Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(4):626-635. doi:10.7150/jca.17579 This issue Cite
Review
The NOR1/OSCP1 proteins in cancer: from epigenetic silencing to functional characterization of a novel tumor suppressor
1. Department of Dermatology, Xiangya Hospital, The Central South University, Changsha, 410008, Hunan, China;
2. The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health, Xiangya Hospital, Central South University, Changsha, Hunan410078, China;
3. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan410078, China;
4. Department of Laboratory Medicine and Pathology, Masonic Cancer Center, University of Minnesota, Minneapolis, Minnesota 55455, USA.
Received 2016-9-14; Accepted 2016-11-27; Published 2017-2-25
Abstract
NOR1 (Oxidored-nitro domain-containing protein 1), also known as OSCP1, was first identified in nasopharyngeal carcinoma (NPC) cells in 2003. NOR1 is evolutionarily conserved among species with its expression is restricted to brain, testis and respiratory epithelial cells. NOR1 was downregulated in NPC and the downregulation associates with poor prognosis. Previous study demonstrated that hypermethylation of NOR1 promoter was observed in NPC and hematological malignancies, which has been believed to be the main epigenetic cause for NOR1 silencing in these cancers. Recently, the NOR1 tumor suppressor status has been fully established. NOR1 inhibited cancer cell growth by disturbing tumor cell energe metabolism. NOR1 also promote tumor cells apoptosis in oxidative stress and hypoxia by inhibition of stress induced autophagy. Moreover, NOR1 suppressed cancer cell epithelial-mesenchymal transition, invasion and metastasis via activation of FOXA1/HDAC2-slug regulatory network. Deciphering the molecular mechanisms underlying NOR1 mediated tumor suppressive role would be helpful to a deeper understanding of carcinogenesis and, furthermore, to the development of new therapeutic approaches. Here we summarize the current knowledge on NOR1 focusing on its expression pattern, epigenetic and genetic association with human cancers and its biological functions. This review will also elucidate the potential application of NOR1/OSCP1 for some human malignancies.
Keywords: NOR1, OSCP1, tumor suppressor, epigenetic
Introduction
Tumor suppressor genes (TSGs) has been defined as of a second class of genes involved in cancer which oppose oncogene function and restrain cancer development. Aberrant activation of oncogenes and inactivation of TSGs is essential for cancer development [1]. It has been proposed that a physiological balance between tumour suppressors and oncogenes in normal cells maintains tissue homeostasis and inhibits malignant transformation [2]. Nasopharyngeal carcinoma is a specific kind of head and neck cancers with rare p53 TSG mutations and retinoblastoma susceptibility gene alterations [3-13]. In order to identify new TSGs involved in NPC development, Nie et al. utilized the cDNA array technique to compare the differential expressed genes between a NPC cell line HNE1 and normal nasopharyngeal epithelial cells and found a novel EST showed dramatically decreased in HNE1. He further isolated the novel gene which this downregulated EST derived. It has been shown that the protein encoded by this downregulated gene exhibits high similarity to the “classical” nitroreductase of Salmonella typhimurium, so it was named as the oxidored nitro domain containing protein 1 (NOR1) [14]. Human NOR1 has also been identified as a solute carrier protein named as human solute carrier protein 1(hOSCP1) which mediates various kinds of organic solutes in a pH-dependent and sodium- independent manner on the basal membrane of the placenta [15]. The previous study demonstrated that NOR1 was downregulated in various cancer types, such as in nasopharynx, lung, gastric, colon, rectum and cervix cancers [16-19], which suggests that its low expression may contribute to the pathogenesis of these cancers. It has been shown that DNA hypermethylation mainly account for NOR1 silencing in NPC and hematological malignancies [20-22]. Moreover, hypermethylation of NOR1 is associated with resistance to imatinib and shortened survival in chronic myelogenous leukemia [23], suggesting NOR1 is a potential epigenetic biomarker for the diagnosis, prognosis and treatment of cancers. Recently, NOR1 was shown to be a tumor suppressor gene and plays an important role in lots of cellular processes such as cell proliferation and survival via modulating cell cycle S phase delay and energe metabolism of tumor cells [18, 19, 24-26]. It has been shown that NOR1 induces ER stress and apoptosis of tumor cells by modulating expression mitochondrial Bax and Bcl-2 family members [18, 19, 24]. Moreover, NOR1 was found to be an antagonist of epithelial-mesenchymal transition (EMT) in tumor cells by activation of FOXA1 and HDAC2 thus in turn inhibit slug transcription [27, 28]. Accordingly, this review will summarize the current literatures on NOR1 and aims to putting forward an assumption of NOR1 as gene therapy strategy for malignant cancers.
Gene structure, evolution, and alternative splicing of NOR1
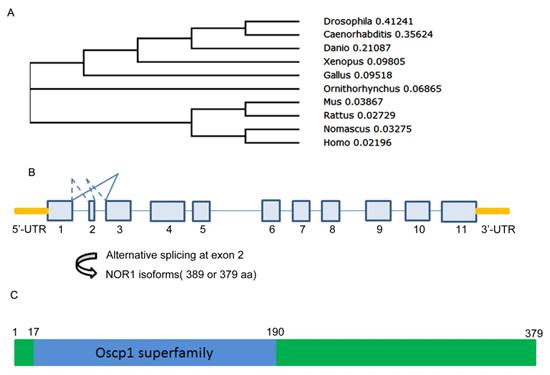
NOR1 is evolutionarily conserved among species. At present, except for human NOR1 gene, mouse, rat and Drosophila orthologue of NOR1 was successively isolated and well characterized[29-31] (Figure 1A) (Table 1). Among these orthologues, mouse NOR1 (mNOR1) and Drosophila NOR1 (dNOR1) share 86% and 58% homology with its human counterpart, respectively [17, 31]. Human NOR1/OSCP1 gene consists of 11 exons which is mapped to chromosome 1p34.2 and spans 33.4 kb of the genomic DNA. Among the 11 exons, exon 2 is only 30-bp length which encoded for 10 amino acid residues and located between two large introns [32]. Alternative splicing at the exon 2 resulted two isoforms of NOR1/OSCP1 gene products, which one is 389 aa residues and the other is 379aa residues (Figure 1B). It has been shown that both isoforms were simultaneously detected in brain. However, the shorter isoform was the sole one expressed in other tissues out of brain [32]. Alternative splicing on exon 2 seem to be evolutionarily conserved among species since mouse NOR1 isoforms resulted from alternative splicing on exon 2 exhibit similar tissue distribution pattern [33].
A Evolution, gene structure and alternative splicing, protein structure of NOR1/OSCP1 protein. A. polygenetic tree of NOR1 orthologues in multiple species. B. Diagram of alternative splicing on human NOR1 exon 2. C. Protein structure of human NOR1 isoform.
NOR1 homologues in multiple species
| Gene locus and Gene ID | Full length protein | Tissue distribution | Subcellular localization | Ref | |
|---|---|---|---|---|---|
| Human sapien NOR1 | 1p34.2(Gene ID: 127700) | 389 aa | selectively in Testis, Brain, nasopharynx, trachea, placenta | cytoplasmic, membrane, mitochondria, ER | [16],[15],[18],[24] |
| Mouse NOR1 | 4 D2.2 (Gene ID: 230751) | 389 aa | selectively in testis, Brain, trachea | cytoplasmic, | [17],[29],[33],[34] |
| Rat NOR1 | 5q36 (Gene ID: 362595) | 379 aa | selectively in testis, Brain | cytoplasmic, | [30],[49] |
| Drosophila NOR1 | 48E2-48E2 (Gene ID: 36300) | 302 aa | ubiquitously in brain lobes, leg discs, wing discs, eye discs, fat bodies, salivary glands | membrane, ER, GA, mitochondria | [31] |
Abbreviation: ER:endoplasmic reticulum,GA:Golgi apparatus.
An evolutionary conserved region called oscp1 superfamily was found in human NOR1 protein from amino acid residue 17 to 190 (Figure 1C). Since the shorter isoform without the 10 aa encoded by exon2 is the sole isoform detected in the most tissues such as testis and nasopharynx, it is likely that exon 2 is dispensable for NOR1/OSCP1 functions in tissues except for brain. However, the exon 2-encoded 10-amino acid insert is restricted to the human and mouse brain [32, 33], thus it may be important for NOR1/OSCP1 function in the central nervous system (CNS). Actually, a potential casein kinase II phosphorylation site was found in the TEVD motif of this 10-amino acid insert [33], which imply that the longer NOR1/OSCP1 isoform in CNS is a candidate target of casein kinase II in brain.
Tissue distribution, subcellular localization of NOR1/OSCP1
Human NOR1/OSCP1 mRNA is highly expressed in testis [15, 32], moderately in nasopharynx, trachea and parotid gland, and weakly or absent in heart, liver, skeletal muscle [32]. By use of homemade anti-serum against human NOR1 protein, Xiang et al. found endogenouse human NOR1 protein was clearly detected in nasopharynx, trachea and brain homogenate [16]. Although NOR1 mRNA was very weak in brain, strong cytoplasmic staining for NOR1 protein was observed in neurons of central nervous system (CNS) but not in glia cells[16]. Normally, NOR1 mRNA and protein was weakly or absent in heart, liver, skeletal muscle, stomach and colon[16, 32], but it is possible that NOR1 expression may be induced by stress during pathological conditions since human NOR1 was shown to be induced by oxidative stress in NPC cells[20].
The tissue distribution pattern of mouse and rat NOR1/OSCP1 homologues is similar to its human counterpart, of which testis have the highest mRNA level, heart, liver, spleen, stomach, muscle and colon have no expression of NOR1 in normal condition [17, 29, 30, 33]. However, Drosophila orthologue of OSCP1 (dOSCP1) exhibits a more ubiquitous tissue distribution pattern including brain lobes, leg discs, wing discs, eye discs, fat bodies and salivary glands[31]. Thus, the tissue expression pattern of NOR1 orthologues seems to be different across multiple species.
Subcellular localization of NOR1 protein is controversial. Although human OSCP1 protein has been reported to localize in the basal membrane of the syncytiotrophoblast in the human placenta [15]. It has been also reported to mainly distribute in cytoplasm of nasopharyngeal epitheliums and neurons [16]. Immune electron microscopic analysis also revealed exogenously expressed human NOR1 protein is localized in mitochondrion and endoplasmic reticulum(ER) in HeLa cells [18]. Mouse NOR1/OSCP1 protein is a 45-kDa cytosolic protein [33]. However, it was also found to distribute in pre- and post-synaptic compartments, but not associated with synaptic membranes or mitochondria [34]. Surprisingly, it has been shown that dOSCP1 distributes not only in ER, Golgi apparatus and mitochondria, but also to the plasma membrane and even translocate into the nucleus [31]. Thus, the subcellular location of NOR1/OSCP1 protein may be variable among species and different cell types (Table 1).
Developmental expression of NOR1/OSCP1 in brain and spermatogenesis
Human NOR1 protein is broadly distributed in the CNS. Intensive immuno-staining of NOR1 was observed in neurons of cerebrum, cerebellum (purkinje cells), corpus striatum, globus pallidus, cingulate gyrus and spinal cord [16]. Mouse NOR1 orthologue is also highly expressed in the CNS. Immunohistochemistry assay revealed that mouse NOR1 protein is broadly distributed in brain, of which the pyramidal cells in the CA1 regions of the hippocampus and dopamine neurons in the midbrain showed the most high expression level [17]. Dynamic analysis revealed that the protein level of NOR1 in mouse brain is temporally changed after birth, which low at the first day after birth and then increase to reach maximal level at 28 days [17, 34]. The dynamic of NOR1 expression was similar to those of synapsin I during postnatal development of the mouse brain, but was inversely correlated with that of MAP2c expression, which imply that NOR1 protein is associated with neuronal maturation after birth [34]. However, little is known about NOR1 function in neuron. Since deletion of NOR1 in mouse fail to produce any visible morphological changes of brain (unpublished data), NOR1 seem to be dispensable for neural morphogenesis.
NOR1/OSCP1 protein seem to be expressed in germ cells but not in Sertoli cells[17]. Mouse NOR1/OSCP1 protein expression during spermatogenesis exhibits a stage- and cell type-specific manner. Its expression is undetectable in the stages I-VI (pachytene spermatocytes) until started from stage IX (leptotene spermatocytes), then followed by an increase in stages II-IV (pachytene spermatocytes) and further reached the maximal level in stages V-X (pachytene spermatocytes), stage XI (diplotene spermatocytes), stage XII (meiotic cells). Its expression retained at submaximal level in steps 1-14 spermatids until finally was disappear in spermatids of step 16 [33]. The developmental expression of OSCP1 protein in testis suggested it might play important role in spermatogenesis. Interestingly, OSCP1 mRNA was found significantly decreased or even disappear in severe disorders of spermatogenesis [35].
NOR1/OSCP1 functions as an organic solute transporter
Membrane transporters on the cell's surface plays a central role in the elimination of a wide range of environmental toxicants and drugs. It has been shown that OSCP1 protein was localized at basal membrane of human placental syncytiotrophoblast and mediates the transport of substrate in a pH-dependent but Na+-independent manner [15]. However, it still remains controversial whether OSCP1 could act as a membrane carrier at all. Firstly, unlike the typical membrane localization of classical transporter family members, OSCP1 protein mostly distributes in cytoplasm [33]. Secondly, OSCP1 protein showed little effect on the transportation for E1S (estrone-3-sulfate) when stably over-expressed in HEK293 cells. Even after expression of human OSCP1 in Xenopus laevis oocytes, the authors failed to get the evidence supporting the transport activity of OSCP1 for steroid sulfates [35].
Genetics, Epigenetics and Expression of NOR1 in human cancers
Abnormal genetic alteration represent a key important mechanism which lead to oncogene activation and tumor suppressor gene inactivation [36-42]. DNA sequence change in the code region of the gene usually lead to amino acid residue replacement or substitution and subsequently result in gain or loss of function of proteins. One of the most frequent types of genetic variations is the single nucleotide polymorphisms (SNPs). A pilot works studied the association between NOR1 genetic variation and risk for NPC. According to the data, two single nucleotide polymorphisms (SNPs), rs1416840 (formerly ss2220003, 656T>C) and rs2275477 (formerly ss3211583, 919G>A), of NOR1 gene is associated with increased risk for NPC. Compared to control, NPC patients displayed a statistically significant increase in genotype CC of NOR1 gene rs1416840 and in genotype AG+AA of NOR1 gene rs2275477. In addition, haplotype analysis revealed haplotype AC consist of these two SNVs rs1416840-rs2275477 might increase the risk of NPC [43].
The expression of hOSCP1 mRNA exhibited quite low level in the human liver [15], However, some liver cancer samples showed intensive immuno-staining for NOR1 protein [17]. This implies genetic variation of NOR1/OSCP1 might be related to the development and/or progression of liver cancer. Toda et al. analyzed 41 single SNPs of NOR1/OSCP1 gene in 18 Japanese patients with non-viral liver carcinoma. As compared with database of dbSNP, they found the genotype frequencies for 2 non-synonymous SNPs [rs34409118 (Thr131→ Ala) and rs1416840 (Ile219→Thr)] and 1 synonymous SNP [rs16822954 (Ser193 → Ser)] to be statistically significant [44]. To date, how these genetic variants of NOR1 gene involved in carcinogenesis is not well studied. One possibility is that genetic variants in NOR1 gene lead to change of amino acids which might affect NOR1 protein structure or biological functions.
Epigenetic modification plays a critical role in gene regulation. In contrast to genetic alterations lead to DNA sequence change, epigenetic alterations affect the chromatin structure and gene transcription without changing the genome sequence [45]. NOR1 epigenetic alteration was firstly found to be associated with acute myeloid leukemia (AML)[21]. By use of bisulfite pyrosequencing, Kroeger et al. found 7 gene of which NOR1 included were hypermethylated in AML. Methylation of NOR1 is correlated with Nucleophosmin/nucleoplasmin 2(NPM2), which is a core histones chaperone involved in chromatin reprogramming [46]. Hypermethylation of NOR1 was also found occurred in chronic myelogenous leukemia (CML). More important, the methylation intensity of NOR1 significantly increased when the disease was progressed from chronic phase (CP) to the accelerated phase (AP) and eventually to the blastic phase (BP). Hypermethylation of NOR1 was associated with imatinib resistance and shorter median survival of CML patients. Thus, aberrant NOR1 methylation is a negative risk factor in the pre-imatinib era patients [23]. Li et al. also found hypermethylation of NOR1 promoter region and concomitant loss of expression occurred in NPC development [20, 47].
Human NOR1 protein is highly expressed in normal nasopharynx epithliums but dramatically decreased in NPC tissues [16, 20]. By use of NPC tissue array, Wang et al. found that loss of NOR1 protein expression was associated with poor overall and event-free survival of NPC patients [27]. Sengupta et al. performed genome-wide expression profiling of laser-microdissected NPC tissues and normal nasopharynx epitheliums [48], from these data, NOR1 mRNA was found to decrease in NPC tissues. NOR1 protein was high in normal testis but it was markedly decreased testicular germ cell tumors (TGCTs), of which seminoma tissues exhibited the lowest level of NOR1 protein. However, NOR1 immunoreactivity was remain relative high level in embryonal carcinoma tissues [49]. Thus, down-regulation of NOR1 protein is strongly associated with histologic types of human testicular cancer. It has been shown that human NOR1 gene was down-regulated in cervical cancer (CCA) and prostate cancer (PC) tissues[18, 19]. Furthermore, Xiang et al. analyzed NOR1 protein expression level by searching of The human protein Atlas database and found NOR1 protein was weak or absent in most malignant cells such as glioma, but increased in heptocellular carcinoma (HCC) tissues [17]. Recently, Li et al. found that HCC samples exhibits elevated NOR1 expression and NOR1 expression is associated with a poor differentiation grade and metastasis [50]. However, our unpublished data suggested overexpression NOR1 showed little effect on HCC HepG2 cells proliferation. The heterogeneity of NOR1 expression in different cancer tissues implies functional diversity of NOR1 in different cells. Thus, the role of NOR1 on HCC development needs to be clarified by more detailed study.
Role of NOR1 in cancer development
Aberrant silencing TSGs play a critical role in cancer development [51-53]. As mentioned above, hypermethylaion of NOR1/OSCP1 promoter region occurs in NPC and hematopoietic malignancy, and down-regulation of NOR1 protein level in NPC, CCA, PC and TGCTs samples, which indicate that NOR1/OSCP1 may be involved in cancer development.
NOR1 suppresses tumor cells proliferation and promotes apoptosis
It has been shown that re-expression of NOR1 significantly inhibited NPC cells and human cervix cancer HeLa cells proliferation and greatly reduced its capacity to form colonies [18, 20, 54, 55]. NOR1 mediated proliferation inhibition in both NPC cells and HeLa cells is associated with cell cycle S phase arrest [18]. Similar to this, an in vivo study also revealed that overexpression of dOSCP1 in Drosophila eye imaginal discs induced a delay in S phase progression and led to a rough eye phenotype in adult flies [26]. Knockdown of NOR1 in HeLa cells enhanced cell viability and attenuated H2O2 induced apoptosis [56]. NOR1 also inhibited cells proliferation and induced apoptosis in prostate cancer cells [19].
More importantly, NOR1 has been observed to promote apoptosis in NPC, CCA and PC cells by mediating mitochondrial apoptosis pathway. Re-expression of NOR1 in HeLa cells up-regulates p53 and Bax protein level, contrastly down-regulates Bcl-2 protein level, consequently activated caspase 9[18]. Knockdown of NOR1 in HeLa cells resulted in increase of Bcl-2 mRNA and protein level, and then attenuated H2O2 induced apoptosis [56]. NOR1 also promotes apoptosis in prostate cancer PC3 cells by up-regulation of pro-apoptotic Bax and Bak but down-regulation of Bcl-2 and Bcl-xL. It has been shown that NOR1 mediated proliferation inhibition and induction apoptosis via activating MAPK signaling pathway [19]. Recently, the tumor suppressor function of NOR1 was further convinced by an in vivo model [25, 26]. When overexpressed in Drosophila eye imaginal discs, dOSCP1 induced cell cycle S phase arrest and caspase 3-dependent cell death [26]. However, dOSCP1 induced endoplasmic reticulum stress and unfolded protein response (UPR) activation in salivary gland cells, which imply dOSCP1 function in vivo is context dependent. Thus, both in vitro and in vivo evidences indicated NOR1 functions as a TSG in cancer development by inhibiting cell proliferation and inducing apoptosis. Interestingly, knockdown of dOSCP1 in the Drosophila eye imaginal discs also caused mitochondrial fragmentation and caspase-dependent apoptosis [25], which suggested that NOR1 expression is also required for maintenance of intact mitochondrion.
NOR1 regulates metabolism reprogramming in NPC cells
In normal cells, the vast majority of cellular ATP is produced through mitochondrial oxidative phosphorylation (OXPHOS). Unlike its normal counterparts, tumor cells use glycolysis rather than OXPHOS to meet energy demands. Even in normoxia condition, tumor cells favour glycolysis rather than OXPHOS to produce ATP to accelerate cell proliferation and tumor growth which is called aerobic glycolysis or Warburg effect [57, 58]. The solid tumor cells is heavily dependent on glycolysis to accelerate cell proliferation and tumor growth. However, the mitochondrion and OXPHOS is not fundamentally impaired in cancer cells as otto Warburg previously expected [59, 60]. For example, in a thymic lymphomas mouse model drived by homogenous deletion of the tumor suppressor p53, both glycolysis and mitochondrial respiration were greatly increased [61]. Accumulated evidences indicate that tumor mitochondria are functionally competent as compared to normal cells. Notably, increased glutamine consumption through mitochondrial tricarboxylic acid cycle (TCA) was observed in many human cancer models[62, 63]. Xiang et al. found exogenously expressed NOR1 protein partially distributes in mitochondrion, which permit it spatially close to mitochondrial energe producer. By use of the yeast two hybrid system, they also found NOR1 interact with mitochondrion ATP synthase subunit ATP5O [64], which imply that NOR1 might be involved in energe metabolism. Li et al. further revealed that re-expression of NOR1 in NPC HNE1 cells resulted in decreased glucose consumption and lactate production. Interestingly, they also found decreased mitochondrial oxygen consumption, reactive oxygen species (ROS) level and consequently slowdown of cell proliferation in NOR1 expressing cancer cells. Thus indicating simultaneous inhibition of both glycolysis and mitochondrial OXPHOS in NPC cells by NOR1 [24]. These observations indicate that glycolysis and mitochondrial respiration are not always mutually exclusive in cancer development.
NOR1 regulates oxidative stress, autophagy apoptosis crosstalk
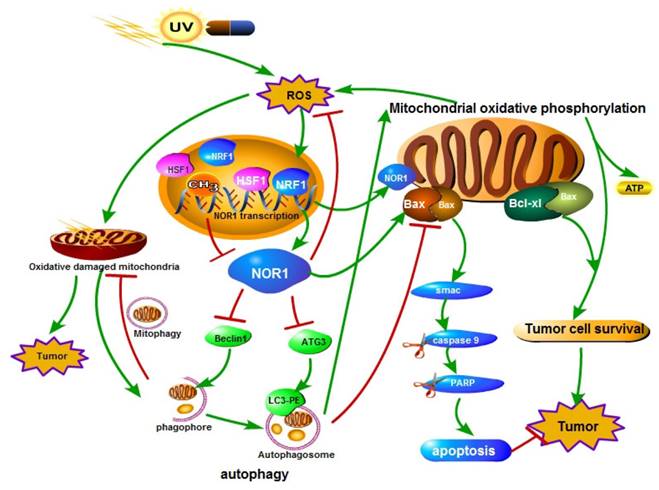
Oxidative stress reflects an imbalance between the production of free radicals and the ability of biological system to remove or detoxify the harmful effects of reactive species (RS). Oxidative stress plays a Janus-face like effect in cancer development and therapy [65]. On one hand, reactive species can chemically interact with biological macromolecules such as DNA, protein or lipid and result in DNA damage, mutations, or loss of enzyme activities, consequently lead to increase of genome instability and cancer formation[66, 67]. On the other hand, sustained oxidative stress or too many free radicals can injure or kill cells [68, 69]. Tumor cells have a increased tolerance to oxidative stress induced cell death although there was high and persistent oxidative stress occurs in tumors [70, 71]. Cells respond to oxidative stress by orchestrating various defense mechanisms such as increase of anti-oxidants, autophagy [72], remove impaired protein by the ubiquitin-proteasome system [73, 74]. Autophagy refers to a regulated cellular catabolic process for delivery, degradation and recycling of unnecessary or dysfunctional cytoplasmic materials such as macromolecules and organelles in lysosomes of eukaryotic cells [71, 75-77]. Li et al. identified a functional promoter of NOR1 gene and characterized the NOR1 promoter is HSF1 and NRF1 regulated [20]. Both HSF1 [78-80] and NRF1 [81, 82] play essential role in oxidative stress response. Li et al. further found that NOR1 expression is induced by H2O2 through activating HSF1 and NRF1. They also found NPC cells respond to oxidative stress by activating autophagy to prevent further damage. Re-expression of NOR1 in NOR1 deficient NPC cells suppresses the production of ROS and the basal level of autophagy and H2O2 induced autophagy, thus lead to apoptotic cell death in stress conditions[24](Figure 2). Recently, it has been reported that knockdown of Knockdown of Drosophila orthologue of human NOR1 dNOR1/dOSCP1 in the eye imaginal discs results a compensatory proliferation and ROS generation[25], suggesting a conserve role of NOR1 on ROS production. NOR1 mediated autophagy/apoptosis switch of tumor cells is associated with suppression ATG3, Beclin 1 and imbalance of mitochondrial Bax/Bcl-xL ratio in NPC cells. NOR1 protein was found partially distributes in mitochondrion which permit it spatially close to mitochondrial Bcl-2 family apoptosis modulators. Under oxidative stress conditions, re-expression of NOR1 facilitates Bax translocation to mitochondrion and further release of death protein Smac/Diablo, which in turn activates cytoplasmic caspase cascade [24] (Figure 2). Furthermore, NOR1 also enhanced cisplatin cytotoxicity and cisplatin-induced apoptosis in NPC tumor cells [24].
A schematic illustration of the role of NOR1 in nasopharyngeal carcinoma cells autophagy-apoptosis crosstalk in oxidative stress. Oxidative stress (ROS) stimulates the expression and activity of HSF1 and NRF1 transcription factor, which bind to NOR1 promoter and promote NOR1 expression. Elevated NOR1 protein inhibits Beclin 1 and ATG3 protein level and led to dysfunction of autophagy. NOR1 protein also translocate to mitochondrion and increase Bax/Bcl-xL ratio of mitochondrion, thus led to increase of the permeability of mitochondrion outer membrane and lease of cell death inducing protein Smac which in turn lead to activation of caspase cascade.
NOR1 mediates tumor cells adaptation to hypoxia
Hypoxic microenvironment represent a characteristic feature of solid tumors, which was believed is mainly due to more aggressive, rapidly growth rate of tumor cells than endothelial cells and decreased perfusion ability of oxygen in intratumoral architecture [83]. Hypoxia is associated with attenuation of proliferation and induction of differentiation. Severe hypoxia even leads to necrosis or apoptosis. On the other hand, tumor cells could adapt to hypoxia environment by switching its metabolism. It has been considered that successful adaptation to hypoxia and acidosis is essential for developing a more aggressive phenotype of cancer [84]. Tumor cells respond to hypoxia mainly by activating hypoxia-inducible factor 1 which is usually hydroxylated and further degraded by ubiquitin-proteasome system under normal oxygen tension [85-87]. It has been reported that HIF1 mediated up-regulation of mitochondrial pyruvate dehydrogenase kinase 1 (PDK1) to rescues tumor cells from hypoxia-induced apoptosis [88]. PDK1 was found to be upregulated in head and neck squamous cancer (HNSCC) and was strongly associated with poor outcome [89]. Xiang et al. also found that PDK1 protein increased in NPC cell lines and tumor biopsies [90]. By use of microarray, Xiang et al. observed that re-expression of NOR1 in NPC cell lines decreases mitochondrial PDK1 expression and strongly inhibits tumor cells proliferation and cell survival under hypoxia. NOR1 causes tumor cells to undergo apoptosis in hypoxic was correlated with a pro-apoptotic increase in the ratio of mitochondrial Bax/Bcl2 ratio and corresponding mitochondrial cyto c release. There was also a corresponding increased activation of the caspase-9/caspase-3/PARP signal cascade in NOR1 expressing NPC cell lines. Overexpression of dNOR1/dOSCP1 also induces of caspase-dependent apoptosis in adult flies eye imaginal discs [26]. It has been proposed that cells adapts to hypoxia by activating the unfolded protein response (UPR)[91]. Huu NT et al. found that dNOR1/dOSCP1 localizes in endoplasmic reticulum (ER) in salivary gland cells[31]. Interestingly, overexpression of dNOR1/dOSCP1 caused ER stress in Drosophila salivary gland cells, as evidenced by increase of ER chaperones marker KDEL immunostaining [26]. Thus, our data and others collectively indicate that NOR1 act as a tumor suppressor by altering PDK1 expression and hampering tumor cells adaptation to hypoxia [90].
NOR1 suppresses epithelial-to-mesenchymal transition and metastasis via FOXA1-HDAC2/slug axis
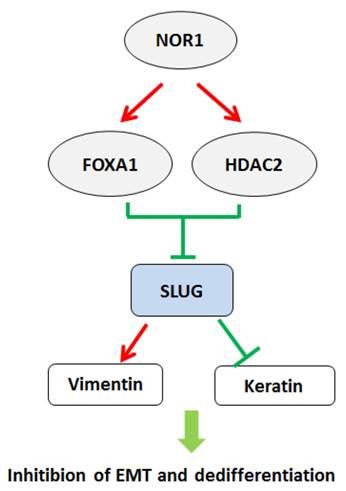
The epithelial-to-mesenchymal transition (EMT) is a biological process characterized epithelial cells lose their cell polarity and cell-cell adhesion but acquire a migratory, mesenchymal properties [92-94]. It has been considered that EMT is a prerequisite for tumor infiltration and metastasis [95, 96]. Several transcriptional factors, including SNAI1, SNAI2 (SLUG), and TWIST were initially recognized as master factors of EMT by suppressing CDH1 expression [97-99]. NPC cells exhibit early tendency of locally spreading to the parapharyngeal space bulky regardless of the size of the primary tumors [100]. Wang et al. found overexpression of NOR1 in NPC cells resulted in inhibition of EMT and decreased mobility. Mechanistically, NOR1-mediated inhibition of EMT via suppressing slug but not snail. NOR1 induced cytokeratin 4 and cytokeratin 13 expression without alteration of E-cadherin [27]. Overexpression of NOR1 in high metastatic potential NPC 5-8F cells induced significant morphological changes characterized by disturbed the actin cytoskeleton and less lamellipodia formation [101]. Overexpression of slug in NOR1 expressing NPC cells rescued cell mobility and invasive behavior. Inhibition on slug by NOR1 is likely through alteration of slug associated histone remodeling by disturbing balance of Slug-associated H3K9 acetylation and tri-methylation, since overexpression of NOR1 led to increased tri-methylation H3K9(H3K9Me3; repressed chromatin marker) but concurrent decreased Slug-associated acetylation of H3K9 (H3K9Ace; active chromatin marker). Conversely, a decrease H3K9 tri-methylation levels and an increase in Slug-associated H3K9 acetylation occurred in NOR1 stable knockdown HeLa cells. The repressive effect of NOR1 on Slug is dependent on upregulation of transcription factors FOXA1 and HDAC2, which suppress slug transcription by directly binding to slug promoter (Figure 3). Silencing either FOXA1 or HDAC2 in NOR1 expressing cells lead to recovery of slug expression and elevated cell mobility and invasive behavior [28]. Thus, it has been proposed that dysfunction of the NOR1-FOXA1/HDAC2-Slug network is an essential step in the EMT program during NPC progression.
Conclusions
There is an emerging role of NOR1 in cancer biology and developmental biology. However, since NOR1 is mainly a cytoplasmic protein, one of the most challenging is to figure out how NOR1 transduce cell signal from cytoplasm to nuclear transcription activation. It's important to identify cellular interaction partners of NOR1 protein in cytoplasm to mediate cell signal transduction by NOR1. To date, most works about NOR1 were conducted in vitro system, it's urgent to further deciphering NOR1 function by using in vivo models such as gene knockout mice.
A schematic illustration of NOR1-FOXA1/HDAC2-SLUG regulatory network in the EMT and dedifferentiation process of NPC. High levels of NOR1 expression lead to optimal expression of pioneering factor FOXA1, which exhibit inhibitory effect on Slug promoter activity. Expression of also lead to increase of histone deacetylase HDAC2 level in NPC cells, which cooperate with FOXA1 to suppress slug transcription via de-acetylation of slug associated H3K9. Repression of slug eventually resulted decrease of vimentin but increase of epithelial keratins, thus lead to inhibition of EMT and dedifferentiaon of NPC cells.
Acknowledgements
This study was supported in part by Grants from The National Natural Science Foundation of China (81272254, 81372304, 81572667), the National “111” Project (Project #111-2-12), The Natural Science Foundation of Hunan Province, China (2015JJ2178).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
2. Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163-9
3. Effert P, McCoy R, Abdel-Hamid M, Flynn K, Zhang Q, Busson P. et al. Alterations of the p53 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3768-75
4. Sun Y, Hegamyer G, Colburn NH. Nasopharyngeal carcinoma shows no detectable retinoblastoma susceptibility gene alterations. Oncogene. 1993;8:791-5
5. Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683-95
6. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
7. Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729-37
8. Zhou Y, Liao Q, Li X, Wang H, Wei F, Chen J. et al. HYOU1, Regulated by LPLUNC1, Is Up-Regulated in Nasopharyngeal Carcinoma and Associated with Poor Prognosis. J Cancer. 2016;7:367-76
9. Xu K, Xiong W, Zhou M, Wang H, Yang J, Li X. et al. Integrating ChIP-sequencing and digital gene expression profiling to identify BRD7 downstream genes and construct their regulating network. Mol Cell Biochem. 2016;411:57-71
10. Zeng Z, Fan S, Zhang X, Li S, Zhou M, Xiong W. et al. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2016;18:206-11
11. He B, Li W, Wu Y, Wei F, Gong Z, Bo H. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353
12. Yang Y, Zhou H, Yang Y, Li W, Zhou M, Zeng Z. et al. Lipopolysaccharide (LPS) regulates TLR4 signal transduction in nasopharynx epithelial cell line 5-8F via NFkappaB and MAPKs signaling pathways. Mol Immunol. 2007;44:984-92
13. Lian Yu LX, Tang Yanyan, Yang Liting, Li Xiaoling, Xiong Wei, Li Guiyuan, Zeng Zhaoyang. Long Non-coding RNAs Function as Competing Endogenous RNAs to Regulate Cancer Progression. Prog Biochem Biophys. 2016;43:219-25
14. Nie X, Zhang B, Li X, Xiang J, Xiao B, Ma J. et al. Cloning, expression, and mutation analysis of NOR1, a novel human gene down-regulated in HNE1 nasopharyngeal carcinoma cell line. J Cancer Res Clin Oncol. 2003;129:410-4
15. Kobayashi Y, Shibusawa A, Saito H, Ohshiro N, Ohbayashi M, Kohyama N. et al. Isolation and functional characterization of a novel organic solute carrier protein, hOSCP1. J Biol Chem. 2005;280:32332-9
16. Xiang B, Yi M, Wang L, Liu W, Zhang W, Ouyang J. et al. Preparation of polyclonal antibody specific for NOR1 and detection of its expression pattern in human tissues and nasopharyngeal carcinoma. Acta Biochim Biophys Sin (Shanghai). 2009;41:754-62
17. Xiang B, Wang W, Li W, Li X, Li G. Differential expression of oxidored nitro domain containing protein 1 (NOR1), in mouse tissues and in normal and cancerous human tissues. Gene. 2012;493:18-26
18. Ouyang J, Wu M, Huang C, Cao L, Li G. Overexpression of oxidored-nitro domain containing protein 1 inhibits human nasopharyngeal carcinoma and cervical cancer cell proliferation and induces apoptosis: Involvement of mitochondrial apoptotic pathways. Oncol Rep. 2013;29:79-86
19. Shan Z, Hou Q, Zhang N, Guo L, Zhang X, Ma Y. et al. Overexpression of oxidored-nitro domain containing protein 1 induces growth inhibition and apoptosis in human prostate cancer PC3 cells. Oncol Rep. 2014;32:1939-46
20. Li W, Li X, Wang W, Tan Y, Yi M, Yang J. et al. NOR1 is an HSF1- and NRF1-regulated putative tumor suppressor inactivated by promoter hypermethylation in nasopharyngeal carcinoma. Carcinogenesis. 2011;32:1305-14
21. Kroeger H, Jelinek J, Estecio MR, He R, Kondo K, Chung W. et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood. 2008;112:1366-73
22. Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X. et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605-13
23. Jelinek J, Gharibyan V, Estecio MR, Kondo K, He R, Chung W. et al. Aberrant DNA methylation is associated with disease progression, resistance to imatinib and shortened survival in chronic myelogenous leukemia. PLoS One. 2011;6:e22110
24. Li W, Li X, Wang W, Yi M, Zhou Y, Zheng P. et al. Tumor suppressor gene Oxidored-nitro domain-containing protein 1 regulates nasopharyngeal cancer cell autophagy, metabolism, and apoptosis in vitro. Int J Biochem Cell Biol. 2013;45:2016-26
25. Huu NT, Yoshida H, Yamaguchi M. Tumor suppressor gene OSCP1/NOR1 regulates apoptosis, proliferation, differentiation, and ROS generation during eye development of Drosophila melanogaster. FEBS J. 2015
26. Huu NT, Yoshida H, Yamaguchi M. Overexpression of tumor suppressor protein OSCP1/NOR1 induces ER stress and apoptosis during development of Drosophila melanogaster. Am J Cancer Res. 2015;5:1718-29
27. Wei Wang, Xiaoling Li, Wenling Zhang, Wenjuan Li, Jianbo Yang, Mei Yi. et al. Oxidored-nitro domain containing protein 1 (NOR1) expression suppresses Slug/vimentin but not Snail in nasopharyngeal carcinoma: inhibition of EMT in vitro and in vivo in mice. Cancer Letters. 2014
28. Wang W, Yi M, Chen S, Li J, Li G, Yang J. et al. Significance of the NOR1-FOXA1/HDAC2-Slug regulatory network in epithelial-mesenchymal transition of tumor cells. Oncotarget. 2016;7:16745-59
29. Kobayashi Y, Tsuchiya A, Hayashi T, Kohyama N, Ohbayashi M, Yamamoto T. Isolation and characterization of polyspecific mouse organic solute carrier protein 1 (mOscp1). Drug Metab Dispos. 2007;35:1239-45
30. Izuno H, Kobayashi Y, Sanada Y, Nihei D, Suzuki M, Kohyama N. et al. Rat organic solute carrier protein 1 (rOscp1) mediated the transport of organic solutes in Xenopus laevis oocytes: isolation and pharmacological characterization of rOscp1. Life Sci. 2007;81:1183-92
31. Huu NT, Yoshida H, Umegawachi T, Miyata S, Yamaguchi M. Structural characterization and subcellular localization of Drosophila organic solute carrier partner 1. BMC Biochem. 2014;15:11
32. Xiang B, Wang W, Yi M, Li W, Zhou M, Li X. et al. Distribution and expression of alternative splice isoforms of NOR1 in human tissues and cell lines. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2012;36:597-603
33. Hiratsuka K, Yin SA, Ohtomo T, Fujita M, Ohtsuki K, Isaka H. et al. Intratesticular localization of the organic solute carrier protein, OSCP1, in spermatogenic cells in mice. Mol Reprod Dev. 2008;75:1495-504
34. Hiratsuka K, Momose A, Takagi N, Sasaki H, Yin SA, Fujita M. et al. Neuronal expression, cytosolic localization, and developmental regulation of the organic solute carrier partner 1 in the mouse brain. Histochem Cell Biol. 2011;135:229-38
35. Fietz D, Bakhaus K, Wapelhorst B, Grosser G, Gunther S, Alber J. et al. Membrane transporters for sulfated steroids in the human testis-cellular localization, expression pattern and functional analysis. PLoS One. 2013;8:e62638
36. Liang F, Li Q, Li X, Li Z, Gong Z, Deng H. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046-56
37. Duan Z, Zheng H, Xu S, Jiang Y, Liu H, Li M. et al. Activation of the Ig Ialpha1 promoter by the transcription factor Ets-1 triggers Ig Ialpha1-Calpha1 germline transcription in epithelial cancer cells. Cell Mol Immunol. 2014;11:197-205
38. Hu D, Duan Z, Li M, Jiang Y, Liu H, Zheng H. et al. Heterogeneity of aberrant immunoglobulin expression in cancer cells. Cell Mol Immunol. 2011;8:479-85
39. Hu D, Zheng H, Liu H, Li M, Ren W, Liao W. et al. Immunoglobulin expression and its biological significance in cancer cells. Cell Mol Immunol. 2008;5:319-24
40. Li M, Zheng H, Duan Z, Liu H, Hu D, Bode A. et al. Promotion of cell proliferation and inhibition of ADCC by cancerous immunoglobulin expressed in cancer cell lines. Cell Mol Immunol. 2012;9:54-61
41. Zheng H, Li M, Ren W, Zeng L, Liu HD, Hu D. et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol Immunol. 2007;44:2221-7
42. Zhao R, Liu Y, Wang H, Yang J, Niu W, Fan S. et al. BRD7 plays an anti-inflammatory role during early acute inflammation by inhibiting activation of the NF-small ka, CyrillicB signaling pathway. Cell Mol Immunol. 2016
43. Xiong Wei, Zeng Zhaoyang, Xiao Bingyi, Xiong Fang, Nie Xinmin, Fan Songqing. et al. Studies of association between nasopharyngeal carcinoma and single-nucleotide polymorphisms in NOR1, a novel oxidored-nitro domain-containing protein gene. Prog Biochem Biophys. 2003;30:401-5
44. Toda M, Kobayashi Y, Koizumi T, Saito K, Ohbayashi M, Kohyama N. et al. Genetic polymorphism of the human organic solute carrier protein 1 (hOSCP1) gene in Japanese patients with non-viral liver carcinoma. Meta Gene. 2014;2:686-93
45. Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673-91
46. Platonova O, Akey IV, Head JF, Akey CW. Crystal structure and function of human nucleoplasmin (npm2): a histone chaperone in oocytes and embryos. Biochemistry. 2011;50:8078-89
47. Xiang B, Li W, Yi M, Wang W, Li X, Li G. [Effect of DNA hypermethylation on NOR1 promoter activity and expression]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2012;37:765-70
48. Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M. et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999-8006
49. Wang W, Li X-L, Li W-J, Yi M, Tang K, Zheng P. et al. Testis Selective Expression of NOR1 Gene in Rat and Down Regulation of NOR1 Protein in Human Testicular Germ Cell Tumors. Prog Biochem Biophys. 2013;40:652-61
50. Li DQ, Qiu M, Nie XM, Gui R, Huang MZ. Oxidored-nitro domain-containing protein 1 expression is associated with the progression of hepatocellular carcinoma. Oncol Lett. 2016;11:3003-8
51. Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2016
52. Li Q, Chen P, Zeng Z, Liang F, Song Y, Xiong F. et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37:12503-12
53. Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783-99
54. Xinmin N, Rong G, Dengqing L, Ming Z, Zufa H, Guiyuan L. The Effects of NOR1 on Cells From Human Nasopharyngeal Carcinoma Cell Line HNE1. Prog Biochem Biophys. 2005;32:777-80
55. Wang W, Yi M, Chen S, Li J, Zhang H, Xiong W. et al. NOR1 Suppresses Cancer Stem-Like Cells Properties of Tumor Cells via the Inhibition of the AKT-GSK-3beta-Wnt/beta-Catenin-ALDH1A1 Signal Circuit. J Cell Physiol. 2016 [Epub ahead of print]
56. Tan Y, Li W, Yi M, Wang W, Zheng P, Zhang H. et al. [Effect of NOR1 gene knockdown on the biological behavior of HeLa cells]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2014;39:757-63
57. Koppenol WH BP, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325-37
58. Yi M, Xiang B, Li X, Li G. [Metabolic reprogramming in cancer: the art of balance]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013;38:1177-87
59. Dang CV. p32 (C1QBP) and cancer cell metabolism: is the Warburg effect a lot of hot air? Mol Cell Biol. 2010;30:1300-2
60. Vaupel P, Mayer A. Availability, not respiratory capacity governs oxygen consumption of solid tumors. Int J Biochem Cell Biol. 2012;44:1477-81
61. Samper E, Morgado L, Estrada JC, Bernad A, Hubbard A, Cadenas S. et al. Increase in mitochondrial biogenesis, oxidative stress, and glycolysis in murine lymphomas. Free Radic Biol Med. 2009;46:387-96
62. Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M. et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788-93
63. Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL. et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827-37
64. Bo X, Li W, Mei Y, Jue O, Xiayu L, Zuping Z. et al. Screen and Identification of The Protein-protein Interactors of NOR1, a Novel Gene Down-regulated in Nasopharyngeal Carcinoma. Prog Biochem Biophys. 2009;36:709-14
65. Kardeh S, Ashkani-Esfahani S, Alizadeh AM. Paradoxical action of reactive oxygen species in creation and therapy of cancer. Eur J Pharmacol. 2014;735:150-68
66. Linhart K, Bartsch H, Seitz HK. The role of reactive oxygen species (ROS) and cytochrome P-450 2E1 in the generation of carcinogenic etheno-DNA adducts. Redox Biol. 2014;3C:56-62
67. Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer. 2014;14:709-21
68. Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401:1-11
69. Yu JH, Cho SO, Lim JW, Kim N, Kim H. Ataxia telangiectasia mutated inhibits oxidative stress-induced apoptosis by regulating heme oxygenase-1 expression. Int J Biochem Cell Biol. 2015
70. Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1-3
71. Klaunig JE, Xu Y, Isenberg JS, Bachowski S, Kolaja KL, Jiang J. et al. The role of oxidative stress in chemical carcinogenesis. Environ Health Perspect. 1998;106(Suppl 1):289-95
72. Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152-62
73. Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305:709-18
74. Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422-7
75. Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679-88
76. Marino G, Lopez-Otin C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61:1439-54
77. Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159-62
78. Dayalan Naidu S, Kostov RV, Dinkova-Kostova AT. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol Sci. 2015;36:6-14
79. Choi YJ, Om JY, Kim NH, Chang JE, Park JH, Kim JY. et al. Heat shock transcription factor-1 suppresses apoptotic cell death and ROS generation in 3-nitropropionic acid-stimulated striatal cells. Mol Cell Biochem. 2013;375:59-67
80. Yan LJ, Rajasekaran NS, Sathyanarayanan S, Benjamin IJ. Mouse HSF1 disruption perturbs redox state and increases mitochondrial oxidative stress in kidney. Antioxid Redox Signal. 2005;7:465-71
81. Digaleh H, Kiaei M, Khodagholi F. Nrf2 and Nrf1 signaling and ER stress crosstalk: implication for proteasomal degradation and autophagy. Cell Mol Life Sci. 2013;70:4681-94
82. Wan X, Gupta S, Zago MP, Davidson MM, Dousset P, Amoroso A. et al. Defects of mtDNA replication impaired mitochondrial biogenesis during Trypanosoma cruzi infection in human cardiomyocytes and chagasic patients: the role of Nrf1/2 and antioxidant response. J Am Heart Assoc. 2012;1:e003855
83. Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225-39
84. Gatenby RA, Smallbone K, Maini PK, Rose F, Averill J, Nagle RB. et al. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br J Cancer. 2007;97:646-53
85. Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343-54
86. Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH. et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149-62
87. Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda). 2004;19:176-82
88. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177-85
89. Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML, Harris AL. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br J Cancer. 2008;98:1975-84
90. Bo X, Mei Y, Wenjuan L, Wei W, Pan Z, Xiaoling L. et al. Expression of oxidored nitro domain containing protein 1(NOR1) impaires tumor cell adaptation to hypoxia and inhibits PDK1 expression. Molecular and Cellular Biochemistry. 2014
91. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K. et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127-41
92. Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740-6
93. Burdsal CA, Damsky CH, Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829-44
94. Nieto MA. The early steps of neural crest development. Mech Dev. 2001;105:27-35
95. Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319-26
96. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15-33
97. Bolós V PH, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499-511
98. Jouppila-Matto A, Narkio-Makela M, Soini Y, Pukkila M, Sironen R, Tuhkanen H. et al. Twist and snai1 expression in pharyngeal squamous cell carcinoma stroma is related to cancer progression. BMC Cancer. 2011;11:350
99. Wang C, Liu X, Huang H, Ma H, Cai W, Hou J. et al. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer. 2012;130:2249-58
100. Chiesa F, De Paoli F. Distant metastases from nasopharyngeal cancer. ORL J Otorhinolaryngol Relat Spec. 2001;63:214-6
101. Bo X, Wei W, Wenjuan L, Ke T, Zhaoyang Z, Xiaoling L. et al. NOR1 Regulates Morphogenetic Cell Behavior in vitro Coincident With Inhibition of a Non-canonical Wnt-signaling Cascade. Prog Biochem Biophys. 2012;39:887-92
Author contact
![]() Corresponding author: Bo Xiang, The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan410078, China. Tel: +86-731-83867186; Fax: +86-731-84805383; E-mail: xiangbolinedu.cn
Corresponding author: Bo Xiang, The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan410078, China. Tel: +86-731-83867186; Fax: +86-731-84805383; E-mail: xiangbolinedu.cn

 Global reach, higher impact
Global reach, higher impact