS1 Promotes Adenoid Cystic Carcinoma Lung Metastasis. Cancer Res. 2020;80:2273-2285
26. Dong S, Qu X, Li W, Zhong X, Li P, Yang S. et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8:43
27. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045
28. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407
29. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24:430-447
30. Zhou X, Xu X, Gao C, Cui Y. XIST promote the proliferation and migration of non-small cell lung cancer cells via sponging miR-16 and regulating CDK8 expression. Am J Transl Res. 2019;11:6196-6206
31. Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y. et al. The Long Non-Coding RNA XIST Controls Non-Small Cell Lung Cancer Proliferation and Invasion by Modulating miR-186-5p. Cell Physiol Biochem. 2017;41:2221-2229
32. Chen SS, Peng M, Zhou GZ, Pu YC, Yi MC, Zhu Y, Jiang B. Long non-coding RNA HOTAIR regulates the development of non-small cell lung cancer through miR-217/DACH1 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:670-678
33. Li H, Cui Z, Lv X, Li J, Gao M, Yang Z. et al. Long Non-coding RNA HOTAIR Function as a Competing Endogenous RNA for miR-149-5p to Promote the Cell Growth, Migration, and Invasion in Non-small Cell Lung Cancer. Front Oncol. 2020;10:528520
34. Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233:6679-6688
35. Wang D, Zhang S, Zhao M, Chen F. LncRNA MALAT1 accelerates non-small cell lung cancer progression via regulating miR-185-5p/MDM4 axis. Cancer Med. 2020;9:9138-9149
36. Guo L, Gu J, Hou S, Liu D, Zhou M, Hua T. et al. Long non-coding RNA DANCR promotes the progression of non-small-cell lung cancer by inhibiting p21 expression. Onco Targets Ther. 2019;12:135-146
37. Yu JE, Ju JA, Musacchio N, Mathias TJ, Vitolo MI. Long Noncoding RNA DANCR Activates Wnt/beta-Catenin Signaling through MiR-216a Inhibition in Non-Small Cell Lung Cancer. Biomolecules. 2020;10:1646
38. Jin M, Ren J, Luo M, You Z, Fang Y, Han Y. et al. Long non-coding RNA JPX correlates with poor prognosis and tumor progression in non-small-cell lung cancer by interacting with miR-145-5p and CCND2. Carcinogenesis. 2020;41:634-645
39. Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233:7164-7172
40. Zhang YL, Li XB, Hou YX, Fang NZ, You JC, Zhou QH. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38:371-381
41. Wu JL, Meng FM, Li HJ. High expression of lncRNA MEG3 participates in non-small cell lung cancer by regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci. 2018;22:5938-5945
42. Xiufen Z, Junying Z, Tian F, Xiaoxiao W, Siwei W, Zhifei M. et al. The long non-coding RNA PIK3CD-AS2 promotes lung adenocarcinoma progression via YBX1-mediated suppression of p53 pathway. Oncogenesis. 2020;9:34
43. He W, Binbin L, Shengnan R, Fubin W, Xinxing W, Caiyun Y, Zhaoxia W. Long Noncoding RNA LINC01116 Contributes to Gefitinib Resistance in Non-small Cell Lung Cancer through Regulating IFI44. Mol Ther Nucleic Acids. 2019;19:218-227
44. Ming L, Minke S, Chaoyue H, Baojun C, Shufeng L. MALAT1 modulated FOXP3 ubiquitination then affected GINS1 transcription and drived NSCLC proliferation. Oncogene. 2021;40:3870-3884
45. Xiaoyang H, Hua J, Jianni Q, Jiamei L, Jinghan Y, Yingying T. et al. Novel lncRNA UPLA1 mediates tumorigenesis and prognosis in lung adenocarcinoma. Cell Death Dis. 2020;11:999
46. Jinglu W, Haibo C, Zhaoxia D, Gang W. Down-regulation of lncRNA XIST inhibits cell proliferation via regulating miR-744/RING1 axis in non-small cell lung cancer. Clin Sci (Lond). 2019;133:1567-1579
47. Xueyan Z, Jianmei G, Jiayin Z, Hui S, Sinan H, Xueying X. et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11:215
48. Zhiguang Y, Xingyu L, Peng Z, Yunpeng L, Zihao L, Benxin Q. et al. RETRACTED: Long non-coding RNA LINC00525 promotes the non-small cell lung cancer progression by targeting miR-338-3p/IRS2 axis. Biomed Pharmacother. 2020;124:109858
49. Panqi F, Hao C, Zhifei M, Chencheng H, Wenda Y, Siwei W. et al. LncRNA LINC00525 suppresses p21 expression via mRNA decay and triplex-mediated changes in chromatin structure in lung adenocarcinoma. Cancer Commun (Lond). 2021;41:596-614
50. Mei Y, Xiaohua H, Xiaoxian H, Jiyang W, Yuxin H, Li W. LncRNA MIR4435-2HG-mediated upregulation of TGF-β1 promotes migration and proliferation of nonsmall cell lung cancer cells. Environ Toxicol. 2019;35:582-590
51. Yong C, Qiongya W, Yu L, Jiying W. AZIN1-AS1, A Novel Oncogenic LncRNA, Promotes the Progression of Non-Small Cell Lung Cancer by Regulating MiR-513b-5p and DUSP11. Onco Targets Ther. 2020;13:9667-9678
52. Rui-Qi W, Xiao-Ran L, Ning-Ning Z, Dong-Ni C, Mei-Yin Z, Zhe-Sheng W. et al. Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging mir-26a-5p to activate PTEN signaling in non-small cell lung cancer. J Exp Clin Cancer Res. 2021;40:9
53. Chenchen L, Wei Z, Xuan P, Xiaoyou L, Fei Y, Siwen L. et al. LncRNA KTN1-AS1 promotes the progression of non-small cell lung cancer via sponging of miR-130a-5p and activation of PDPK1. Oncogene. 2020;39:6157-6171
54. Changmin L, Xiaoming L, Yanzhang H, Feng W, Zhiwen C, Haitao G, Dianzhong G. STAT1-induced upregulation of lncRNA KTN1-AS1 predicts poor prognosis and facilitates non-small cell lung cancer progression via miR-23b/DEPDC1 axis. Aging (Albany NY). 2020;12:8680-8701
55. Yuxing Z, Jingjing L, Hao B, Dong H, Mengqing X, Liang X. et al. LINC00467 is up-regulated by TDG-mediated acetylation in non-small cell lung cancer and promotes tumor progression. Oncogene. 2020;39:6071-6084
56. Jilin Y, Yuedong L, Xiaoshi M, Shun L, Li J, Xiang T. STAT1-induced upregulation of LINC00467 promotes the proliferation migration of lung adenocarcinoma cells by epigenetically silencing DKK1 to activate Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2019;514:118-126
57. Yanxiang C, Lisheng Y. LINC00467 promotes cell proliferation and stemness in lung adenocarcinoma by sponging miR-4779 and miR-7978. J Cell Biochem. 2019;121:3691-3699
58. Hang Y, Lin C, Shiqi P, Yiru W, Zhange L, Yuan L. et al. M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 2021;30:605-617
59. Cheng-Cao S, Wei Z, Shu-Jun L, Wei H, Jian Z, Yue Z. et al. Correction to: FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Med. 2021;13:25
60. Geng H, Li S, Xu M. Long Noncoding RNA SNHG6 Functions as an Oncogene in Non-Small Cell Lung Cancer via Modulating ETS1 Signaling. Onco Targets Ther. 2020;13:921-930
61. Li K, Jiang Y, Xiang X, Gong Q, Zhou C, Zhang L. et al. Long non-coding RNA SNHG6 promotes the growth and invasion of non-small cell lung cancer by downregulating miR-101-3p. Thorac Cancer. 2020;11:1180-1190
62. Chen Z, Lei T, Chen X, Gu J, Huang J, Lu B, Wang Z. Long non-coding RNA in lung cancer. Clin Chim Acta. 2020;504:190-200
63. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ. et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461
64. Lv D, Bi Q, Li Y, Deng J, Wu N, Hao S, Zhao M. Long non-coding RNA MEG3 inhibits cell migration and invasion of non-small cell lung cancer cells by regulating the miR-21-5p/PTEN axis. Mol Med Rep. 2021;23:191
65. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1-E12
66. Dong L, Li G, Li Y, Zhu Z. Upregulation of Long Noncoding RNA GAS5 Inhibits Lung Cancer Cell Proliferation and Metastasis via miR-205/PTEN Axis. Med Sci Monit. 2019;25:2311-2319
67. Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X. et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558-25575
68. Tian Y, Zhang N, Chen S, Ma Y, Liu Y. The long non-coding RNA LSINCT5 promotes malignancy in non-small cell lung cancer by stabilizing HMGA2. Cell Cycle. 2018;17:1188-1198
69. Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J. et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. J Exp Clin Cancer Res. 2017;36:54
70. Liu D, Shi X. Long non-coding RNA NKILA inhibits proliferation and migration of lung cancer via IL-11/STAT3 signaling. Int J Clin Exp Pathol. 2019;12:2595-2603
71. Ziyang G, Yingchu D, Wentao H, Yongsheng Z, Zhifei C, Weiwei P. et al. The long noncoding RNA CRYBG3 induces aneuploidy by interfering with spindle assembly checkpoint via direct binding with Bub3. Oncogene. 2021;40:1821-1835
72. Baorui T, Xiaoyang H, Guanzhen L, Hua J, Jianni Q, Jiamei L. et al. A Long Intergenic Non-coding RNA, LINC01426, Promotes Cancer Progression via AZGP1 and Predicts Poor Prognosis in Patients with LUAD. Mol Ther Methods Clin Dev. 2020;18:765-780
73. Xiaoli L, Zuwei Y, Linping X, Huaimin L, Lifeng J, Shuochuan L, Xu S. Upregulation of LINC01426 promotes the progression and stemness in lung adenocarcinoma by enhancing the level of SHH protein to activate the hedgehog pathway. Cell Death Dis. 2021;12:173
74. Dexin J, Ying X, Yuning Z, Mengru C, Fanglin T, Weina F. et al. LINC02678 as a Novel Prognostic Marker Promotes Aggressive Non-small-cell Lung Cancer. Front Cell Dev Biol. 2021;9:686975
75. Zhenyao C, Xin C, Tianyao L, Yu G, Jinyao G, Jiali H. et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther. 2020;28:1479-1493
76. Zhenyao C, Xin C, Binbin L, Yu G, Qinnan C, Tianyao L. et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13:7
77. Fang Z, Jing L, ChangJu M, XiaoJuan T, Qing T, JingJing W. et al. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J Cell Mol Med. 2020;24:5578-5592
78. Qian H, Mingming J, Baoming M, Fei X, Tian L, Li Z. et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol. 2019;12:91
79. Dalleau S, Baradat M, Gueraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615-1630
80. Li J, Lian Y, Yan C, Cai Z, Ding J, Ma Z. et al. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017;50:e12312
81. Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49-63
82. Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532-542
83. Jiang N, Zhang X, Gu X, Li X, Shang L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021;7:30
84. Han B. LncRNA LINC02418 regulates proliferation and apoptosis of non-small cell lung cancer cells by regulating miR-4677-3p/SEC61G. Eur Rev Med Pharmacol Sci. 2019;23:10354-10362
85. Wang T, Tang X, Liu Y. LncRNA-ATB promotes apoptosis of non-small cell lung cancer cells through MiR-200a/beta-Catenin. J BUON. 2019;24:2280-2286
86. Yang Z, Lin X, Zhang P, Liu Y, Liu Z, Qian B. et al. RETRACTED: Long non-coding RNA LINC00525 promotes the non-small cell lung cancer progression by targeting miR-338-3p/IRS2 axis. Biomed Pharmacother. 2020;124:109858
87. Pei H, Dai Y, Yu Y, Tang J, Cao Z, Zhang Y. et al. The Tumorigenic Effect of lncRNA AFAP1-AS1 is Mediated by Translated Peptide ATMLP Under the Control of m(6) A Methylation. Adv Sci (Weinh). 2023;10:e2300314
88. Lu Z, Fang Z, Guo Y, Liu X, Chen S. Cisplatin resistance of NSCLC cells involves upregulation of visfatin through activation of its transcription and stabilization of mRNA. Chem Biol Interact. 2022;351:109705
89. Blach J, Wojas-Krawczyk K, Nicos M, Krawczyk P. Failure of Immunotherapy-The Molecular and Immunological Origin of Immunotherapy Resistance in Lung Cancer. Int J Mol Sci. 2021;22:9030
90. Li C, Fan K, Qu Y, Zhai W, Huang A, Sun X, Xing S. Deregulation of UCA1 expression may be involved in the development of chemoresistance to cisplatin in the treatment of non-small-cell lung cancer via regulating the signaling pathway of microRNA-495/NRF2. J Cell Physiol. 2020;235:3721-3730
91. Junbin W, Jin G, Qinnan C, Weiyan Z, Fen Y, Chenchen W, Zhaoxia W. LncRNA LINC01116 Contributes to Cisplatin Resistance in Lung Adenocarcinoma. Onco Targets Ther. 2020;13:9333-9348
92. Bin S, Zhenxiang L, Meng L, Shujuan J, Zhen F, Zhixin C, Hui W. Silencing LINC01116 suppresses the development of lung adenocarcinoma via the AKT signaling pathway. Thorac Cancer. 2021;12:2093-2103
93. Jie Y, Xiao C, Xiao L, Rui L, Xijia Z, Yiqing Q. Characterization of a ferroptosis and iron-metabolism related lncRNA signature in lung adenocarcinoma. Cancer Cell Int. 2021;21:340
94. Yan P, Jing C, Leilei T, Kai Z, Rui W, Xiaoyuan C, Longbang C. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2017;8:33144-33158
95. Hui P, Tao J, Ningning C, Qi W, Shengxiang R, Xuefei L. et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948-49960
96. Chee-Seng T, David G, Simon P. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16:e447-e459
97. Pei M, Meiling Z, Fengqi N, Zebo H, Jing H, Wei L, Liang H. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother. 2017;87:20-26
98. Wei N, Hui-juan G, Xiao-qun Y, Xiangjie S, Hai H, Xia T. et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2015;371:99-106
99. Lun Z, Minghong B, Haoran Z, Mohan S. Downregulation of NEAT1 Suppresses Cell Proliferation, Migration, and Invasion in NSCLC Via Sponging miR-153-3p. Cancer Biother Radiopharm. 2020;35:362-370
100. Zhenyao C, Qinnan C, Zhixiang C, Jingyao G, Wenyan F, Tianyao L. et al. Long non-coding RNA CASC9 promotes gefitinib resistance in NSCLC by epigenetic repression of DUSP1. Cell Death Dis. 2020;11:858
101. Tianwei X, Shuai Y, Mengwei W, Lihua J, Pei M, Binbin L. et al. LncRNA UCA1 Induces Acquired Resistance to Gefitinib by Epigenetically Silencing CDKN1A Expression in Non-small-Cell Lung Cancer. Front Oncol. 2020;10:656
102. Yan W, Lei Z, Jiasheng Y, Ruilin S. LncRNA KCNQ1OT1 promotes cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis in non-small cell lung cancer. Cancer Cell Int. 2020;20:144
103. Yan K, Yaoli J, Qilong W, Qianru Z, Meng S, Ran N, Jing W. Long Noncoding RNA KCNQ1OT1 Promotes the Progression of Non-Small Cell Lung Cancer via Regulating miR-204-5p/ATG3 Axis. Onco Targets Ther. 2019;12:10787-10797
104. Huanyu H, Xinmao S, Zuozhang Y, Yuchi M, Kunming Z, Yuanyuan W. et al. Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell Death Dis. 2020;11:883
105. I-Lu L, Ya-Sian C, Wen-Ling C, Ya-Ting L, Ju-Chen Y, Chin-An Y. et al. Male-Specific Long Noncoding RNA TTTY15 Inhibits Non-Small Cell Lung Cancer Proliferation and Metastasis via TBX4. Int J Mol Sci. 2019;20:3473
106. Pengfei J, Junna H, Mengying Y, Jing W, Gaofei R. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed Pharmacother. 2019;117:109164
107. Hao Z, Bing F, Mubalake A, Yongting L, Xinrong L, Chuan T. et al. The functional role of long non-coding RNAs and their underlying mechanisms in drug resistance of non-small cell lung cancer. Life Sci. 2020;261:118362
108. Fangchao G, Dong D, Ting Z, Weiling X. Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur J Pharmacol. 2019;853:345-352
109. Yuta N, Kazutoshi I, Hiroshi K, Kyohei K, Takuma I, Susumu S. et al. Clinical importance of long non-coding RNA LINC00460 expression in EGFR-mutant lung adenocarcinoma. Int J Oncol. 2019;56:243-257
110. Xuehao L, Xin Z, Chunlu Y, Su C, Qiming S, Shun X. The lncRNA RHPN1-AS1 downregulation promotes gefitinib resistance by targeting miR-299-3p/TNFSF12 pathway in NSCLC. Cell Cycle. 2018;17:1772-1783
111. Jiayuan H, Banzhou P, Guohao X, Jingni Z, Chenchen L, Jifeng F. LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. Cell Death Dis. 2020;11:525
112. Zhongxing B, Jiashu H, Zhibo Z, Naixin L. FOXO3-induced oncogenic lncRNA CASC9 enhances gefitinib resistance of non-small-cell lung cancer through feedback loop. Life Sci. 2021;287:120012
113. Jing H, Shidai J, Wei Z, Deqin W, Jun L, Jing X, Wen G. Long non-coding RNA LOC554202 promotes acquired gefitinib resistance in non-small cell lung cancer through upregulating miR-31 expression. J Cancer. 2019;10:6003-6013
114. Degui S, Yufen X, Wenyu C. Knockdown of lncRNA BLACAT1 reverses the resistance of afatinib to non-small cell lung cancer via modulating STAT3 signalling. J Drug Target. 2019;28:300-306
115. Zewu L, Huanfu N, Qianqian Q, Sanhui Y, Qin W, Chunna Y. et al. lncRNA UCA1 Mediates Resistance to Cisplatin by Regulating the miR-143/FOSL2-Signaling Pathway in Ovarian Cancer. Mol Ther Nucleic Acids. 2019;17:92-101
116. Chaoyi L, Kai F, Yue Q, Wei Z, Ai H, Xiangfu S, Shijie X. Deregulation of UCA1 expression may be involved in the development of chemoresistance to cisplatin in the treatment of non-small-cell lung cancer via regulating the signaling pathway of microRNA-495/NRF2. J Cell Physiol. 2019;235:3721-3730
117. Jun F, Hangmei C, Yan W, Sanyou F, Daofeng W. Elevation of FGD5-AS1 contributes to cell progression by improving cisplatin resistance against non-small cell lung cancer cells through regulating miR-140-5p/WEE1 axis. Gene. 2020;755:144886
118. Z-S J, B S, D B, X-F Z. Effect of lncRNA-BLACAT1 on drug resistance of non-small cell lung cancer cells in DDP chemotherapy by regulating cyclin D1 expression. Eur Rev Med Pharmacol Sci. 2020;24:9465-9472
119. Zhaolong Z, Guofang Z, Huangkai Z, Liangqin N, Lifeng H, Jiangtao L. et al. LncRNA FOXD3-AS1 promoted chemo-resistance of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. Cancer Cell Int. 2020;20:350
120. Daolu Y, Wenyan F, Yu Z, Junxia L, Zhenqing F, Tianwei X. et al. Long non-coding RNA linc00665 inhibits CDKN1C expression by binding to EZH2 and affects cisplatin sensitivity of NSCLC cells. Mol Ther Nucleic Acids. 2021;23:1053-1065
121. Tayvia B, Jamie A J, Abby D I, Brandon M H, Emily S W, Michael T W. et al. Y Chromosome LncRNA Are Involved in Radiation Response of Male Non-Small Cell Lung Cancer Cells. Cancer Res. 2020;80:4046-4057
122. Zhiwu D, Ping Y, Xiaojian Q, Shuang L, Bing G, Haisheng Y. et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol. 2018;234:11304-11314
123. Zhanwu Y, Gebang W, Chenlei Z, Yu L, Wei C, Haoyou W, Hongxu L. LncRNA SBF2-AS1 affects the radiosensitivity of non-small cell lung cancer via modulating microRNA-302a/MBNL3 axis. Cell Cycle. 2020;19:300-316
124. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-1055
125. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167-179
126. Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491
127. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625-639
128. Wen D, Danquah M, Chaudhary AK, Mahato RI. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J Control Release. 2015;219:237-247
129. Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA Biomarkers: Emerging Frontiers in Colorectal and Other Human Cancers. Expert Rev Mol Diagn. 2016;16:553-567
130. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188502
131. Feichtenschlager V, Zheng YJ, Ho W, Chen L, Callanan C, Chen C. et al. Deconstructing the role of MALAT1 in MAPK-signaling in melanoma: insights from antisense oligonucleotide treatment. Oncotarget. 2023;14:543-560
132. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y. et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653-668
133. Silva-Fisher JM, Dang HX, White NM, Strand MS, Krasnick BA, Rozycki EB. et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun. 2020;11:2156
134. Islam Khan MZ, Law HKW. RAMS11 promotes CRC through mTOR-dependent inhibition of autophagy, suppression of apoptosis, and promotion of epithelial-mesenchymal transition. Cancer Cell Int. 2021;21:321
135. Li C, Wang S, Xing Z, Lin A, Liang K, Song J. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106-119
136. Zhuo W, Kang Y. Lnc-ing ROR1-HER3 and Hippo signalling in metastasis. Nat Cell Biol. 2017;19:81-83
137. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270-288
138. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629-651
Author contact
![]() Corresponding authors: wsj1088edu.cn (S. Wang), wangzhao1396edu.cn (Z. Wang); Tel.: +86-518-80689650; +86-512-65223637.
Corresponding authors: wsj1088edu.cn (S. Wang), wangzhao1396edu.cn (Z. Wang); Tel.: +86-518-80689650; +86-512-65223637.

 Global reach, higher impact
Global reach, higher impact