Impact Factor
ISSN: 1837-9664
J Cancer 2011; 2:165-176. doi:10.7150/jca.2.165 This volume Cite
Research Paper
Protein Signatures in Human MDA-MB-231 Breast Cancer Cells Indicating a More Invasive Phenotype Following Knockdown of Human Endometase/Matrilysin-2 by siRNA
1. Department of Chemistry and Biochemistry and Institute of Molecular Biophysics, Florida State University, Tallahassee, Florida 32306-4390, USA;
2. Department of Biomedical Sciences, College of Medicine, Florida State University, Tallahassee, Florida 32306-4390, USA;
3. Department of Pathology, Comprehensive Cancer Center, and National Foundation for Cancer Research Center for Metastasis Research, University of Alabama at Birmingham, Birmingham, Alabama, USA.
* Present address: Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, 21205, USA.
Received 2011-2-25; Accepted 2011-3-14; Published 2011-3-16
Abstract
Human matrix metalloproteinase-26 (MMP-26/endometase/matrilysin-2) is a putative biomarker for carcinomas of breast, prostate, and other cancers of epithelial origin. MMP-26 expression was silenced using small interfering RNA (siRNA) in the human breast cancer cell line MDA-MB-231. Immunological and proteomics approaches, including two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization time-of-flight mass spectrometry, were employed to identify differential protein expression in MMP-26 knockdown cells. A comparison of the protein expression profiles of control and MMP-26 knockdown cells revealed nine differentially regulated proteins. Five of the proteins (heat shock protein 90, glucose-regulated protein 78 (GRP78), annexin V, tropomyosin, and peroxiredoxin II) were up-regulated, while alpha-tubulin, cystatin SA-III, breast cancer metastasis suppressor 1 (BRMS1) and beta-actin were down-regulated. This decrease of BRMS1 expression is concomitant with an increase of invasion through matrix-coated membranes. These results suggest an important role for MMP-26 in the regulation of proteins involved in invasive and metastatic breast cancers.
Keywords: Matrix metalloproteinase (MMP), MMP-26, breast cancer metastasis suppressor 1, putative protein biomarkers, invasion and metastasis, mass spectrometry, proteomics
Introduction
During past decades, studies have indicated that matrix metalloproteinases (MMPs) participate in a number of physiological processes including reproduction, development, morphogenesis, and tissue remodeling as well as several pathological conditions including arthritis, cardiovascular diseases, and cancer metastasis [1-3]. MMPs were believed to be involved in these processes solely by their ability to degrade extracellular matrix (ECM). Later however, it was demonstrated that in few cases MMPs might not be required for tumor cells to invade tissue as shown by their ability to invade and move through an intact ECM by adopting amoeba-like movements, a process independent of MMPs [4, 5]. Complexed by the number of studies involving this family of enzymes, MMP activities today include the release and processing of cryptic fragments, neo-epitopes, growth factors, growth factor receptors, cytokines, chemokines, and precursor proteins from matrix and non-matrix substrates, as well as microenvironmental-dependent modification of the cell-ECM interface [1, 2, 6-8]. Most recently, the expression of MMPs can be a prognostic marker correlated with aggressive stages of cancer [9-11].
Human matrix metalloproteinase-26 (MMP-26/endometase/matrilysin-2), comprised of only pro- and catalytic domains, is the smallest member of the MMP family with its tertiary structure and enzymatic activity regulated by a calcium ion [12-15]. MMP-26 promotes invasion of a highly metastatic and tumorigenic prostate cancer cell [16]. Expression of MMP-26 is significantly higher in preinvasive human breast ductal carcinoma in situ (DCIS) and high-grade prostatic intraepithelial neoplasia (HGPIN) when compared with that in normal human breast tissue samples and non-neoplastic ducts [11, 17, 18]. Levels of MMP-26 expression are also high in early stage invasive carcinoma (I, II), whereas in stage III invasive carcinomas the level of MMP-26 decreases [11, 17]. A similar expression pattern has been discovered in squamous cell cancer (SCC) where low-grade SCC correlated to increased MMP-26 expression and lowered expression in dedifferentiated grade III tumors (9). MMP-26 has been postulated as a putative biomarker for human carcinomas of breast, prostate, and other cancers of epithelial origin [9, 11, 17, 18].
We undertook a proteomic approach to identify proteins that are regulated directly or indirectly by MMP-26. In the present study, we utilized siRNA to knockdown expression of endogenous MMP-26 in the human breast cancer cell line MDA-MB-231. Changes of the protein expression pattern in MDA-MB-231 were investigated by two-dimensional gel electrophoresis (2-DE). The proteins in question were identified by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and Western blots were utilized to confirm changes of protein expression upon silencing MMP-26 expression.
Material and Methods
Construction of MMP-26 Specific siRNA containing vector- The construction of the siRNA-expression plasmids was based on siSTRIKE™ U6 Hairpin Cloning Systems (Promega Corporation). The vector includes a human U6 promoter, Ampr / Neomycinr genes, and facilitated sticky ends with downstream overhang PstI partial sites. The inserted hairpin sequence, which is sense nucleotides, a loop-creating region, and anti-sense nucleotides, was designed by using the siRNA Target Designer Program (www.promega.com/siRNADesigner/). The sequences generated by this program were compared to all sequences in Genbank by using NCBI BLAST to confirm the specificity for MMP-26. Among those sequences, only the MMP-26 specific target sequence was selected and also a scrambled sequence was designed in the same manner as the control. Forward and reverse sequences of siRNA target insert and scramble insert were, respectively, as follows: MMP-26 target, 5'-ACCGGAAGATGCAAGTGGAATAAAGTTCTCTTATTCCACTTGCATCTTCCTTTTTC -3' and 5'-TGCAGAAAAAGGAAGATGCAAGTGGAATAAGAGAACTTTATTCCACTTGCATCTTC -3'; scrambled target, 5'-ACCGATAGTGAACGGTAAGAAGAAGTTCTCTCTTCTTACCGTTCACTATCTTTTTC -3' and 5'-TGCAGAAAAAGATAGTGAACGGTAAGAAGAGAGAACTTCTTCTTACCGTTCACTAT -3'.
After annealing, the DNA fragment was ligated. The one new PstI site produced by ligation and already existing PstI site were used for the selection of siSTRIKE™/MMP-26 or siSTRIKETM/scrambled plasmids.
Cell culture, transfection of MDA-MB-231 cells and isolation of cell lines containing MMP-26 specific siRNA- MDA-MB-231 (ATCC), an established human breast carcinoma cell line, was routinely grown in polystyrene tissue culture dishes (100 x 20 mm, Becton Dickinson Labware) with high-glucose Dulbecco's modified Eagle's Medium (DMEM, Gibco Invitrogen Corporation) supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 units/mL penicillin, and 100 μg/mL streptomycin (Cambrex) in a humidified atmosphere containing 5% CO2 at 37ºC. MDA-MB-231 cells were transfected with siSTRIKE™/MMP-26 or siSTRIKETM/scrambled using Transfectol (GeneChoice, PGC Scientifics Corporation). Transfectol-mediated DNA transfections into MDA-MB-231 cells were performed following the instructions provided by GeneChoice. Transfected cell lines were maintained in the presence of 550 μg/mL Geneticin (G-418, Fisher Science), and were screened on the basis of down-regulation of MMP-26 expression. Selected stably transfected cell lines were maintained in G-418 due to rapid loss of vector in the absence of selection pressure.
Immunoblotting- At confluence, cells were washed three times with phosphate buffered saline (PBS). Each dish was treated with 1 mL of 0.25% trypsin solution (GIBCO Invitrogen Corporation), mixed with 9 mL of serum free DMEM and cell number was determined using a hemacytometer. Cell lysates were collected according to the manufacturer's protocol (Pierce). Briefly, cells were washed three times with PBS, an appropriate amount of M-PER (Pierce) reagent and protease inhibitor cocktail (Sigma) was added to adjust 107 cells/mL. After 5 min gentle shaking, cells were removed with a scraper and transferred to a microfuge tube. The protein concentration (1.58 mg/mL) was determined by using the BCA protein assay kit (Pierce). Protein (35 μg) was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% polyacrylamide gel and was then transferred to a BioTrace NT nitrocellulose membrane (Pall Life Science). The membrane was blocked with 5% w/v bovine serum albumin (BSA, Sigma-Aldrich) in TBST (100 mM Tris/HCl, pH 7.4, 150 mM NaCl, 20 mM KCl, 0.05% Tween 20) for 2 h at room temperature and then incubated with primary antibody for another 2 h at room temperature. The primary antibodies used were as follows: rabbit anti-human MMP-26 polyclonal and mouse anti-human BRMS1 monoclonal antibodies were prepared as described before [11, 19]; mouse anti-human cystatin SA monoclonal antibody (R&D Systems); all other primary antibodies (rabbit anti-human 90kDa HSP polyclonal antibody, goat anti-human GRP78 polyclonal antibody, goat anti-human peroxiredoxin II (PRX II) polyclonal antibody, goat anti-human annexin V polyclonal antibody, and goat anti-human tropomyosin polyclonal antibody) were purchased from Santa Cruz Biotechnology. After three washes with TBST, the membrane was incubated with alkaline phosphatase conjugated secondary antibody for 30 min at room temperature. The bands were visualized by NBT/BCIP substrates.
Two-dimensional gel electrophoresis- Cells grown to confluence were washed three times with PBS. A single 100-mm cell culture dish was lysed by adding an appropriate amount of 2D-lysis buffer containing 7 M urea, 2 M thiourea, 2% w/v CHAPS, 0.25% w/v Biolyte 3-10 ampholyte (Bio-Rad) and protease inhibitor cocktail (Sigma), and 1% w/v DTT to the dish containing the cells (107 cells/mL). Cells were removed with a scraper, transferred to a centrifuge tube, and sonicated for 2 min in an ice bath followed by centrifugation at 17,000 x g for 20 min/4ºC. The supernatant was transferred to a clean tube, and protein concentration was determined using the BCA protein assay kit (Pierce). After addition of 2D-lysis buffer (ca. 50 μL) containing a trace amount of bromophenol blue to cell lysates (200 μg), the sample was vortexed for 1 h at room temperature and centrifuged (1,000 x g for 5 min). The resultant supernatant was applied to immobilized pH gradient (IPG) strips (pH 4-7, 11 cm, Bio-Rad). After 12 h of active rehydration (50 V) at 20ºC, the proteins in the sample were focused at 250 V for 15 min with a final voltage of 8000 V for a total of 60,000 volt-hours. The strips were subsequently equilibrated for 10 min in 2 mL of solution consisting of 6 M urea, 2% w/v SDS, 375 mM Tris/HCl (pH 8.8), 2% w/v DTT and 20% glycerol followed by another 10 min equilibration with 2 mL of solution containing 375 mM Tris/HCl (pH 8.8), 6 M urea, 2% w/v SDS, 2.5% w/v iodoacetamide, and 0.1% w/v bromophenol blue. Equilibrated IPG strips were transferred onto 10% polyacrylamide gels casted in-house in 2D gel Criterion cassette (Bio-Rad) with the proteins separated by SDS-PAGE (100 V for 5 h) at 4ºC. Separated proteins were visualized by EZ-Blue staining solution (Sigma). SeeBlue Plus2 pre-stained standard (Invitrogen) was used to determine approximate molecular weight of protein spot while spot size was measured by Integrated Morphometry Analysis (IMA) followed by statistical analyses. Data were expressed as means ± standard deviation (SD). P-values of < 0.05 were considered statistically significant.
Protein identification by in-gel trypsin digestion and MALDI-TOF MS- The protein spots of interest were manually excised with cut pipette tips from the stained 2D gels and transferred to siliconized tubes (0.6 mL). The gel plugs were trypsin digested using in-gel trypsin digestion kit (Pierce). Briefly, the gel plugs were dehydrated (15 min) by adding 50 μL of acetonitrile (ACN). ACN was then removed and gel plugs were dried. The gel plugs were digested with trypsin (10 μL of 10 ng/μL trypsin solution) at 30 ºC overnight with constant shaking. Thereafter, the supernatant containing tryptic peptides was transferred to another siliconized tube. The peptides in the gel plug were extracted (5 min) with 1% trifluoroacetic acid (10 μL) and the supernatant was pooled. Tryptic peptides were desalted with ZipTipC18 (Millipore Corporation), mixed with alpha-cyano-4-hydroxycinnamic acid (CHCA, Sigma), and spotted onto a MALDI target plate. The tryptic peptides were analyzed with a MALDI-TOF mass spectrometer (Axima CFRplus, Shimadzu Corporation) by Kompact v.2.4.1 software. The acquired masses were calibrated with ProteoMassTM Peptide MALDI-MS calibration kit (Sigma) as external reference masses and trypsin auto-digested peptides (m/z 515.33, 842.51, 2211.10) as internal references masses. Spectra were acquired in the reflection mode. The tryptic peptide ((M+H)+) masses were used to search the database. The database search was performed online with Mascot server (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF). Carbamidomethylation of cysteine was selected for the fixed modification. The NCBInr database was searched within a mass tolerance of ± 100 ppm for human proteins, allowing for one missed cleavage. Proteins were considered to be successfully identified by the following parameters - peptide tolerance lower than ± 100 ppm. Mowse score greater than 68, low probability that the observed match between the experimental data and the database sequence is a random event, number of matched peptides and percent coverage. A false positive identification was evaluated by searching a decoy database.
Modified Boyden chamber invasion assays- The invasiveness of wild type and siRNA transfected MDA-MB-231 cells through reconstituted ECM was determined as described previously [12]. Briefly, invasion inserts containing polycarbonate filters with 8 μm pores (BD Biosciences) were coated with 70 μL of 250 μg/mL of Type IV collagen (Sigma). The Boyden chambers were dried and sterilized in a laminar flow hood under ultraviolet radiation overnight. To commence the assay, 500 μL of DMEM high-glucose culture medium containing 10% v/v FBS was added to the lower chambers, and 300 μL of prepared cell suspensions (1.5 x 105 cells/mL) in serum free DMEM high glucose media was added to each insert. After 12 h of incubation, invasive cells that had passed through the filter to the lower surface of the membrane were stained using a 0.1% w/v crystal violet solution. Cells remaining inside the chamber were removed with a cotton swab, and the filters were removed and mounted on a microscope slide. The membranes were photographed with an Olympus DP10 digital camera (Melville) under a Nikon FX Microscope (Melville). Cells were counted by IMA. For statistical analyses wild type MDA-MB-231 cell line was assumed to reflect 100% cell invasion. The ratio of the number of invaded cells of each cell line to the invaded cells of the wild type MDA-MB-231 cell line was used for subsequent comparative analyses. Values are mean ± SD of triplicate experiments. A difference in ratio was considered statistically significant at p < 0.05.
Results
Silencing expression of MMP-26 by using small interfering RNA in MDA-MB-231 cells
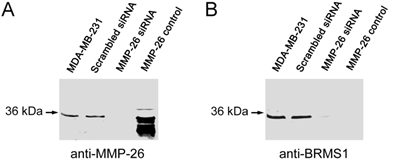
Utilizing siRNA the endogenous expression of MMP-26 was knocked-down in the MDA-MB-231 human breast carcinoma cell line. Cell clones in which MMP-26 expression was knocked-down most completely by siSTRIKE™/MMP-26 was assessed by immunoblot (Figure 1). A scrambled siRNA transfected cell line was used to control for off target siRNA effects. Furthermore, MMP-26 knock-down cell line also showed a dramatic decrease of BRMS1 expression.
Blocking expression of MMP-26 in MDA-MB-231 cell line silenced expression of BRMS1. (A) 10% SDS-PAGE followed by immunoblot analyses using anti-MMP-26. siRNA, interfering with the mRNA of MMP-26, silenced expression of MMP-26 compared with parental and scrambled siRNA transfected cell lines. Purified recombinant MMP-26 control, which was expressed in BL21 (DE3)-competent E. coli cells and refolded as described previously (Lee et al 2007). (B) 10% SDS-PAGE followed by immunoblot analyses using anti-BRMS1 antibody. Blocking of MMP-26 mRNA silenced expression of BRMS1 compared with parental and scrambled siRNA transfected cell lines. Purified recombinant MMP-26 control.
Identification of protein expression changes by MALDI-TOF spectroscopy and immunoblotting in MMP-26 knockdown MDA-MB-231 cells
2-DE of MMP-26 or scrambled siRNA transfected MDA-MB-231 cells showed changes of the expression pattern. Only identified protein spots were designated by arrows (Figure 2A). Silencing of MMP-26 modified the expression of eight different spots. Spots designated by si1/scr1, si2/scr2, si4/scr4. si5/scr5 and si6 where 5 proteins shown to increase their expression. While si3/scr3, si7/scr7 and si8/scr8 were down-regulated by MMP-26 knockdown. Figure 2B shows detailed spot areas. Detailed area for si4/scr4 was observed from another set of 2-DE gels. Figure 2C shows statistically analyzed protein expression level. MMP-26 knockdown caused five significantly increased expression levels of proteins (si1/scr1, si2/scr2, si4/scr4, si5/scr5, and si6) and three significantly decreased expression of proteins (si3/scr3, si7/scr7, and si8/scr8).
Representative images of two dimensional gel electrophoresis of cell lysate. (A) 250 μg of whole MDA-MB-231 cell lysate using 11 cm, 4 to 7 pH range IPG strips in the first dimension and 10 % polyacrylamide gel in the second dimension were used. Gels were visualized by using EZ Blue. Blocking expression of MMP-26 by siRNA showed a differential expression pattern. Spots identified successfully by MALDI-TOF MS were the only ones labeled. (B) Cropped images were obtained from same 2D gel except for si7/scr7 which were obtained from another pair of 2D gels. (C) Spot sizes were measured by IMA. MMP-26 knockdown MDA-MB-231 cell line showed significantly increased expression of HSP90 (si1/scr1, p < 0.0005), GRP78 (si2/scr2, p < 0.0005), annexin V (si4/scr4, p < 0.0005), tropomyosin (si5/scr5, p < 0.05), and PRX II (si6, no p value due to no spot for scramble detected), and significantly decreased expression of α-tubulin (si3/scr3, p < 0.0005), cystatin SA (si7, p < 0.05), and β-actin (si8/scr8, p < 0.005).
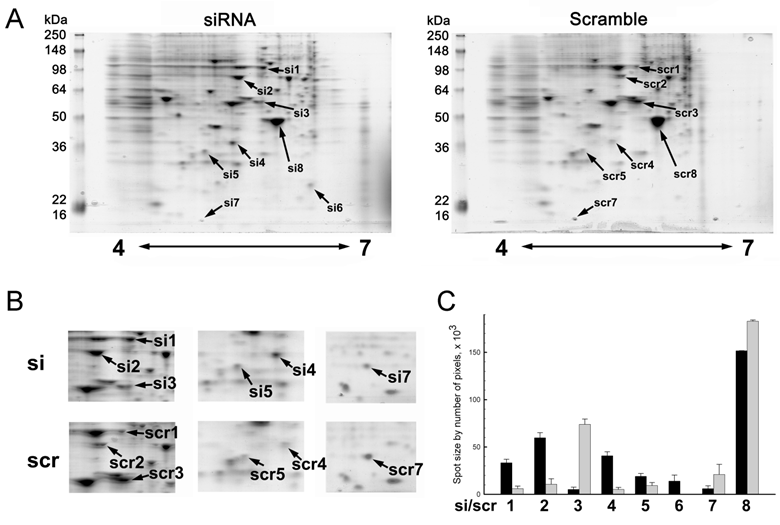
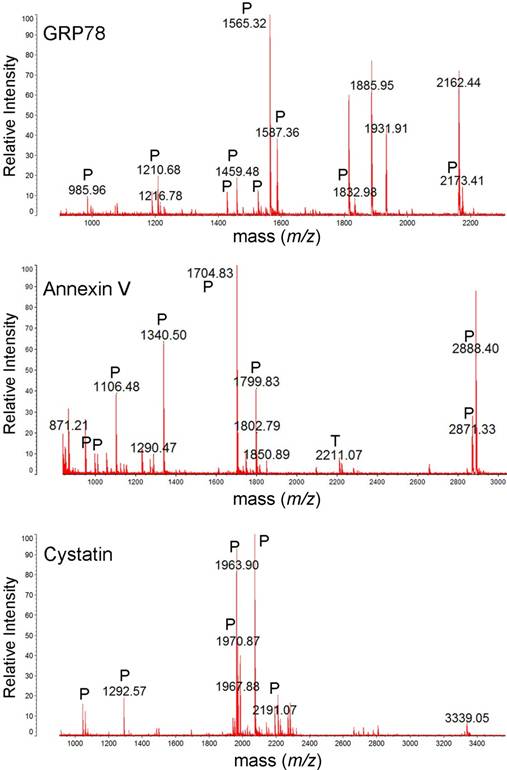
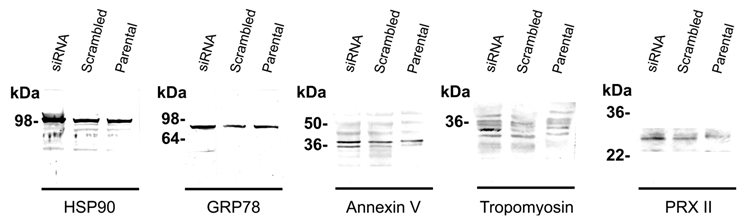
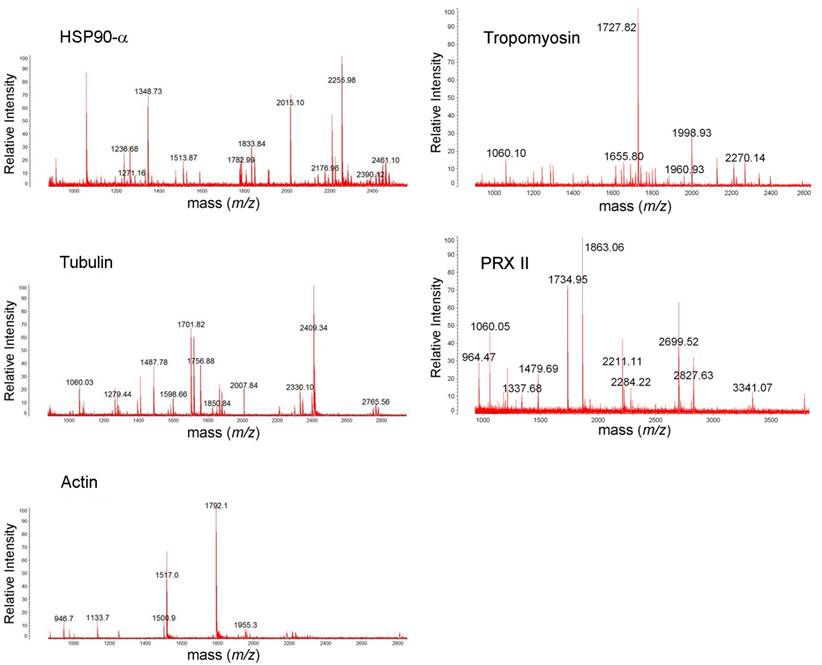
Table 1 shows identified protein spots by MALDI-TOF spectroscopy (si1/scr1, 90 kDa heat shock protein A (HSP90A), Mowse score = 143, sequence coverage = 36%; si2/scr2, GRP 78, Mowse score = 185, sequence coverage = 31%; si3/scr3, α-tubulin, Mowse score = 140, sequence coverage = 49%; si4/scr4, annexin V, Mowse score =243, sequence coverage = 68%; si5/scr5, tropomyosin, Mowse score = 117, sequence coverage = 31%; si6, peroxiredoxin II (PRX II), Mowse score = 77, sequence coverage = 26%; si7/scr7, cystatin SA, Mowse score = 117, sequence coverage = 68%; si8/scr8, β-actin, Mowse score =88, sequence coverage = 26%). Figure 3 and Figure S1 shows the mass spectra of identified proteins. Western blot analysis supported 2-DE analysis data. MMP-26 knock-down MDA-MB-231 cell line showed increased expression of HSP90, GRP78, annexin V, tropomyosin, and PRX II (Figure 4). Validation of down-regulated proteins could not be determined by western blot due to the presence of nonspecific bands.
Identification of proteins which are differentially expressed upon treatment with MMP-26 specific siRNA. Protein spots were excised from 2D gels, in-gel trypsin digested, desalted using ZipTip C18, and identified by MALDI-TOF MS. Five identified proteins (HSP90, GRP78, annexin V, tropomyosin, PRX II) were upregulated and three identified proteins (α-tubulin, cystatin, β-actin) were downregulated in MMP-26 specific siRNA transfected MDA-MB-231 cells compared with control (scrambled siRNA transfected cells).
| Spot | Identification | NCBInr Accession # | Theoretical Mr (103) /pI | Experimental Mr (103) /pI | Score | Expect | Peptide # | % Coverage |
|---|---|---|---|---|---|---|---|---|
| si1/scr1 | HSP90A | NP_001017963 | 85/4.94 | 100/5.74 | 143 | 1e-10 | 24 | 36 |
| si2/scr2 | GRP78 | P07823 | 72.4/5.07 | 80/5.47 | 185 | 6.5e-15 | 20 | 31 |
| si3/scr3 | α-Tubulin | CAA25855 | 50.8/5.02 | 55/5.68 | 140 | 2.2e-09 | 17 | 49 |
| si4/scr4 | Annexin V | NP_001145 | 36.0/4.94 | 35/5.32 | 243 | 1.1e-19 | 20 | 68 |
| si5/scr5 | Tropomyosin | NP_001036817 | 29.1/4.79 | 32/5.05 | 117 | 3.1e-07 | 10 | 31 |
| si6 | PRX II | NP_005800 | 21.9/5.67 | 25/6.22 | 77 | 2.9e-3 | 5 | 26 |
| si7/scr7 | Cystatin-SA | NP_001890 | 16.5/4.95 | 14/4.8 | 117 | 4.1e-08 | 10 | 68 |
| si8/scr8 | β-Actin | AAH12854 | 40.5/5.55 | 45/5.95 | 88 | 2.8e-4 | 8 | 26 |
MALDI-TOF mass spectra of GRP78, annexin V and cystatin. The protein spots were excised from 2DE gel, in-gel trypsin digested, ZipTip purified, and were analyzed with MALDI-TOF MS. Tryptic (P), and trypsin autodigestion (T) peptides are indicated in the spectrum.
Western blot analysis. The MALDI-TOF MS identified proteins were validated by immunoblotting. Silencing MMP-26 expression showed the differential expression level of the proteins, HSP90 (100 kDa), GRP78 (82 kDa), annexin V (double bands around 36 kDa), tropomyosin (32 kDa), and PRX II (24 kDa). Only up-regulated proteins were identified by western blot. It was not be able to determine down regulated protein because of the heavy nonspecific bands.
Silencing expression of MMP-26 increases invasion by MDA-MB-231 cells
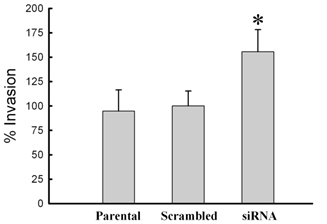
To initially assess biological changes associated with siRNA knock-down of MMP-26, three cell lines (parental, MMP-26 target- or scrambled siRNA- transfected MDA-MB-231 cell lines) were evaluated for invasion through type IV collagen-coated filters. Lack of MMP-26 in MDA-MB-231 cells showed a statistically significant increase of invasion potential through a type IV collagen-coated membrane; whereas, scrambled siRNA-transfected cells showed insignificant change of invasion potential when compared with the parental cell line (wild vs. siRNA, p < 0.005; scramble vs. siRNA, p < 0.005; wild vs. scramble, p > 0.05) (Figure 5).
Change of invasive potential of MDA-MB-231 cells according to the transfection with MMP-26 siRNA. Invasion assays were performed with modified Boyden chambers, and the percentage of invading cells was quantified as described under “Experimental Procedures”. All error bars shown are SD from the mean of triplicate experiments for each group. MMP-26 siRNA transfected cell line showed statistically significant increase in invasion potential (p < 0.005 compared with wild type and control (scrambled siRNA transfected cell line)). The asterisk denotes statistically significant.
Discussion
Human endometase, also known as matrilysin-2/MMP-26, is a putative biomarker for various preinvasive cancers [9, 11, 17, 18]. Past studies have demonstrated that in contrast with the other secretory MMPs, MMP-26 is predominantly intracellular, despite the presence of the signal peptide in MMP-26's prodomain [13-16, 20]. Western blot analysis of MDA-MB-231 cell lysate indicated high levels of intracellular MMP26, while expression of enzyme was not detected in the cell culture medium (data not shown). We began to explore MMP-26 as an anti-tumor or anti-invasion protein. Selectively blocking of MMP-26 expression by siRNA resulted in increased invasiveness and changes in expression of several proteins associated with invasion and/or metastasis, especially silencing BRMS1 expression.
BRMS1 is a suppressor of metastasis for human breast cancer [21-23], melanoma [24], ovarian carcinoma [25] and non-small cell lung cancer [26]. Microarray and proteomics analyses showed multiple changes in gene and protein expression when BRMS1 was re-expressed [27, 28]. BRMS1 has been proposed to regulate gene transcription through interaction with large SIN3:HDAC complexes [21, 29] and through negative regulation of NF-κB [30]. Importantly, BRMS1 reduced invasion through Matrigel-coated filters [23].
The 2D-gel, MALDI-TOF MS analyses revealed that silencing MMP-26 expression also modifies the regulation of additional proteins which have been implicated in tumor growth, invasion, motility and metastasis. For example, the five up-regulated proteins include HSP90, GRP78, annexin V, tropomyosin and PRXII, while three down-regulated proteins are composed of β-actin, α-tubulin and cystatin SA-III.
Heat shock protein (HSP) and glucose regulated protein (GRP) are molecular chaperones involved in proper folding of nascent polypeptides, post-translational regulation of signaling molecules and reducing apoptosis under the stress [31-34]. However, these characteristics of chaperones endow tumor cells to survive under stressful microenvironments [35, 36]. HSP90's client proteins are involved in malignant phenotypes including uncontrolled proliferation, immortalization, impaired apoptosis, and angiogenesis [36, 37]. However, not all client proteins are involved in cancer progression. Yet, BRMS1 was identified as a client protein of HSP90 [19]. Recent research revealed that HSP90 stabilizes and activates cell surface receptors by its interactions with extracellular and cytoplasmic domain of receptors to facilitate cell migration [38-41]. HSP90 has been recognized as a target for the treatment of cancer [36, 42]. Elevated expression of HSP90 has been reported in various leukemia cases and a number of tumor types including breast, lung along with other high-grade malignant tumors [43, 44]. GRP78 also has been reported to be up-regulated in a variety of cancer cell lines, solid tumors, and human cancer biopsies correlating with malignancy [45-48].
Peroxiredoxins (PRXs), a novel group of cysteine-containing proteins with efficient antioxidant capacity [49], are involved in signal transduction, cell proliferation, differentiation, apoptosis, and gene expression [50, 51]. PRX II, a member of PRX family, is a cytosolic protein. PRX II is known to not only protect cells from oxidative damage caused by H2O2, but also to endow cancer cells with resistance to both H2O2 and cisplatin granting them radioresistance [52]. The expression level of PRX II is very high in malignant mesothelioma compared with healthy pleural mesothelium [53, 54]. Recent proteomic profiling shows that progressive cancer cells overexpress PRX II compared with regressive cancer cells [52].
Annexins are a family of proteins which bind to negatively charged phospholipids in a calcium-dependent manner. Annexin V forms voltage sensitive channels [55] and mediates calcium flux, phospholipid-dependent inhibition of blood coagulation, modulation of protein kinase C, and the inhibition of phospholipase A2 activity [55, 56]. Cancer related research shows that the expression of annexin V is augmented in growth hormone-secreting carcinomas [57] while proteomics profiling shows that progressive cancer cells overexpress annexin V compared with regressive cancer cells [52].
Tropomyosins encompass a large family of actin regulatory proteins that stabilize the actin cytoskeleton and regulate actin-myosin interactions during cell migration [58-60]. Tropomyosin destabilizes actin filaments and is expressed more in highly metastatic cells than low ones. Moreover, suppression of tropomyosin in the highly metastatic cell line results in suppression of cell motility [61].
Actin and its associated proteins play important structural and functional roles, such as maintaining cell morphology, cell adhesion, cell motility, exocytosis, endocytosis, and cell division [62, 63]. When mammalian cells are transformed or progressed to acquire metastatic potential, the expression of actin and/or actin-related proteins becomes modified. β-actin is present in non-muscle cells, especially in submembrane, and participates in active cell movement and wound healing [64].
Tubulin, the building block of microtubules, is a heterodimmer comprising alpha and beta subunits, each approximately 50 kDa. Microtubules are the essential components of all eukaryotic cells and are involved in a diverse range of cellular functions, including motility [65, 66]. Disruption of the microtubule network through incorporation of nitrotyrosinated α-tubulin results in apoptosis and inhibition of myogenic differentiation. Modification of the tubulin may play a role in multi-drug resistance to chemotherapy and it is a key molecular target for cancer therapy [67]. MDA-MB-231 breast cancer cells contain only tyrosinated tubulin and low level monoglutamylation in certain isoforms [65]. Tubulin detyrosination also has been reported in breast cancer and is linked to tumor aggressiveness [68].
Cystatins are endogenous cysteine proteinase inhibitors that are found in body fluids and tissues. They are generally tight-binding inhibitors of cysteine proteinases such as papain, ficin, and cathepsins. The physiological functions of cystatins are regulation of protein metabolism, protection of cells and tissues against unfavorable proteolysis, and tissue damage [69]. Cystatin SA is abundant in human saliva and known to be expressed tissue-specifically [70].
Recent proteomics profiling shows that progressive cancer cells overexpress HSP90, annexin V, PRX II, and tropomyosin compared with regressive cancer cells [52]. Other individual researchers have shown the role of these in carcinomas. HSP90 promotes invasive potential through activating proMMP-2 [71], and expression level of PRX II is very high in malignant mesothelioma compared with healthy pleural mesothelium [49, 53]. Suppression of tropomyosin in the highly metastatic cell line results in suppression of cell motility [61]. Therefore, previous results show that HSP90, annexin V, PRX II, tropomyosin, and β-actin are all involved in invasive potential and/or metastatic phenotypes of cancer cells. Anti-tumor properties of MMP-26 also have been studied. MMP-26-mediated, intracellular pathway targets ERβ and MMP-26 contributes favorably to the survival of the ERα/β-positive cohort of breast cancer patients [72]. Taken together with our results, MMP-26 down-regulates several proteins enabling cancer cell to be invasive supporting anti-tumor function of MMP-26.
In summary, silencing MMP-26 in MDA-MB-231 cells increased invasion commensurate with changes in invasion-associated protein expression. These results directly correspond to our previously reported studies showing that MMP-26 expression is significantly higher in preinvasive DCIS and HGPIN, compared with normal breast, non-neoplastic ducts, and invasive carcinomas [11, 18]. While there may be context-dependent differences in the role(s) of MMP-26 [16], the data suggest that MMP-26 may be a useful biomarker for premalignant carcinomas.
Acknowledgements
This work was supported in part by grants from the Susan G. Komen Breast Cancer Foundation (BCTR0504465), the Florida Breast Cancer Coalition Research Foundation, the Elsa U. Pardee Foundation, and Florida State University, and grants DAMD17-02-1-0238 and W81XWH-07-1-0225 from DOD US Congressionally Directed Medical Research Programs (to Dr. Q.-X. Sang); United States Public Health Service NIH Grants CA87728 (to Dr. D.R. Welch) and F32CA113037 (to Dr. D.R. Hurst), and a grant from the National Foundation for Cancer Research (to Dr. D.R. Welch). We thank the Biomedical Proteomics Laboratory, College of Medicine, FSU, for MALDI-TOF mass spectrometer access.
Abbreviations
MMP: Matrix metalloproteinase; siRNA: small interfering RNA; BRMS1: breast cancer metastasis suppressor 1; HSP: heat shock protein; GRP: glucose-regulated protein; PRX: peroxiredoxin; ECM: extracellular matrix; SCC: squamous cell cancer; DCIS: ductal cancinoma in situ; HGPIN: high-grade prostatic intraepithelial neoplasia; DMEM: Dulbecco'c modified Eagle's medium; FBS: fetal bovine serum; IMA: Integrated Morphometry Analysis.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. DeClerck YA, Mercurio AM, Stack MS. et al. Proteases, extracellular matrix, and cancer: a workshop of the path B study section. Am J Pathol. 2004;164:1131-9
2. Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004;16:558-64
3. Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562-73
4. Friedl P, Brocker EB. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. 2000;57:41-64
5. Wolf K, Mazo I, Leung H. et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267-77
6. DeClerck YA. Interactions between tumour cells and stromal cells and proteolytic modification of the extracellular matrix by metalloproteinases in cancer. Eur J Cancer. 2000;36:1258-68
7. Jodele S, Blavier L, Yoon JM, DeClerck YA. Modifying the soil to affect the seed: role of stromal-derived matrix metalloproteinases in cancer progression. Cancer Metastasis Rev. 2006;25:35-43
8. McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore!. Curr Opin Cell Biol. 2001;13:534-40
9. Ahokas K, Skoog T, Suomela S. et al. Matrilysin-2 (matrix metalloproteinase-26) is upregulated in keratinocytes during wound repair and early skin carcinogenesis. J Invest Dermatol. 2005;124:849-56
10. Gontero P, Banisadr S, Frea B, Brausi M. Metastasis markers in bladder cancer: a review of the literature and clinical considerations. Eur Urol. 2004;46:296-311
11. Lee S, Desai KK, Iczkowski KA. et al. Coordinated peak expression of MMP-26 and TIMP-4 in preinvasive human prostate tumor. Cell Res. 2006;16:750-8
12. Lee S, Park HI, Sang QX. Calcium regulates tertiary structure and enzymatic activity of human endometase/matrilysin-2 and its role in promoting human breast cancer cell invasion. Biochem J. 2007;403:31-42
13. Marchenko GN, Ratnikov BI, Rozanov DV, Godzik A, Deryugina EI, Strongin AY. Characterization of matrix metalloproteinase-26, a novel metalloproteinase widely expressed in cancer cells of epithelial origin. Biochem J. 2001;356:705-18
14. Park HI, Ni J, Gerkema FE, Liu D, Belozerov VE, Sang QX. Identification and characterization of human endometase (Matrix metalloproteinase-26) from endometrial tumor. J Biol Chem. 2000;275:20540-4
15. Uria JA, Lopez-Otin C. Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity. Cancer Res. 2000;60:4745-51
16. Zhao YG, Xiao AZ, Newcomer RG. et al. Activation of pro-gelatinase B by endometase/matrilysin-2 promotes invasion of human prostate cancer cells. J Biol Chem. 2003;278:15056-64
17. Savinov AY, Remacle AG, Golubkov VS. et al. Matrix metalloproteinase 26 proteolysis of the NH2-terminal domain of the estrogen receptor beta correlates with the survival of breast cancer patients. Cancer Res. 2006;66:2716-24
18. Zhao YG, Xiao AZ, Park HI. et al. Endometase/matrilysin-2 in human breast ductal carcinoma in situ and its inhibition by tissue inhibitors of metalloproteinases-2 and -4: a putative role in the initiation of breast cancer invasion. Cancer Res. 2004;64:590-8
19. Hurst DR, Mehta A, Moore BP. et al. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Commun. 2006;348:1429-35
20. Li W, Savinov AY, Rozanov DV. et al. Matrix metalloproteinase-26 is associated with estrogen-dependent malignancies and targets alpha1-antitrypsin serpin. Cancer Res. 2004;64:8657-65
21. Hurst DR, Xie Y, Vaidya KS. et al. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438-44
22. Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809-17
23. Samant RS, Seraj MJ, Saunders MM. et al. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2000;18:683-93
24. Shevde LA, Samant RS, Goldberg SF. et al. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229-39
25. Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer. 2006;16:522-31
26. Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196-203
27. Champine PJ, Michaelson J, Weimer BC, Welch DR, DeWald DB. Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis. 2007;24:551-65
28. Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006-18
29. Meehan WJ, Samant RS, Hopper JE. et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562-9
30. Samant RS, Clark DW, Fillmore RA. et al. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6
31. Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232-5
32. Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504-10
33. Liu H, Bowes RC3rd, van de Water B, Sillence C, Nagelkerke JF, Stevens JL. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J Biol Chem. 1997;272:21751-9
34. Wegele H, Muller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1-44
35. Takayama S, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;22:9041-7
36. Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761-72
37. Beere HM. "The stress of dying": the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117:2641-51
38. Annamalai B, Liu X, Gopal U, Isaacs JS. Hsp90 is an essential regulator of EphA2 receptor stability and signaling: implications for cancer cell migration and metastasis. Mol Cancer Res. 2009;7:1021-32
39. Sidera K, Gaitanou M, Stellas D, Matsas R, Patsavoudi E. A critical role for HSP90 in cancer cell invasion involves interaction with the extracellular domain of HER-2. J Biol Chem. 2008;283:2031-41
40. Tsutsumi S, Scroggins B, Koga F. et al. A small molecule cell-impermeant Hsp90 antagonist inhibits tumor cell motility and invasion. Oncogene. 2008;27:2478-87
41. Woodley DT, Fan J, Cheng CF. et al. Participation of the lipoprotein receptor LRP1 in hypoxia-HSP90alpha autocrine signaling to promote keratinocyte migration. J Cell Sci. 2009;122:1495-8
42. Waza M, Adachi H, Katsuno M. et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088-95
43. Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92:1564-72
44. Zuo DS, Dai J, Bo AH, Fan J, Xiao XY. Significance of expression of heat shock protein90alpha in human gastric cancer. World J Gastroenterol. 2003;9:2616-8
45. Fernandez PM, Tabbara SO, Jacobs LK. et al. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat. 2000;59:15-26
46. Little E, Ramakrishnan M, Roy B, Gazit G, Lee AS. The glucose-regulated proteins (GRP78 and GRP94): functions, gene regulation, and applications. Crit Rev Eukaryot Gene Expr. 1994;4:1-18
47. Shuda M, Kondoh N, Imazeki N. et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605-14
48. Takashima M, Kuramitsu Y, Yokoyama Y. et al. Proteomic profiling of heat shock protein 70 family members as biomarkers for hepatitis C virus-related hepatocellular carcinoma. Proteomics. 2003;3:2487-93
49. Lehtonen ST, Markkanen PM, Peltoniemi M, Kang SW, Kinnula VL. Variable overoxidation of peroxiredoxins in human lung cells in severe oxidative stress. Am J Physiol Lung Cell Mol Physiol. 2005;288:L997-1001
50. Karihtala P, Mantyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9:3418-24
51. Kim H, Lee TH, Park ES. et al. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. J Biol Chem. 2000;275:18266-70
52. Hayashi E, Kuramitsu Y, Okada F. et al. Proteomic profiling for cancer progression: Differential display analysis for the expression of intracellular proteins between regressive and progressive cancer cell lines. Proteomics. 2005;5:1024-32
53. Kinnula VL, Lehtonen S, Sormunen R. et al. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316-23
54. Lehtonen ST, Svensk AM, Soini Y. et al. Peroxiredoxins, a novel protein family in lung cancer. Int J Cancer. 2004;111:514-21
55. Kirsch T, Nah HD, Demuth DR. et al. Annexin V-mediated calcium flux across membranes is dependent on the lipid composition: implications for cartilage mineralization. Biochemistry. 1997;36:3359-67
56. Wang W, Xu J, Kirsch T. Annexin V and terminal differentiation of growth plate chondrocytes. Exp Cell Res. 2005;305:156-65
57. Mulla A, Christian HC, Solito E, Mendoza N, Morris JF, Buckingham JC. Expression, subcellular localization and phosphorylation status of annexins 1 and 5 in human pituitary adenomas and a growth hormone-secreting carcinoma. Clin Endocrinol (Oxf). 2004;60:107-19
58. Cooper JA. Actin dynamics: tropomyosin provides stability. Curr Biol. 2002;12:R523-5
59. Marston S, Burton D, Copeland O. et al. Structural interactions between actin, tropomyosin, caldesmon and calcium binding protein and the regulation of smooth muscle thin filaments. Acta Physiol Scand. 1998;164:401-14
60. O'Neill GM. The coordination between actin filaments and adhesion in mesenchymal migration. Cell Adh Migr. 2009;3:355-7
61. Miyado K, Kimura M, Taniguchi S. Decreased expression of a single tropomyosin isoform, TM5/TM30nm, results in reduction in motility of highly metastatic B16-F10 mouse melanoma cells. Biochem Biophys Res Commun. 1996;225:427-35
62. Lu QY, Jin YS, Pantuck A. et al. Green tea extract modulates actin remodeling via Rho activity in an in vitro multistep carcinogenic model. Clin Cancer Res. 2005;11:1675-83
63. Taniguchi S. Suppression of cancer phenotypes through a multifunctional actin-binding protein, calponin, that attacks cancer cells and simultaneously protects the host from invasion. Cancer Sci. 2005;96:738-46
64. Nowak D, Skwarek-Maruszewska A, Zemanek-Zboch M, Malicka-Blaszkiewicz M. Beta-actin in human colon adenocarcinoma cell lines with different metastatic potential. Acta Biochim Pol. 2005;52:461-8
65. Rao S, Aberg F, Nieves E, Band Horwitz S, Orr GA. Identification by mass spectrometry of a new alpha-tubulin isotype expressed in human breast and lung carcinoma cell lines. Biochemistry. 2001;40:2096-103
66. Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41-7
67. Idriss HT. Three steps to cancer: how phosphorylation of tubulin, tubulin tyrosine ligase and P-glycoprotein may generate and sustain cancer. Cancer Chemother Pharmacol. 2004;54:101-4
68. Mialhe A, Lafanechere L, Treilleux I. et al. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 2001;61:5024-7
69. Kato T, Imatani T, Minaguchi K, Saitoh E, Okuda K. Salivary cystatins induce interleukin-6 expression via cell surface molecules in human gingival fibroblasts. Mol Immunol. 2002;39:423-30
70. Sabatini LM, Warner TF, Saitoh E, Azen EA. Tissue distribution of RNAs for cystatins, histatins, statherin, and proline-rich salivary proteins in humans and macaques. J Dent Res. 1989;68:1138-45
71. Eustace BK, Sakurai T, Stewart JK. et al. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507-14
72. Strongin AY. Mislocalization and unconventional functions of cellular MMPs in cancer. Cancer Metastasis Rev. 2006;25:87-98
Figures
MALDI-TOF mass spectra of HSP90A, tropomyosin, tubulin, PRX II, and actin. The protein spots were excised from 2DE gel, in-gel trypsin digested, ZipTip purified, and were analyzed with MALDI-TOF MS.
Author contact
![]() Corresponding author: Professor Qing-Xiang Amy Sang, Department of Chemistry and Biochemistry, Florida State University, Chemical Sciences Laboratory building, Rm. 3007, Tallahassee, FL 32306-4390. Tel.: 850-644-8683; Fax: 850-644-8281; E-mail: sangfsu.edu.
Corresponding author: Professor Qing-Xiang Amy Sang, Department of Chemistry and Biochemistry, Florida State University, Chemical Sciences Laboratory building, Rm. 3007, Tallahassee, FL 32306-4390. Tel.: 850-644-8683; Fax: 850-644-8281; E-mail: sangfsu.edu.

 Global reach, higher impact
Global reach, higher impact