Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(4):523-530. doi:10.7150/jca.17510 This issue Cite
Research Paper
Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma
1. Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China;
2. Department of Otorhinolaryngology Head and Neck Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan, China;
3. The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health, Cancer Research Institute, Central South University, Changsha, Hunan, China;
4. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Xiangya Hospital, Central South University, Changsha, Hunan, China;
5. Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China.
6. Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Received 2016-9-8; Accepted 2016-11-10; Published 2017-2-11
Abstract
Altered expression of long non-coding RNAs (lncRNAs) associated with human carcinogenesis and might be used as diagnosis and prognosis biomarkers. However, the expression of lncRNAs in tongue squamous cell carcinoma (TSCC) and their relevance on the diagnosis, progression and prognosis of TSCC have not been thoroughly elucidated. To discover novel TSCC-related lncRNAs, we analyzed the lncRNA expression patterns in two sets of previously published TSCC gene expression profile data (GSE30784 and GSE9844), and found that long intergenic non-coding RNA 152 (LINC00152) was significantly upregulated in TSCC samples. We then detected LINC00152 expression in two other cohorts of TSCC samples. Quantitative Real time PCR (qRT-PCR) results indicated that LINC00152 was highly expressed in 15 primary TSCC biopsies when compared with 14 adjacent non-tumor tongue squamous cell epithelium samples. The expression of LINC00152 was also measured in 182 paraffin-embedded human TSCC tissues by in situ hybridization, increased expression of LINC00152 was significantly correlated with TSCC progression, such as T stage (p = 0.009), N stage (p = 0.036), TNM stage (p = 0.017), and associated with relapse (p < 0.001), and invasion (p < 0.001). Kaplan-Meier analysis demonstrated that increased LINC00152 expression contributed to both poor overall survival (p = 0.006) and disease-free survival (p = 0.007) of TSCC patients. These findings suggest that LINC00152 might serve as a potential biomarker for early detection and prognosis prediction of TSCC.
Keywords: Long non-coding RNA (LncRNA), long intergenic non-coding RNA 152 (LINC00152), tongue squamous cell carcinoma (TSCC), metastasis, prognosis.
Introduction
Oral cancer is one of the most common head and neck malignant tumors all around the world. Among them, tongue squamous cell carcinoma (TSCC) accounts for approximately 25% to 40% of oral cancers [1]. Surgical resection combined with chemotherapy and radiotherapy has been used as the primary clinic treatment for TSCC. In spite of recent progresses of operation, chemotherapy and radiotherapy, TSCC has a high risk of developing secondary or recurrent tumors in the surrounding area. So the overall mortality rate remains high, and the 5-year survival rate of TSCC patients is less than 50% [2]. Local trauma, tobacco and areca hobbies play important roles in the development of TSCC [3, 4]. The same as other human tumors, the occurrence and development of TSCC is a multi-step process that involves oncogene activation and tumor suppressor silencing [5-15]. Increasing evidences suggest a close association between genetic and epigenetic alterations and TSCC [16]. Discovering suitable biomarkers is a key to monitor cancer recurrence or screening high-risk TSCC populations, which will be helpful to guide adjuvant or neoadjuvant therapy of TSCC.
Long non-coding RNAs (lncRNAs) are newly-identified class of RNAs that are more than 200 nucleotides in length and have been considered to be the “noise” of gene transcription because of their absence of protein-encoding ability [17]. However, accumulating evidence indicates that they participate in many physiological processes, such as carcinogenesis [18-29], by modulating gene expression at the epigenetic, transcriptional and posttranscriptional levels. LncRNAs appear to participate in all stages of cancer development, including tumor initiation, progression and metastasis, and the dysregulation of lncRNAs are a primary feature of some human cancers [21]. Profiling LncRNA expression is a powerful strategy to screen dysregulated lncRNAs and to identify novel prognostic factors in cancer [18]. To date, more than 50,000 lncRNAs have been reported in the human genome; however, the function of most of these lncRNAs remains unknown.
In this study, we performed gene expression profiling analysis by mining two online TSCC data sets (GSE9844 [30] and GSE30784 [31]), based on Affymetrix gene expression microarray platform. One lncRNA, long intergenic non-coding RNA 152 (LINC00152, NCBI accession number: NR_024204, Affymetrix probe set: 225799_at) was significantly overexpressed in TSCC. Then, we examined its expression in fresh TSCC biopsies and paraffin-embedded tissues by Quantitative Real time PCR (qRT-PCR) and in situ hybridization, respectively. LINC00152 was highly expressed TSCC, and increased expression of LINC00152 was significantly correlated with TSCC progression, relapse and poor prognosis. These findings provided a novel insight concerning the role of LINC00152 in the progression of TSCC.
Material and methods
Tissue samples
Two sets of TSCC samples were collected for this study: Set 1 for qRT-PCR, containing 15 TSCC and 14 non-tumor lingual mucous membrane biopsies; Set 2 for in situ hybridization, including 182 paraffin-embedded TSCC and 46 non-tumor lingual mucous membrane tissue samples to confirm the expression of LINC00152. In brief, after surgical removal of the tongue cancer tissues, the tissues were washed three times in glucose-containing Hank's Balanced Salt Solution (HBSS). The fat and fiber tissue were trimmed out [32]. All specimens were confirmed by histopathological examination. All of patients were complete remission after surgical resection combined with chemotherapy. All tissue samples were acquired with approval of patients and the protocol was reviewed and approved by the Ethics Committee at the Affiliated Cancer Hospital of Central South University.
Data mining and analysis
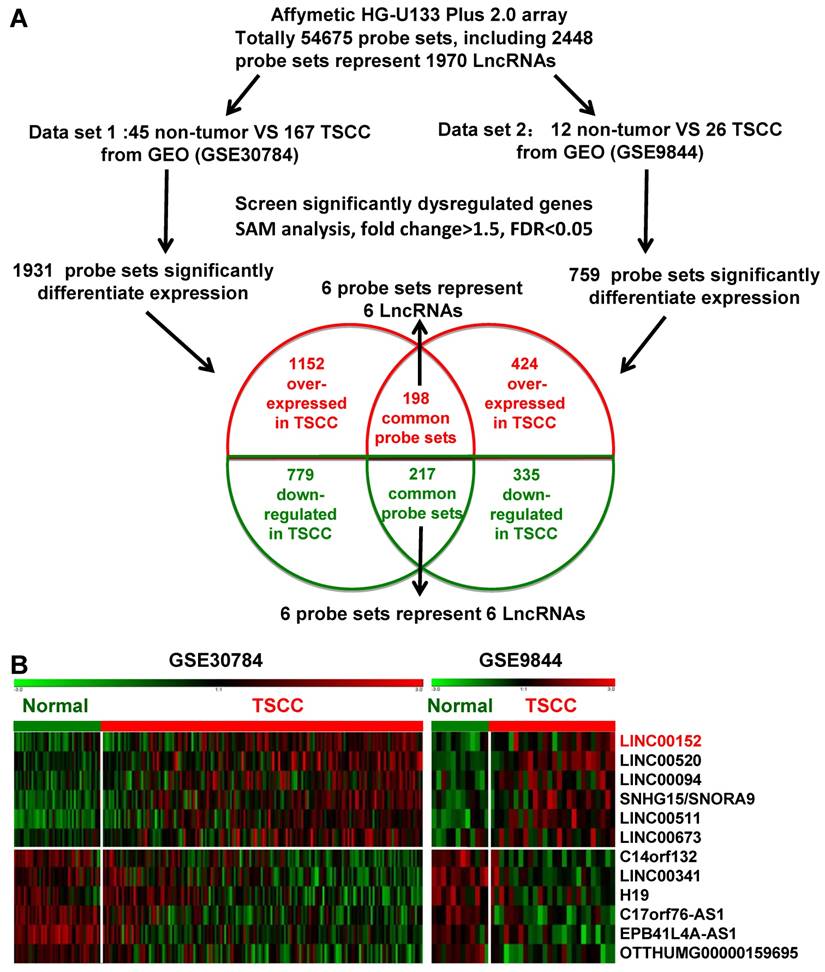
Two independent cohorts of primary TSCC gene expression profiling (GEP) data based on the Affymetrix Human Genome U133 Plus 2.0 platform and their correlated clinic data, GSE9844 [30] and GSE30784 [31] were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). GSE30784 has 167 cancer tissues and 45 normal adjacent tissues. GSE9844 contain 26 cancer tissues and 12 normal adjacent tissues. Significant Analysis of Microarray (SAM) software [33-35] were used to analyze normal lingual mucous membrane and TSCC tissue samples for differences in the expression of lncRNAs in these two published TSCC dataset. The cut-off value for differentially expressed lncRNA was set at ≥ 1.5-fold or ≤ 0.67-fold change and the false discovery ratio (FDR) was < 0.05. The data analysis procedures are shown in Figure 1A.
RNA isolation and qRT-PCR
Total RNA was extracted from frozen tissues using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA according to the manufacturer's instructions [36-42]. One μg of total RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (Biorad, Hercules, CA, USA). qRT-PCR was performed with SYBR Green (Biorad) in the CFX96 Real-Time PCR Detection System (Bio-Rad) to determine the relative expression levels of target genes. The primers used were LINC00152: 5'-TTGATGGCTTGAACATTTGG-3' and 5'-TCG TGATTTTCGGTGTCTGT-3'; ACTB (β-actin): 5'-TCACCAACTGGGACGACATG -3' and 5'-GTCACCGGAGTCCATCACGAT-3'. ACTB was used as the reference and normalization control [43, 44]. The average of three independent analyses for each gene was calculated. Relative expression was calculated using the equation: ΔCt = Ct (LINC00152) - Ct (ACTB), fold expression = 2-(ΔCt(tumor) - ΔCt(normal)).
Dysregulated lncRNA expression analysis using two independent TSCC cohorts and cDNA microarray analysis. A. Schematic overview of the workflow used to identify dysregulated lncRNAs in two TSCC microarray data cohorts (GSE9844 and GSE30784). B. Heatmap of 12 dysregulated lncRNAs mined from the GEO data sets.
In situ hybridization
In situ hybridization was performed to detect LINC00152 expression as previously described [45-48]. Three probes from different LINC00152 regions (5'-CTATGTCTTAATCCCT TGTCCTTCATTAAAAGC-3', 5'-CTTCATTGAACA GTTTGTATATTGGAAACTTGCC-3', and 5'-GCTGCTTTTAAGTTTCAAATTGACATTCCAGAC-3') were synthesized and labeled with DIG-dUTP at the 3' end (Invitrogen, Shanghai, China). Three GAPDH probes used as positive controls were 5'-CCACTTTACCAGAGTTAAAAGCAGCCCTGG-3', 5'-CAGTAGAGGCAGGG ATGATGTTCTGGAGAG-3', and 5'-GTCAGAGGAGACCACCTGGTGCTCAGT GTA-3'. A semi-quantitative scoring criterion for in situ hybridization was used in which both the staining intensity and the number of positive cells were recorded. The scoring was graded as 0 (negative), 1 (< 10% positive), 2 (10% - 50% positive), or 3 (> 50% positive) in accordance with the staining proportion and intensity. The final scores were regarded as low expression (0-1) and high expression (2-3). The scores corresponding to the overall distribution of LINC00152 were averaged across the different tumor plugs in each case. All sections were independently scored by two pathologists who were blinded to the clinicopathological features and the clinical data.
Statistical analysis
Statistical analysis was performed using SPSS software, version 19.0 (SPSS, Chicago, IL, USA). Student's t-tests were used to evaluate significant differences between any two groups of data and one way ANOVA were used to evaluate significant differences for multiple comparisons. Overall survival (OS) or relapse-free survival (RFS) were calculated using the Kaplan-Meier method, and the results of the analysis were considered significant in a log-rank test if p < 0.05. All data are represented as means ± standard deviation. A two-tailed p value of 0.05 or less was considered statistically significant. The graphs were plotted using Graph-Pad Prism 6.0 (Graph-Pad Software, La Jolla, CA).
Results
LncRNA expression profiles in TSCC tissues
Firstly, we analyzed the profiles of TSCC patient from Gene Expression Omnibus (GEO) [33], datasets GSE9844 and GSE30784, and found that there were 1931 differentially expressed genes of the GSE30784 dataset and 759 of the GSE9844 dataset in TSCC patients compared with normal lingual mucous membrane (Figure 1A). Through aggregation of differentially expressed lncRNAs signatures from these two GEO datasets, 12 overlapping probe sets, representing 12 lncRNAs genes were identified, which included 6 upregulated and 6 downregulated lncRNAs (Figure 1B). Most of them were unknown and not well identified, such as SNHG5, LINC00520, LINC00094, LINC00511, EPB41L4A-AS1, and LINC00341. H19, a well-known lncRNA that has been identified in multiple cancers, was also down-regulated in TSCC samples among these overlapping probe sets.
Expression of LINC00152 in TSCC and non-cancerous lingual mucous membrane
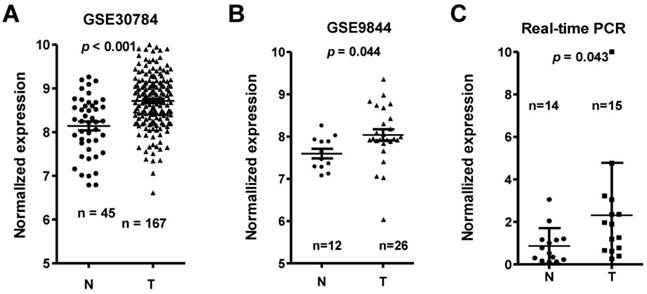
Among the differentially expressed lncRNAs, LINC00152 expression was one of the most significantly overexpressed in the TSCC tissues compared to adjacent non-tumor tissues according to the GS30784 and GSE9844 datasets (Figure 2A & 2B; p < 0.001 and p = 0.044, respectively). To confirm the role of LINC00152 in TSCC progression, we detected the LINC0152 expression levels in 15 TSCC tissues and 14 adjacent non-tumor tissues using by qRT-PCR, and normalizing to β-actin. Results showed that the transcript levels of LINC00152 in TSCC tissues was significantly high compared to that in adjacent normal tissues (p = 0.043, Figure 2C), which was consistent with the GEO datasets.
Correlations between aberrant expression of LINC00152 and TSCC clinical pathological parameters
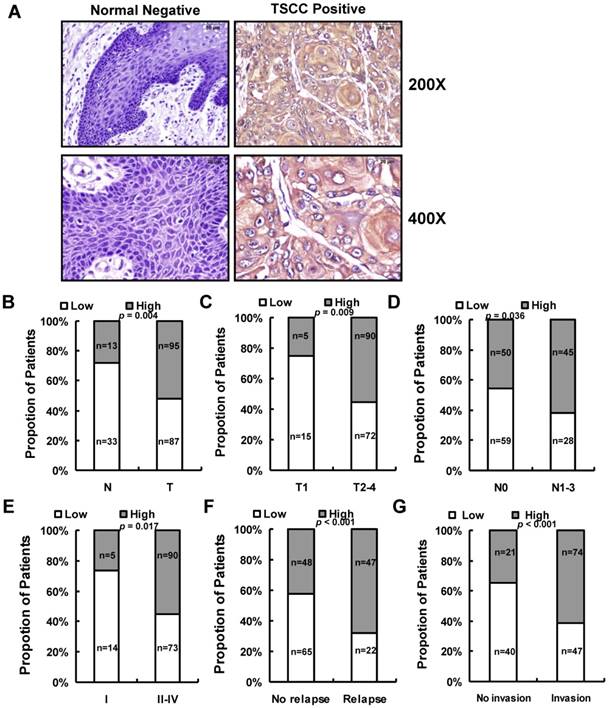
We next assessed LINC00152 expression in 182 paraffin embedded TSCC samples and 46 adjacent non-tumor tissues via in situ hybridization. Representative images of LINC00152 signals are shown in Figure 3A. The data showed that LINC00152 was highly expressed in 52.2% (95 of 182) cancerous tissues compared with 28.3% (13 of 46) adjacent non-tumor tissues (p = 0.004, Figure 3B). Then we examined the correlation of LINC00152 expression level with patients' clinical features in TSCC, such as gender, age, smoking, chewing areca, pathological stage, tumor size (T stage), lymph-vascular invasion (N stage), invasion muscles of tongue, and relapse. As shown in Figure 3, high LINC00152 expression was positively correlated with T stage (p = 0.009, Figure 3C), lymph node metastasis (p = 0.036, Figure 3D), higher TNM stage (p = 0.017, Figure 3E), relapse (p < 0.001, Figure 3F), and invasion muscles of tongue (p < 0.001, Figure 3G). However, other clinical parameters, such as gender, age, smoking and chewing areca, were found not to be significantly correlated with LINC00152 expression in this study.
LINC00152 expression was upregulated in three independent cohorts of TSCC biopsies. T, tumor; N, non-tumor lingual mucous membrane. LINC00152 expression, as measured by Affymetrix microarray, was upregulated TSCC biopsies when compared with non-tumor lingual mucous membrane tissues in GSE30784 (A) and GSE9844 (B). C. LINC00152 mRNA expression was measured in 15 TSCC tissues and 14 adjacent non-tumor tissues using quantitative PCR (qRT-PCR).
The relationship between LINC00152 and pathological clinical feature. A. LINC00152 expression was measured by in situ hybridization in paraffin embedded non-tumor lingual mucous membrane (N, n = 46) and TSCC biopsies (T, n = 182). Representative cases of normal lingual mucous membrane (N) and TSCC biopsies were shown. B. LINC00152 expression in tumor compared to adjacent non-tumor lingual mucous membrane. C. Correlation of LINC00152 expression and tumor size in TSCC patients. D. Correlation of LINC00152 expression and lymph node metastasis in TSCC patients. E. Correlation of LINC00152 expression and TNM stage in TSCC patients. F. Correlation of LINC00152 expression and recurrence of TSCC patients. G. Correlation of LINC00152 expression and tongue muscle invasion in TSCC patients.
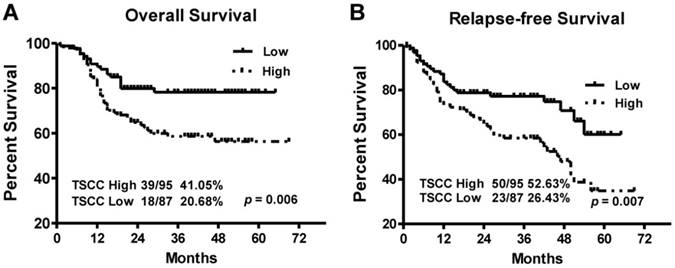
Kaplan-Meier survival curves in TSCC patients according to LINC00152 expression levels. The highly expressed LINC00152 was correlated with shorter overall survival (A) or Relapse free survival (B) of TSCC patients.
LINC00152 upregulation is associated with poor prognosis in TSCC
To explore the relationship between LINC00152 expression and TSCC patients' prognosis, we assessed the correlation between LINC00152 expression and clinical outcomes. The results of Kaplan-Meier analysis showed that the median overall survival (OS) time was 35 and 28 months in TSCC patients with low and high expression of LINC00152, respectively (Figure 4A, p = 0.006). While the median relapse-free survival (RFS) time of TSCC patients with low and high expression of LINC00152 was 29 and 26.5 months, respectively (Figure 4B, p = 0.007).
Discussion
Many studies have reported a close association between lncRNA expression and tumor development and progression. The GEO database is a public gene expression data repository that has collected a large amount of gene expression data and serves as a valuable data source for biomedical research. Mining of published high throughput data is a commonly used and low-cost method to identify novel biomarkers [49]. The Affymetrix HG-U133 plus 2.0 array is one of the most common commercial microarrays used in human cancer profiling and contains 2448 probe sets for lncRNA genes [18]. In the current study, we combined two previously published GEO dataset to identify differentially expressed lncRNAs in TSCC. We found 12 differentially expressed lncRNAs in TSCC tissues when compared with the noncancerous tissue samples. The majority of these lncRNA transcripts have not been well characterized. These lncRNAs might participate in carcinogenesis and progression of TSCC and worth for further study. And more lncRNA microarray analysis may help to identify more valuable and effective lncRNA-based diagnosis, prognosis and therapeutic biomarkers. Among these 12 lncRNAs, H19 is a long non-coding RNA differentially expressed in many tumors and participates in tumorigenesis [50-53], it was also downregulated in TSCC samples compared with the adjacent non-tumor tissues. Until now, there were few reports about lncRNA H19 in head and neck squamous carcinoma [54, 55], and it might participate in the development of TSCC.
In this paper, we chose and focused on LINC00152, since it had the highest fold change among lncRNAs dysregulated in TSCC but never been reported in TSCC previously. We performed qRT-PCR and in situ hybridization to verify our results using existing data in the GEO database. We found LINC00152 was upregulated in TSCC and its high expression was associated with tumor progression, such as tumor size, invasion of muscles, lymph node metastasis, clinical stages, and recurrence. These results indicated the potential role of LINC00152 abnormalities in TSCC progression and potential value as diagnostic and prognostic factor in patients with TSCC. More and more research shows that tobacco and areca hobbies play an important role in the development of oral cancer [56, 57]. However, our result demonstrated that high LINC00152 expression was lacking correlations with smoking and areca hobbies, this may partly owing to the limited sample size.
Recent studies have reported that LINC00152 was upregulated in gastric and hepatocellular cancer [58]. It promoted tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 and participated in cell cycle arrest, apoptosis, epithelial to mesenchymal transition (EMT), cell migration and invasion in gastric cancer [59]. In addition, another research confirmed that LINC00152 contributed proliferation through the EGFR-dependent pathway in gastric cancer [59] or by targeting EpCAM via the mTOR signaling pathway in hepatocellular carcinoma [58]. Further exploring found that LINC00152 could also act as a circulating biomarker for the diagnosis of hepatocellular cancer [60]. To our knowledge, we are the first to report the prognostic values of LINC00152 in prediction of TSCC metastases. Further functional studies of LINC00152 will enrich our knowledge to understand the underlying mechanisms of TSCC metastases.
In conclusion, we found that LINC00152 was the most significantly upregulated lncRNA in TSCC, elevated LINC00152 expression was associated with poor prognosis, suggesting that LINC00152 was promising prognostic biomarkers of TSCC.
Acknowledgements
This study was supported in part by grants from the National Natural Science Foundation of China (81372907, 81301757, 81472531, 81402009, 81572787, 81672683 and 81672993) and the Natural Science Foundation of Hunan Province (14JJ1010 and 2015JJ1022).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kimple AJ, Welch CM, Zevallos JP. et al. Oral cavity squamous cell carcinoma-an overview. Oral Health Dent Manag. 2014;13:877-82
2. Schwam ZG, Judson BL. Improved prognosis for patients with oral cavity squamous cell carcinoma: Analysis of the National Cancer Database 1998-2006. Oral Oncol. 2016;52:45-51
3. Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol. 2010;46:414-7
4. Sciubba JJ. Oral cancer. The importance of early diagnosis and treatment. Am J Clin Dermatol. 2001;2:239-51
5. Stransky N, Egloff AM, Tward AD. et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-60
6. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9-22
7. Liang F, Li Q, Li X. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046-56
8. Li Q, Chen P, Zeng Z. et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37:12503-12
9. Liao Q, Zeng Z, Guo X. et al. LPLUNC1 suppresses IL-6-induced nasopharyngeal carcinoma cell proliferation via inhibiting the Stat3 activation. Oncogene. 2014;33:2098-109
10. Song Y, Li X, Zeng Z. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783-99
11. Xiong W, Zeng ZY, Xia JH. et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972-4
12. Zeng Z, Zhou Y, Zhang W. et al. Family-based association analysis validates chromosome 3p21 as a putative nasopharyngeal carcinoma susceptibility locus. Genet Med. 2006;8:156-60
13. Zhou Y, Liao Q, Li X. et al. HYOU1, Regulated by LPLUNC1, Is Up-Regulated in Nasopharyngeal Carcinoma and Associated with Poor Prognosis. J Cancer. 2016;7:367-76
14. Tu CF, Qi P, Li XY. et al. Tumor Heterogeneity: The Challenge of Precision Medicine. Prog Biochem Biophys. 2015;42:881-90
15. Yu ZY, Song YL, Gong ZJ. et al. The Mechanism and Tumorigenesis of Oncogenic DNA Virus Integration. Prog Biochem Biophys. 2014;41:324-31
16. Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143-54
17. Gong Z, Zhang S, Zhang W. et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120-4
18. Bo H, Gong Z, Zhang W. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
19. Gong Z, Zhang S, Zeng Z. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674
20. He B, Li W, Wu Y. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353
21. Wang Y, Xue D, Li Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
22. Zeng Z, Bo H, Gong Z. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729-37
23. Zeng Z, Fan S, Zhang X. et al. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2016;18:206-11
24. Zhang W, Huang C, Gong Z. et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545-54
25. Yu J, Liu Y, Gong Z. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017 doi: 10.18632/oncotarget.14200
26. Lian Y, Li XY, Tang YY. et al. Long Non-coding RNAs Function as Competing Endogenous RNAs to Regulate Cancer Progression. Prog Biochem Biophys. 2016;43:219-25
27. Li YW, Wang YM, Zhang XY. et al. Progress of Long Noncoding RNA HOTAIR in Human Cancer. Prog Biochem Biophys. 2015;42:228-35
28. Tang K, Wei F, Bo H. et al. Cloning and Functional Characterization of a Novel Long Non-coding RNA Gene Associated With Hepatocellular Carcinoma. Prog Biochem Biophys. 2014;41:153-62
29. Gong ZJ, Huang HB, Xu K. et al. Advances in microRNAs and TP53 Gene Regulatory Network. Prog Biochem Biophys. 2012;39:1133-44
30. Ye H, Yu T, Temam S. et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69
31. Chen C, Mendez E, Houck J. et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152-62
32. Zhang W, Fan S, Zou G. et al. Lactotransferrin could be a novel independent molecular prognosticator of nasopharyngeal carcinoma. Tumour Biol. 2015;36:675-83
33. Xiong W, Wu X, Starnes S. et al. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112:4235-46
34. Liang F, Xu K, Gong ZJ. et al. ChIP-seq: a New Technique for Genome-wide Profiling of Protein-DNA Interaction. Prog Biochem Biophys. 2013;40:216-27
35. Huang HB, Liang F, Xiong W. et al. Bioinformatics Accelerates Drug Repositioning. Prog Biochem Biophys. 2012;39:35-44
36. Xiao K, Yu Z, Li X. et al. Genome-wide Analysis of Epstein-Barr Virus (EBV) Integration and Strain in C666-1 and Raji Cells. J Cancer. 2016;7:214-24
37. Zeng Z, Huang H, Huang L. et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315-26
38. Duan Z, Zheng H, Xu S. et al. Activation of the Ig Ialpha1 promoter by the transcription factor Ets-1 triggers Ig Ialpha1-Calpha1 germline transcription in epithelial cancer cells. Cell Mol Immunol. 2014;11:197-205
39. Hu D, Duan Z, Li M. et al. Heterogeneity of aberrant immunoglobulin expression in cancer cells. Cell Mol Immunol. 2011;8:479-85
40. Hu D, Zheng H, Liu H. et al. Immunoglobulin expression and its biological significance in cancer cells. Cell Mol Immunol. 2008;5:319-24
41. Li M, Zheng H, Duan Z. et al. Promotion of cell proliferation and inhibition of ADCC by cancerous immunoglobulin expressed in cancer cell lines. Cell Mol Immunol. 2012;9:54-61
42. Zhao R, Liu Y, Wang H. et al. BRD7 plays an anti-inflammatory role during early acute inflammation by inhibiting activation of the NF-small ka, CyrillicB signaling pathway. Cell Mol Immunol. 2016
43. Yang Y, Zhou H, Yang Y. et al. Lipopolysaccharide (LPS) regulates TLR4 signal transduction in nasopharynx epithelial cell line 5-8F via NFkappaB and MAPKs signaling pathways. Mol Immunol. 2007;44:984-92
44. Zheng H, Li M, Ren W. et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol Immunol. 2007;44:2221-7
45. Zhang W, Zeng Z, Fan S. et al. Evaluation of the prognostic value of TGF-beta superfamily type I receptor and TGF-beta type II receptor expression in nasopharyngeal carcinoma using high-throughput tissue microarrays. J Mol Histol. 2012;43:297-306
46. Zeng Z, Huang H, Zhang W. et al. Nasopharyngeal carcinoma: advances in genomics and molecular genetics. Sci China Life Sci. 2011;54:966-75
47. Zeng ZY, Zhou YH, Zhang WL. et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol. 2007;38:120-33
48. Zeng Z, Zhou Y, Xiong W. et al. Analysis of gene expression identifies candidate molecular markers in nasopharyngeal carcinoma using microdissection and cDNA microarray. J Cancer Res Clin Oncol. 2007;133:71-81
49. Yang L, Tang Y, He Y. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97-103
50. Kallen AN, Zhou XB, Xu J. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101-12
51. Han D, Gao X, Wang M. et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159-73
52. Chu M, Yuan W, Wu S. et al. Quantitative assessment of polymorphisms in H19 lncRNA and cancer risk: a meta-analysis of 13,392 cases and 18,893 controls. Oncotarget. 2016 doi: 10.18632/oncotarget.12530
53. Yan L, Zhou J, Gao Y. et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076-84
54. el-Naggar AK, Lai S, Tucker SA. et al. Frequent loss of imprinting at the IGF2 and H19 genes in head and neck squamous carcinoma. Oncogene. 1999;18:7063-9
55. Esteves LI, Javaroni AC, Nishimoto IN. et al. DNA methylation in the CTCF-binding site I and the expression pattern of the H19 gene: does positive expression predict poor prognosis in early stage head and neck carcinomas? Mol Carcinog. 2005;44:102-10
56. Li YC, Chang JT, Chiu C. et al. Areca nut contributes to oral malignancy through facilitating the conversion of cancer stem cells. Mol Carcinog. 2016;55:1012-23
57. Majumder KR, Karmakar R, Alam MM. et al. Carcinoma Tongue-Clinicopathological Presentation. Mymensingh Med J. 2015;24:787-93
58. Ji J, Tang J, Deng L. et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813-24
59. Zhao J, Liu Y, Zhang W. et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112-23
60. Li J, Wang X, Tang J. et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687-96
Author contact
![]() Corresponding author: Wei Xiong or Xinming Yang. Tel.: 86-731-8480-5446; Fax: 86-731-8480-5383; Email: xiongweiedu.cn or x16y2003com.
Corresponding author: Wei Xiong or Xinming Yang. Tel.: 86-731-8480-5446; Fax: 86-731-8480-5383; Email: xiongweiedu.cn or x16y2003com.

 Global reach, higher impact
Global reach, higher impact