Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1255-1259. doi:10.7150/jca.12969 This issue Cite
Research Paper
CT120A Acts as an Oncogene in Head and Neck Squamous Cell Carcinoma
1. Cerrahpasa Medical Faculty, Department of Medical Biology, Istanbul University, Istanbul, Turkey
2. Cerrahpasa Medical Faculty, Department of Otorhinolaryngology, Istanbul University, Istanbul, Turkey
Received 2015-6-17; Accepted 2015-8-29; Published 2015-10-20
Abstract
Squamous cell carcinoma of the head and neck (HNSCC) is among the most frequent cancers worldwide. The etiology and pathogenesis of HNSCC are influenced by multiple genetic factors in addition to environmental and lifestyle-related factors. However, the mechanism underlying the HNSCC is still far from clear.
The membrane associated gene CT120 was previously identified from chromosome 17p13.3 as a lung cancer-associated gene. Its function as an activator of the Erk and Akt signaling pathways in human lung cancer cell lines suggested that CT120 has an oncogenic function. However, there is no data in the literature on the role of CT120 in any other cancer type. Therefore, the aim of this study was to determine the expression rate and probable function of CT120 in HNSCC.
Tumor tissues from 50 patients were analyzed by real-time quantitative RT-PCR to investigate the expression rate and by direct sequencing to differentiate the CT120A and CT120B variants. CT120 overexpression was observed in 58% of tumors compared to non-cancerous tissue samples and this up-regulation was directly associated with the upregulation of the CT120A variant and with the stage of the disease (p=0.001).
Our results indicate that the CT120 gene may function in the development of HNSCC.
Keywords: HNSCC, CT120, Variant, Expression
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common form of cancer and represents the third common cause of cancer-related deaths worldwide [1, 2]. It comprises upper aerodigestive tract anatomic sites and is strongly associated with certain risk factors such as tobacco use, alcohol consumption, and viral infections [3-5]. HNSCC develops as a result of multistep processes characterized by the accumulation of genetic and epigenetic alterations causing disruption of molecular mechanisms such as cell cycle control, DNA repair and carcinogen metabolism [6].
In 2002 He et al. have isolated a novel human plasma membrane-associated gene CT120/FAM57A (GenBank accession No: AF 477201) from chromosome 17 by positional cloning and rapid amplification of cDNA ends (RACE) [7]. The CT120A gene spans over 10.288 bp on chromosome 17p13.3 and is composed of 5 exons. The main product is a protein with 257 amino acids. Functional predictions based on its amino acid sequence using the SMART and TMHMM software revealed that the CT120A protein has 7 transmembrane domains. Using the yeast-two hybrid system the SLC3A2 and GGTL3B proteins have been identified as the partners of CT120A [7]. SLC3A2 and GGTL3B function in the amino acid transport and glutathione (GSH) metabolism [8, 9]. In malignant cells increased GSH levels have been associated with a proliferative response and cell cycle progression [10]. Furthermore, in several cancers, the GSH status has been associated with growth, cellular proliferation, and metastatic activity [11]. Homology comparison studies have also revealed that the CT120A protein has been conserved during evolution [7]. These results and its universal expression in all human tissues except lung indicate that CT120A may have essential physiological functions. On the other hand, it has also been shown that CT120A is expressed in various tumor cell lines and its expression is significantly higher in lung cancer than in control tissue or in normal lung tissue [7]. He et al. [12] also reported that ectopic CT120 gene expression in the NIH3T3 cells activates two major signaling pathways which are involved in cell proliferation, survival and apoptosis. Silencing of CT120A results in reduced cell growth rates and tumorigenicity in nude mice and in tumor cell lines [13, 14]. All of these studies have been performed with lung cancer cell lines and depending on these results it has been suggested that CT120A is a novel lung cancer-related gene. An alternative splice variant (CT120B) of CT120A with an opposite (antogonistic) function has also been reported [15]. These results indicate that CT120A may be a key modulator in lung carcinogenesis and might contribute to cancer progression by activation of oncogenic pathways in human lung cancer cells [14].
However, there is no data in the literature on the role of CT120A and B variants in any type of cancer except lung cancer. Therefore, the aim of this study was to investigate the expression of the CT120A and B variants in HNSCC.
Materıals and Methods
A total of 50 tumors and the corresponding normal tissue samples were obtained from patients who underwent surgery at the Cerrahpasa Medical Faculty, Department of Otorhinolaryngology. Clinicopathological characteristics, including patient's age, gender, TNM staging, histological grade and tumor location are shown in Table 1. The Cerrahpasa Medical Faculty Ethics Committee approved the study and all patients provided their written informed consent.
RNA isolation and complementary DNA (cDNA) synthesis
Total RNA was extracted with the PureLinkTM RNA Mini Kit (Ambion, USA) and 400 ng of total RNA was reverse-transcribed using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific, Lithuania) in 20 μL reaction volume with random hexamer primers. Real-Time PCR (RT-PCR) was performed in duplicate using the Light Cycler 480-II (Roche Diagnostics, Mannheim, Germany) system and UPL Hydrolysis probes. The G6PD (glucose-6-phosphate dehydrogenase) transcript was used as the internal control for all samples and the target gene-specific mRNA level was normalized to the level of G6PD. The UPL Hydrolysis probes were labeled with Dark Quencher at the 3'-end, and with FAM or VIC/Hex/Yellow at the 5'-end for CT120A (Probe #64) and G6PD (Universal ProbeLibrary Human G6PD Gene Assay), respectively. The PCR program included a faststart step of 10 min at 95°C followed by 45 cycles of amplification. Each cycle consisted of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec and elongation at 70°C for 1 sec with a final step of 40°C for 30 sec. The target and reference genes were co-amplified in the same reaction. The expression rate of the CT120A mRNA in each sample was determined using the 2-ΔΔCt method [16].
Clinicopathological characteristics and CT120 expression levels in HNSCC patients
| Patients n (%) | FAM57A Expression n (%) | p valuea | ||||
|---|---|---|---|---|---|---|
| Decreased | No Change | Increased | ||||
| Location of the tumor | Larynx | 32 (64%) | 11 (22%) | 2 (4%) | 19 (38%) | 0.976 |
| Pharynx-Oral Cavitiy-Parotis | 16 (32%) | 5 (10%) | 1 (2%) | 10 (20%) | ||
| Unknown | 2 (4%) | 1 (2%) | 1 (2%) | - | NA | |
| Stage | Early (1-2) | 8 (16%) | 1 (2%) | 2 (4%) | 5 (10%) | 0.055 |
| Late (3-4) | 35 (70%) | 13 (26%) | 1 (2%) | 21 (42%) | ||
| Unknown | 7 (14%) | 3 (6%) | 1 (2%) | 3 (6%) | NA | |
| Tumor Histology | SCC | 46 (92%) | 16 (32%) | 3 (6%) | 27 (54%) | 0.169 |
| Other | 3 (6%) | - | 1 (2%) | 2 (4%) | ||
| Unknown | 1 (2%) | 1 (2%) | - | - | NA | |
| Local Invasion | Absent | 2 (4%) | - | - | 2 (4%) | 0.498 |
| Present | 43 (86%) | 15 (30%) | 3 (6%) | 25 (50%) | ||
| Unknown | 5 (10%) | 2 (4%) | 1 (2%) | 2 (4%) | NA | |
| Age | >50 | 46 (92%) | 16 (32%) | 4 (8%) | 26 (52%) | 0.716 |
| ≤50 | 4 (8%) | 1 (2%) | - | 3 (6%) | ||
| Sex | Male | 46 (92%) | 16 (30.6%) | 3 (5.75%) | 27 (51.65%) | 0.423 |
| Female | 4 (8%) | 1 (2%) | 1 (2%) | 2 (4%) | ||
| Smoke | Smoking | 36 (72%) | 11(22%) | 2 (4%) | 23 (46%) | 0.511 |
| Non-Smoking | 7 (14%) | 3 (6%) | 1 (2%) | 3 (6%) | ||
| Unknown | 7 (14%) | 3 (6%) | 1 (2%) | 3 (6%) | NA | |
a Statistical analyses were performed using the Pearson Chi-Square test (NA: Not Available)
Analysis of the Wild Type and Splice Variants
Forward and reverse primers were designed to identify the CT120A and CT120B transcripts. The same forward primer was used to amplify both variants whereas the reverse primers were specific for exon 4 and exon 3-4 junction for CT120A and CT120B, respectively (Table 2). The reaction conditions were 2 µl of cDNA template, 1xPCR buffer, 200 µM dNTP mix, 6 mM (CT120A) or 3 mM (CT120B) MgCl2, 10 pmol of each primer, and 2U of Taq DNA Polymerase Recombinant (Invitrogen, Brasil) in 25 µl final volume. Amplification was carried out at 95°C for 10 minutes and 35 cycles of 94°C for 30 seconds, 63°C (CT120A) or 61°C (CT120B) for 30 seconds, and 72°C for 30 seconds followed by a final extension for 10 minutes at 72°C. The size of the PCR products were 233 bp and 165 bp for CT120A and CT120B, respectively. All products were analyzed by electrophoresis on 2% agarose gels using the BioID analysis software (Vilber Lourmat, France).
Sequences of the primers for variant analysis and RT-PCR
| Gene | Primer | Sequence |
|---|---|---|
| CT120A | Forward | 5'- CATGATCTATGACTCGTACGCCAT -3' |
| Reverse | 5'- ACACAAACGGAGTGCTCAGTT -3' | |
| CT120B | Forward | 5'- CATGATCTATGACTCGTACGCCAT -3' |
| Reverse | 5'- TTTAGCTGTGCGACTGGCACAA -3' | |
| CT120 RT-PCR | Forward | 5'-CACCGACTGCGTGATGAT -3' |
| Reverse | 5'-GAGCGAATGATGACGATCC -3' |
cDNA Sequence Analysis
To confirm whether the amplified product is wild-type or the splice variant we purified the PCR products which have been obtained by amplification of the cDNA using primers specific for CT120A and CT120B with the High Pure PCR Product Purification Kit (Roche, Germany). Sequencing reactions were performed using the ABI Prism BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA) and the Applied Biosystem ABI Prism 3100-AvantTM Genetic Analyzer.
Results
Expression of CT120 in HNSCC tumor cells was compared to the expression in the corresponding normal tissue samples. We detected the CT120 transcript in 44 (88%) of the tumors and corresponding normal tissues. In five of the remaining 6 samples the transcript was only detected in the tumor tissue, and in one sample CT120 was not present either in the tumor or normal tissue. Up-regulation of CT120 expression was observed in 29 of the 50 HNSCC tumor samples when compared to non-cancerous tissue. Reduced expression was observed in 17 tumor samples and there was no change in 4 samples. A borderline significant correlation was present between the up-regulation of expression and the stage of the disease (p=0.055). No association was found with any other clinicopathological characteristic such as the age, gender, smoking status, histology and invasion (Table 1).
Previously Pan et al. [14] have reported that a splice variant of CT120, the CT120B acts as a tumor suppressor exerting opposite effects to its oncogenic full variant. It was not possible to differentiate the transcripts with the primers and probes used in the RT-PCR reaction. Therefore, we analyzed the amplification products using specific primers designed for exon 4 and exon 4-5 junction. As a result of this analysis we could not detect the full variant (CT120A) in 21 of the normal tissue samples (Figure 1). On the other hand, we detected CT120A in 46 of 50 tumor tissues and the total CT120 mRNA level was increased in 26 of these tumor tissues compared to non-cancerous tissue. In two samples the wild-type variant was not present either in the normal or in tumor tissue. In 13 of the remaining 29 samples CT120 mRNA level was higher in the tumors compared to normal tissue samples. These results show a significant association between the increase of CT120 mRNA expression and increase the CT120A mRNA level in tumor tissues (Table 3; p= 0.001).
In 27 of the 49 (55.1%) tissues the CT120B level was increased in tumor tissue compared to normal tissues. However, there was no association between the increase of the CT120 and CT120B variants (Table 3; p=0.351). The CT120B splice variant was not detected in 2 of the normal tissue samples and 2 tumors. In one of these samples the splice variant was not present in both normal and tumor tissue.
CT120A and CT120B variants expression levels in tumor tissue
| CT120A level | CT120B level | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Increased | Decreased | No change | p valuea | Increased | Decreased | No change | p valuea | ||
| CT120 mRNA | Increased | 23 | 3 | 1 | 0.001 | 17 | 6 | 6 | 0.351 |
| Decreased | 6 | 3 | 8 | 7 | 7 | 2 | |||
| No change | 1 | 2 | 1 | 3 | 0 | 1 | |||
a Statistical analyses were performed using the Pearson Chi-Square test
The PCR products of the tumor and normal tissue samples indicate the presence of CT120A and CT120B variants, respectively. (T: Tumor; N: Normal)
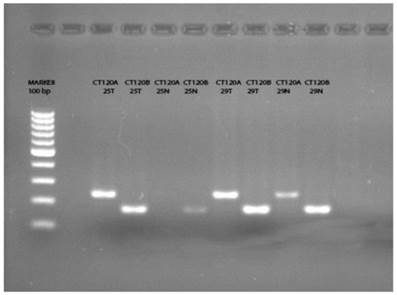
Discussion
Chromosome 17p13.3 is one of the most frequently mutated chromosomal regions in many types of human cancers. This finding has led to the assumption that the chromosome 17p13.3 locus may harbor tumor-associated genes and many scientists aimed to identify these genes. As a result of these studies, in 2002 a novel human plasma membrane-associated gene (CT120) has been isolated using electronic cloning and RACE from this region [7]. The authors also reported that the CT120 mRNA was not expressed in normal human lung tissue whereas it was highly expressed in the SPC-A-1 human lung adenocarcinoma cancer cell line. It has also been shown that knocking down CT120 in SPC-A-1 cells results in reduced in vitro cell growth rate and in a decrease of tumorigenicity in nude mice [7, 14]. In addition to these findings the study revealed that proliferation of the NIH3T3 and A549 cells is promoted by CT120 expression [7, 12].
In 2004 by using Atlas mouse cDNA expression arrays He et al. [12] identified the downstream targets of CT120 in NIH3T3 cells and reported that two major signaling pathways, Raf/Mek/Erk and PI3K/Akt are activated. Both of these pathways are involved in carcinogenesis by affecting cell proliferation, survival and apoptosis. It has been known that activation of Erk in lung cancer cells promotes cell survival and resistance to chemotherapy. He et al also showed that CT120 may also contribute to the overexpression of the metastasis-associated genes such as cathepsins B, D and L and MMP-2/TIMP2 [12]. Silencing of CT120 by antisense oligonucleotides resulted in reduced expression of cyclin D1 and cdk4 and increased expression of p53 and caspase-3 which cause G1 phase arrest and apoptosis [15]. These results indicated that CT120 may be a key regulator in lung carcinogenesis contributing to the activation of oncogenic pathways in human lung cancer. High CT120 expression is also observed in different human tumor cell lines such as liver, ovarian, breast and hepatocellular carcinoma [7]. However, the expression level of CT120 in fresh tumor tissue has been investigated in only one study [15]. In this study, CT120 protein expression was investigated by Western blotting in 8 pairs of matched normal and human lung cancer tissue samples and overexpression of CT120 was detected in 6 of 8 (75%) samples. There is no study in the literature investigating CT120 expression in any other cancer. As shown in lung cancer, the CT120 was upregulated in 58% of the tumor samples in our study. This frequency was lower than reported for lung cancer. The difference may be due to the sample size or the methods used to detect expression. The sample size in the lung study was very small compared to our sample size. On the other hand, in their study Pan et al. [13] have investigated the CT120A protein levels in 16 paired lung cancer tissue samples but not the mRNA levels. Although the protein levels usually reflect the mRNA level, in some cells mRNA is not degraded as soon as it is translated. Post-transcriptional modifications may extend the half-life of the mRNA molecules. Therefore, the difference may also depend on the post-transcriptional modifications of the CT120 mRNA molecule.
Recently, a second variant of the CT120 has been reported by Pan et al. [14] with an antagonistic function. In our study we evaluated the ratio of the two variants and have shown that the A variant represents the significant proportion of CT120 overexpression in the tumor tissues. In accordance with the results reported by Pan et al., our data indicate that the full variant of CT120A has an oncogenic function in cancer and is overexpressed in HNSCC tumor tissues in contrast to the CT120B splice variant.
In conclusion, our findings indicate that CT120 wild-type variant acts as an oncogene in HNSCC and its overexpression is associated with high tumor grade. However, further studies analyzing the function of CT120 in HNSCC are needed to reveal its detailed mechanism of action.
Acknowledgements
The present work was supported by the Research Fund of Istanbul University. (Project No: 50324)
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-49
2. Stell PM. Survival times in end-stage head and neck cancer. Eur J Surg Oncol. 1989;15:407-10
3. Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110:1429-35
4. Hashibe M, Brennan P, Benhamou S. et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777-89
5. Pelucchi C, Gallus S, Garavello W, Bosetti C, La Vecchia C. Alcohol and tobacco use, and cancer risk for upper aerodigestive tract and liver. Eur J Cancer Prev. 2008;17:340-44
6. Wilken B, Veena MS, Wang MB, Srivatsan ES. Curcumin: A review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma. Mol Cancer. 2011;10:12
7. He X, Di Y, Li J. et al. Molecular cloning and characterization of CT120, a novel membrane-associated gene involved in amino acid transport and glutathione metabolism. Biochem Biophys Res Commun. 2002;297:528-36
8. Sies H. Glutathione and its role in cellular functions. Free Radic Biol Med. 1999;27:916-21
9. Mastroberardino L, Spindler B, Pfeiffer R. et al. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature. 1998;395:288-91
10. Traverso N, Ricciarelli R, Nitti M. et al. Role of glutathione in cancer progression and chemoresistance. Oxid Med Cell Longev. 2013;2013:972913
11. Lushchak VI. Glutathione homeostasis and functions: potential targets for medical interventions. Journal of Amino Acids. 2012;2012:736837
12. He XH, Li JJ, Xie YH. et al. Altered gene expression profiles of NIH3T3 cells regulated by human lung cancer associated gene CT120. Cell Research. 2004;14:487-96
13. Pan DN, Wei L, Yao M, Wan D, Gu J. Down-regulation of CT120A by RNA interference suppresses lung cancer cells growth and sensitizes to ultraviolet-induced apoptosis. Cancer Lett. 2006;235:26-233
14. Pan DN, Li JJ, Wei L, Yao M, Wan DF, Gu JR. Inhibitory effect of CT120B, an alternative splice variant of CT120A, on lung cancer cell growth. Acta Biochim Biophys Sin (Shanghai). 2005;37:588-92
15. Li Z, Shao S, Xie S, Jiao F, Ma Y, Shi S. Silencing of CT120 by antisense oligonucleotides could inhibit the lung cancer cells growth. Ir J Med Sci. 2010;179:217-23
16. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-18
Author contact
![]() Corresponding author: Prof. Dr. Nur BUYRU. Address: Istanbul University, Cerrahpasa Medical Faculty, Department of Medical Biology, Kocamustafapasa, 34098, Istanbul, Turkey. Phone: +90-212-414 30 00 ext: 22656; Cellular Phone: +90-505-524 94 71; e-mail: nbuyrucom
Corresponding author: Prof. Dr. Nur BUYRU. Address: Istanbul University, Cerrahpasa Medical Faculty, Department of Medical Biology, Kocamustafapasa, 34098, Istanbul, Turkey. Phone: +90-212-414 30 00 ext: 22656; Cellular Phone: +90-505-524 94 71; e-mail: nbuyrucom

 Global reach, higher impact
Global reach, higher impact