3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(14):2846-2853. doi:10.7150/jca.19897 This issue Cite
Research Paper
ERCC1_202 Is A Prognostic Biomarker in Advanced Stage Non-Small Cell Lung Cancer Patients Treated with Platinum-Based Chemotherapy
1. Medical school of Southeast University, Nanjing 210009, China;
2. Department of Respiratory Medicine, Zhongda Hospital, Southeast University, Nanjing, 210009, China;
3. Center of Clinical Laboratory Medicine, Zhongda Hospital, Southeast University, Nanjing, 210009, China.
Received 2017-3-2; Accepted 2017-6-25; Published 2017-8-23
Abstract
Purpose: To develop a qPCR method to examine the 202 isoform of excision repair cross-complementation group 1 (ERCC1_202) and to evaluate its clinical utility as a predictive biomarker for platinum-based chemotherapy in non-small cell lung cancer (NSCLC).
Methods: The relative complementary DNA (cDNA) quantification for ERCC1_202 was conducted using a fluorescence-based, real-time detection method and β-actin was used as a reference gene.
Results: A strong correlation was observed between ERCC1_202 mRNA and ERCC1 mRNA levels in NSCLC cells (P < 0.001). 28 patients completed this research. Our results implied that as ERCC1_202 levels increased, the risk of progression (HR = 4.296, P = 0.011) and death (HR = 6.503, P = 0.001) increased. At multivariate analysis, high expression of ERCC1_202 was shown to be an independent predictive factor for time to progression (P = 0.047), and progression-free survival (P = 0.014). However, the high expression of ERCC1_202 was not an independent predictive factor for response (P = 0.324).
Conclusions: This study suggests that the efficacy of platinum-based chemotherapy can be improved when customized according to the expression of ERCC1_202.
Keywords: non-small cell lung cancer, ERCC-1, 202 isoform, Cisplatin, resistance.
Introduction
Lung cancer is one of the leading causes of cancer mortality worldwide [1-3]. Among the patients, approximately 80% suffer from non-small-cell lung cancer (NSCLC) [3]. The disease has a poor prognosis and a meager five year survival rate in the disease regardless of the stage [2, 3]. Despite development of new treatments, platinum-based chemotherapy remains a predominant treatment mode for advanced NSCLC patients [2]. However, platinum resistance has become one of the major hindrances to treatment of the disease [4].
Therefore, identifying the molecular biomarkers of platinum resistance to not only screen chemo sensitive patients, but also to select a suitable drug with minimum toxicities synthesized with a cost-effective approach is the need of the hour. This will directly improve patient outcome [5]. There is a scarcity for clinical biomarkers to customize chemo sensitivity. Even though excision repair cross-complementation group 1 (ERCC1) gene expression has been suggested as one of the potential biomarkers for cancer cell response to platinum-based chemotherapy [2, 6], considerable amount of studies have opposed this view about ERCC1 or its role in prognosis of the disease [7-9]. Adding to this, a considerable volume of patients with high ERCC1 expression possess significantly improved survival rate in comparison to patients with low ERCC1 expression through platinum-based chemotherapy [10, 11]. There is no unanimous opinion on whether ERCC1 is a useful marker for platinum resistance now.
Friboulet et al. created the dawn for solving this problem, because they found that among all its four isoforms, 202 isoform of ERCC1 was the only functional gene in DNA repair pathways [12, 13]. It is believed that ERCC1_202 isoform is a useful marker for platinum resistance, however, so far, no solution to examine the ERCC1_202 isoform has been found. Sixteen commercial antibodies were used by Friboulet et al. but none of them could uniquely identify these isoforms, owing to sequence homologies [12, 13].
In this study, a qPCR method to examine ERCC1_202 mRNA was found and applied to NSCLC cells and tumor samples. We analyzed the relationships between ERCC1_202 mRNA expression in tumors and the respective survival time to determine whether its expression could predict progression-free and overall survival in recurrent NSCLC patients treated with platinum-based chemotherapy.
Patients and Methods
Patients and tissue specimens
This prospective study was approved by the Research Ethics Board of Zhongda Hospital affiliated to Southeast University (Nanjing, China). Written informed consent was obtained from all patients. Fresh tumor samples of primary NSCLC and non-cancer tissues were collected during the bronchofiberscope or thoracentesis. All the biopsy tissues were freshly frozen in liquid nitrogen immediately and then stored at -80 °C until analysis was performed. All patients were followed up to September 2016. No patient received preoperative chemotherapy or radiotherapy. The patients were classified in accordance with the NCCN (National Comprehensive Cancer Network) classification of the tumor by two pathologists and independently reviewed by an expert NSCLC pathologist. All patients received performance status (PS) score before chemotherapy; those with scores of 0, 1 or 2 were enrolled in this study. A total of 28 patients received at least one cycle of chemotherapy within five months after diagnosis during the period of May 2015 to September 2016.
Cell culture
Three NSCLC cell lines A549, NCI-H1299 and SK-MES-1 were characterized by Cobioer Biosciences CO., LTD (Nanjing, China) using short tandem repeat (STR) markers. All these cells were cultured in RPMI1640 medium (HyClone, GE Healthcare Life Sciences, USA) with 10% fetal calf serum. Culture plates were incubated at 37 °C with 5% CO2. Cells were collected after drug intervention for RNA extraction. Cisplatin was obtained from Sigma Aldrich.
RNA isolation and RT-PCR
RNA was isolated from cultured cells using TRIzol (Invitrogen) in accordance with the manufacturer's instructions. Total RNA was isolated from fresh lung tumor and normal tissues using TaKaRa MiniBEST universal RNA extraction kit. The RNA quality was confirmed by Nanodrop spectrophotometry. Reverse transcription was performed with 1μg of total RNA following the manufacturer's instructions using an RT Kit (TakaRa, Dalian, China). Real-time PCR analysis was performed on an BioRad CFX96 Detection System using the PCR Master Mix (TakaRa, Dalian, China). The relative expression of each gene was normalized to beta-actin. The primers used for the quantitative RT-PCR are shown in supplementary Table 1 and supplementary Fig. 1A.
Standard plasmid preparation
The plasmids: pUC57-ERCC1_201, pUC57-ERCC1_202, pUC57-ERCC1_203 and pUC57-ERCC1_204 were obtained from Shanghai Generay Biotech Co., Ltd. The plasmid DNA concentration was calculated by using the molecular weight of the plasmid-insert and OD of the extracted plasmid.
Real-time PCR
To distinguish ERCC1_202 from other isoforms, as supplementary Table 2 shows, the forward primer was designed for gene fragment E3, the reverse for E10, and the probe for E8 (supplementary Fig. 1B). Reagents were mixed to get a final volume of 25 μL. Cycling conditions were 95 °C for 30 s, 95 °C for 5 s, and 57 °C for 50 s. Samples were analyzed in triplicate, with the results of each sample normalized to that of β-actin[14]. To determine the least detectable amount of the target in the samples, the dilutions of pUC57-ERCC1_202 plasmid with 10-fold serial dilutions from 101 to 107 copies/μL was prepared, and each sample was measured in triplicates.
Western blot
Western blot was performed as described previously. Samples were collected and denatured at 95 °C for 5 min. The samples were then electrophoresed on a 12% Tris-HCl gel and transferred to nitrocellulose membranes (Millipore). After treating with blocking buffer, the membranes were incubated with a primary antibody overnight at 4°C and then with a secondary antibody conjugated for two hours at room temperature; the signal was detected using a chemiluminescence method. The following primary antibodies were used: anti-GAPDH (Santa Cruz, 1:5000); anti-ERCC1 (3H11: sc-53281, Santa Cruz, 1:200), goat anti-mouse secondary antibodies (Cell Signaling, 1:5000). After final washes, the membranes were visualized using enhanced chemiluminescence solution (ECL). The levels of ERCC1 were normalized to GAPDH.
Statistical methods
Tumor response was assessed after chemotherapy following Response Evaluation Criteria in Solid Tumors (RECIST) [4]. Complete response (CR) or partial response (PR) were classified as “response” and stable disease (SD). progressive disease (PD) was classified to be “non-response” group. The endpoints considered in the pooled analyses were response rate (RR) and Progression-Free-Survival (PFS). PFS was defined as the interval from the date of chemotherapy to the date of death or last follow-up. The association between ERCC1_202 level and PFS was evaluated using the hazard ratio (HR) from multivariate Cox proportional hazards models. All data were analyzed using SPSS 18.0 statistical software package, and probability values of less than 0.05 were considered statistically significant.
Results
Establishment of qPCR method for detection of ERCC1
ERCC1 isoforms expression in A549, NCI-H1299, and SK-MES-1 cell lines were analyzed by reverse transcription (RT)-PCR and agarose gel electrophoresis (Supplementary Fig. 2). ERCC1_202 was accounted for the major proportion of all four isoforms in these cells. To examine ERCC1_202 isoform, we designed many primers and probes based on the nature of the 4 isoforms, until a pair of primers and probe were found. The amplification curve and the standard curve of the real-time PCR were shown in Supplementary Fig. 3 A, B. To determine the analytical sensitivity, a serial dilution of the standard plasmid from 101 to 107 copies/μL was performed. Each standard was tested in triplicate, all of which were detectable by the assay. The linear range is 101 to 106 copies of mRNA with a detection limit of 101 copies of mRNA. The PCR products evaluated by electrophoresis were as expected: a single band at 590 bp (Supplementary Fig. 3 C).
To verify the specificity of the method, the plasmid pUC57-ERCC1_201, pUC57-ERCC1_202, pUC57-ERCC1_203, pUC57-ERCC1_204 were constructed. These plasmids were used as template to amplify the gene of interest. The results show that only the pUC57-ERCC1_202 template was amplified (Supplementary Fig. 3D, E), and its Ct value was 20.88±0.14.
ERCC1_202 expression in NSCLS cells
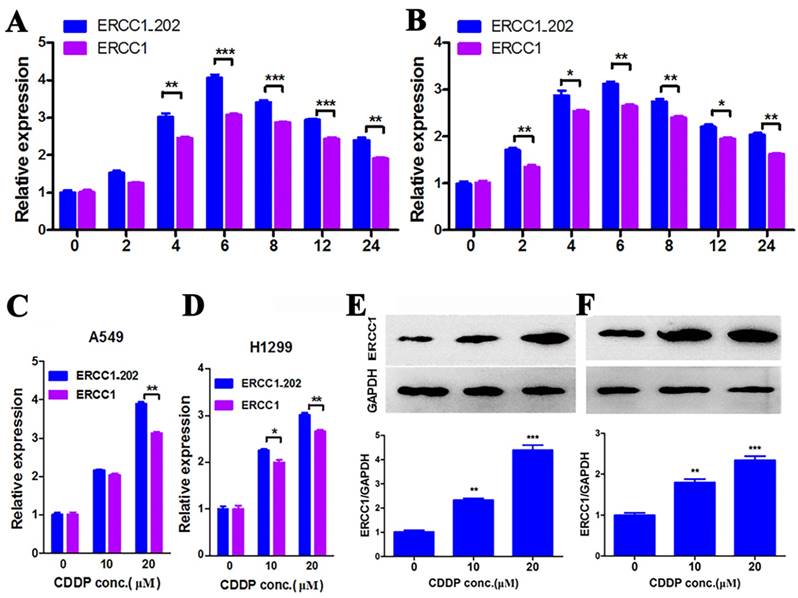
To illuminate the practicability of the method, two NSCLC cells A549 and H1299 were detected after treatment with cisplatin for varying time intervals. The results showed that both ERCC1 and ERCC1_202 reached the highest value at 6 h after cisplatin treatment (Fig. 1 A, B). ERCC1_202 mRNA expression was significantly associated with total ERCC1 mRNA levels (r = 0.995, P < 0.001 for A549; r = 0.991, P < 0.001 for H1299). So 6 h was chosen as a time point for the role of cisplatin in lung cancer cells. Varying concentrations (0, 10, 20 μM) of cisplatin were used for 6 hours against lung cancer cells. The results showed that levels of both ERCC1 and ERCC1_202 increased with the increase in concentration of cisplatin, both of which had the same change trend, but the former was more obvious (Fig. 1 C, D).
To verify whether the ERCC1 protein has the same change trend, we performed Western Blot experiments. Due to unavailability of specific antibodies against ERCC1_202, a much similar anti-ERCC1 antibody (3H11) was chosen. The results showed that the changes of ERCC1 protein in two lung cancer cells with varying concentrations of cisplatin were similar to that of mRNA levels (Fig. 1 E, F).
Patient characteristics and clinical outcomes
A total of 28 patients met the inclusion criteria and were included in this study. The characteristics of these patients are shown in Table 1. The median follow-up time was 344 days (range 274-508), and the median progression-free survival time was 210 days (range 42-479). Of them, 16 were adenocarcinomas and 12 were squamous cell carcinomas. Twelve received “Cisplatin + Etoposide” (Cisplatin 75 mg/m2 D1, Etoposide 100 mg D1~ D5), 4 patients received “Cisplatin + Gemcitabine” (Cisplatin 75 mg/m2 D1, Gemcitabine 1000 mg/m2 D1+ D8) and 10 patients received “Cisplatin + Pemetrexed”. The final 1 patient received “Cisplatin + Vinorelbine” (Cisplatin 75 mg/m2 D1, Vinorelbine 25 mg/m2 D1 + D8). The treatment was received every 3 weeks. Seventeen were stage III and eleven, stage IV disease. The total response rate was 21.42 %, the median TTP 4 months, and overall median survival time was 7 months.
ERCC1_202 and response to treatment
ERCC1_202 mRNA expression levels were calculated in comparison to β-actin levels. Median mRNA expression level was 2.88 (range 0.76- 4.49). Higher level of ERCC1_202 significantly correlated with advanced TNM stage (P = 0.018). However, no significant associations were found between ERCC1_202 and age, gender, histology, smoking status or performance status (all at P ≥ 0.05, supplementary Table 3).
In order to predict response to treatment, a logistic regression model was fitted for the expression of ERCC1_202 as a continuous variable, as showed in Table 2. As ERCC1_202 levels increased, the probability of response did not increase significantly (OR = 2.398, 95% CI = 0.361-15.873, P = 0.324), the high expression of ERCC1_202 was not an independent predictive factor for response. Our results also suggested age, gender, performance status, histology, and TNM stage were not significantly associated with probability of response (P > 0.05 for all).
ERCC1_202 and time to progression
Univariate analysis was carried out to identify these factors (age, sex, histology, smoking status, performance status, response to chemotherapy and TNM stage) significantly associated with TTP. As in Table 3, two clinical variables, namely ERCC1_202 expression and TNM stage, were associated with TTP. The univariate analysis showed that as ERCC1_202 levels increased, the risk of progression increased significantly (HR = 4.296, 95% CI = 1.391-3.266, P = 0.011). In addition, the multivariate model analysis, ERCC1 emerged as a significant factor for TTP (HR = 3.252, 95% CI = 1.014-10.428, P = 0.047).
A. (A549) and B. (NCI-H1299) cell lines were treated with cisplatin at 20 μM for different time durations. ERCC1 and ERCC1_202 were analyzed by qPCR. The data are expressed as the mean ± SD. The values are from the average of three independent experiments. A549 (C) and NCI-H1299 (D) cell lines were treated with cisplatin at different doses for 6 hours, then ERCC1 and ERCC1_202 expression were assessed by qPCR.A549 (E) and NCI-H1299 (F) cell lines were treated with cisplatin at different doses for 24 hours, then ERCC1 and ERCC1_202 expression were assessed by Western blot. The data are expressed as the mean ± SD. The values were from the average of three independent experiments. (* P<0.05, ** P<0.01 and *** P<0.001)
Patient characteristics
| Characteristic | n | Percentage (%) |
|---|---|---|
| Age (years) | ||
| Median | 64 (44-79)a | |
| Sex | ||
| Female | 7 | 25.00 |
| Male | 21 | 75.00 |
| Histology | ||
| Adenocarcinoma | 16 | 57.14 |
| Squamous cell carcinoma | 12 | 42.86 |
| Smoking status | ||
| Never smoker | 14 | 50.00 |
| smoker | 14 | 50.00 |
| Performance status | ||
| 0 | 22 | 78.57 |
| 1 | 5 | 17.86 |
| 2 | 1 | 3.57 |
| Chemotherapy regimen | ||
| cisplatin + etoposide | 12 | 42.86 |
| cisplatin + gemcitabine | 5 | 14.29 |
| cisplatin + pemetrexed | 10 | 35.71 |
| cisplatin + vinorelbine | 1 | 3.57 |
| Treatment response | ||
| Complete response (CR) | 1 | 3.57 |
| Partial response (PR) | 5 | 14.86 |
| Stable disease (SD) | 14 | 50.00 |
| Progressive disease (PD) | 8 | 28.57 |
| Stage | ||
| III | 17 | 60.71 |
| IV | 11 | 39.29 |
a. range
Factors associated with response to treatment
| Factor | Non-responders SD+PD N (%) | Responders CR+PR N (%) | OR (95% CI) | P |
|---|---|---|---|---|
| Age (years) | ||||
| Low (≤60) | 8(28.6) | 3(10.7) | 0.571(0.093-3.530) | 0.653 |
| High (>60) | 14(50) | 3(10.7) | 1 | |
| Sex | ||||
| Female | 6(21.4) | 1(3.6) | 1.875(0.180-19.525) | 0.522 |
| Male | 16(57.1) | 5(17.9) | 1 | |
| Histology | ||||
| Adenocarcinoma | 11(39.3) | 5(17.9) | 0.200(0.20-2.003) | 0.16 |
| Squamous cell carcinoma | 11(39.3) | 1(3.6) | 1 | |
| Smoking status | ||||
| Never smoker | 12(42.9) | 2(7.1) | 2.4(0.361-15.942) | 0.324 |
| smoker | 10(35.7) | 4(14.3) | 1 | |
| Performance status | ||||
| 0 | 17(60.7) | 5(17.9) | 0.680(0.064-7.254) | 0.617 |
| 1 or 2 | 5(17.9) | 1(3.6) | 1 | |
| ERCC1_202 | ||||
| Low | 10(35.7) | 4(14.3) | 1 | |
| High | 12(42.9) | 2(7.1) | 2.398(0.361-15.873) | 0.324 |
| TNM | ||||
| III | 13(46.4) | 4(14.3) | 0.722(0.108-4.820) | 0.561 |
| IV | 9(32.1) | 2(7.1) | 1 |
ERCC1_202 expression and PFS
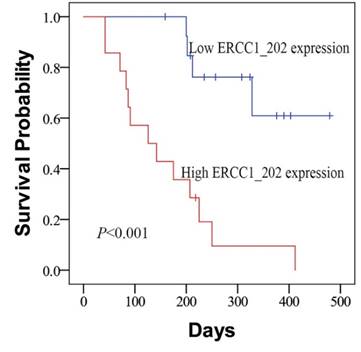
The relationship between clinicopathologic parameters and the PFS was assessed by log-rank test analysis firstly. We found that ERCC1_202 expression level and TNM stage were associated with PFS. Then, the Kaplan-Meier survival curves analysis was carried out and the results showed patients of the low-ERCC1_202 group had a significantly longer median progression-free survival than the high-ERCC1_202 group (450 vs 135 days, Fig. 2, P < 0.001). In the univariate analysis of survival, ERCC1_202 (HR = 6.503, 95% CI = 2.072-20.409, P = 0.001) and TNM stage (HR = 8.324, 95% CI = 2.817-24.595, P = 0.001). In the multivariate model, a high ERCC1_202 level (P = 0.014) together with TNM stage (P = 0.002) are independent prognostic factors for survival (Table 4).
Kaplan-Meier curves for PFS according to the levels of ERCC1_202 in patients with NSCLC
Discussion
In the present study, we firstly measured the four isoforms of ERCC1 in NSCLC cells, and we found that ERCC1_202 was accounted for the major proportion of ERCC1. This result was consistent with Friboulet's [12, 13, 15]. We then used the qPCR method to detect the ERCC1-202 mRNA, thus avoiding the use of antibodies to detect ERCC1-202 protein. Because of the high homology among these 4 isoforms, it was very difficult to design the probe and primers for qPCR. Therefore, the amplification products were extended, meanwhile the former primers were assigned in the E3 region, the reverse primer was assigned to the E10 region, and the probe in E8 region (supplementary Fig. 1 B). From the methods, we were able to directly detect ERCC1_202 in vitro and in NSCLC patients. In vitro, the trend of ERCC1_202 and ERCC1 were consistent, but the former was more obvious, so it can explain why the former was better than the latter. Our clinical results indicated ERCC1_202 was significantly associated with time to progression and progression-free survival, and may be a predictive biomarker for platinum based chemotherapy in NSCLC patients. The 28 patients analysed were treated with cisplatin in combination with one of four other chemotherapeutic agents (etoposide, gemcitabine, pemetrexed, vinorelbine). Etoposide exerts the anti-tumor efficiency through DNA topoisomerase II. Gemcitabine is a cytotoxic drug which destroys cell replication. Pemetrexed play a role of anti-tumor by destroying intracellular folic acid. Vinorelbine can inhibit cell division, because it could induce microtubule formation. These four chemotherapeutic agents have not been reported to be associated with ERCC1 and so are unlikely to be a confounding factor in the analysis.
Factors associated with time to progression (TTP)
| Factor | Median TTP months (95% CI) | Log-rank P | Univariate HR (95% CI) | Cox P | Multivariate HR (95% CI) | Cox P |
|---|---|---|---|---|---|---|
| Age (years) | 0.624 | |||||
| Low (≤60) | 181.2(131.9-230.7) | |||||
| High (>60) | 191.1(124.9-257.3) | |||||
| Sex | 0.742 | |||||
| Female | 167.7(111.6-223.8) | |||||
| Male | 182.2(127.6-236.7) | |||||
| Histology | 0.696 | |||||
| Adenocarcinoma | 161.1(124.3-198.0) | |||||
| Squamous cell carcinoma | 214.3(130.8-297.8) | |||||
| Smoking status | 0.152 | |||||
| Never smoker | 230.9(154.2-307.6) | |||||
| smoker | 145.0(101.0-188.9) | |||||
| Performance status | 0.068 | |||||
| 0 | 201.7(144.8-258.6) | |||||
| 1 or 2 | 111.3(74.1-148.5) | |||||
| Treatment response | 0.402 | |||||
| CR or PR | 201.1(155.3-246.9) | |||||
| SD or PD | 194.7(131.9-257.5) | |||||
| ERCC1_202 | 0.006 | 4.296(1.391-13.266) | 0.011 | 3.252(1.014-10.428) | 0.047 | |
| Low | 271.2(181.3-361.1) | |||||
| High | 126.8(89.4-164.1) | |||||
| TNM | 0.002 | 4.323(1.581-11.819) | 0.004 | 3.332(1.177-9.428) | 0.023 | |
| III | 263.7(187.5-339.9) | |||||
| IV | 115.8(75.1-156.5) |
Factors associated with progression-free survival (PFS)
| Factor | Median PFS months (95% CI) | Log-rank P | Univariate HR (95% CI) | Cox P | Multivariate HR (95% CI) | Cox P |
|---|---|---|---|---|---|---|
| Age (years) | 0.532 | |||||
| Low (≤60) | 290.3(199.8-380.8) | |||||
| High (>60) | 257.1(177.6-336.6) | |||||
| Sex | 0.918 | |||||
| Female | 241.9(145.6-338.1) | |||||
| Male | 268.3(200.3-336.3) | |||||
| Histology | 0.684 | |||||
| Adenocarcinoma | 246.1(182.4-309.9) | |||||
| Squamous cell carcinoma | 281.5(187.6-375.4) | |||||
| Smoking status | 0.386 | |||||
| Never smoker | 309.9(221.6-398.3) | |||||
| smoker | 233.1(155.1-311.0) | |||||
| Performance status | 0.549 | |||||
| 0 | 285.2(212.8-357.6) | |||||
| 1 or 2 | 230.7(103.2-358.1) | |||||
| Treatment response | 0.83 | |||||
| CR or PR | 283.3(221.2-345.4) | |||||
| SD or PD | 265.9(193.1-338.7) | |||||
| ERCC1_202 | 0.002 | 6.503(2.072-20.409) | 0.001 | 4.510(1.353-15.034) | 0.014 | |
| Low | 390.6(320.3-460.9) | |||||
| High | 160.6(101.3-220.0) | |||||
| TNM | 0.001 | 8.324(2.817-24.595) | 0.001 | 5.965(1.935-18.390) | 0.002 | |
| III | 366.2(300.9-431.5) | |||||
| IV | 132.9(79.0-186.8) |
Whether ERCC1 can be used as a predictive biomarker for platinum-based chemotherapy in NSCLC remains to be a controversial issue. Many researchers [16-20] found that the expression of ERCC1 was associated with the clinical outcome of NSCLC treated with platinum-based chemotherapy. In a study conducted by Zhang et al. [21] among 297 advanced stage NSCLC patients who were treated with platinum-based chemotherapy, it was found that ERCC1 mRNA expression levels in blood were related to the treatment response and survival time. Patients with low ERCC1 mRNA expression level in blood had a significantly higher rate of CR to chemotherapy, with an odds ratio (OR) of 1.56 (95%CI: 1.03-2.47). In yet another study where the total tumor samples of 214 advanced stage NSCLC patients were evaluated it indicated that low expression levels of ERCC1 were predictive of a better outcome (P=0.014) [16]. In contrast, contradictory results have been obtained by other researchers. Li et al., informed that ERCC1 expression levels were significantly correlated with the sensitivity of cisplatin in vitro (P < 0.01, r = 0.37), but it was not prognostic in terms of survival time in 112 patients with NSCLC [11]. In the study performed by Lafuente-Sanchis et al. [22], ERCC1 mRNA in fresh-frozen tumor samples of 64 stage I NSCLC patients who received chemotherapy was evaluated. They elucidated that ERCC1was not an independent prognostic factor of recurrence in NSCLC patients. Unlike our method, what these studies examined was the total ERCC1 but not the 202 isoform alone. On the other hand, different expression levels, tumor tissues samples, methods and assessment criteria could also have cause disparity in the results. Here, the ERCC1_202 mRNA expression level in fresh tumor samples was examined by real time RT-PCR method, and the median value of relative expression of ERCC1_202 was used as the assessment criteria. The advantages of our method were cheap, fast, simple and easily quantitative.
Our results also suggested that ERCC1_202 was not an independent predictive factor for response (P = 0.324). But when we analysed the data, we found that lower expression of ERCC1_202 had an impact on the response to treatment. Of the 14 patients with low expression of ERCC1_202 four were CR or PR, while only two of the 14 high expression patients were CR or PR. The P values that were not less than 0.05 may partly be because of insufficient sample size.
However, this study also had several potential limitations. This included the examination of ERCC1_202 expression and cisplatin sensitivity in a relatively small number of samples, and clinical follow-up on cisplatin-based chemotherapy was not long enough. We intended to conduct a retrospective study at the very beginning as there were hundreds of samples for the past five years. However, we found that the RNAs in FFPE specimens preserved 2 years before had completely degraded and was considered improper for further study, so we chose to conduct a prospective study. Sixty-five patients were enrolled for this study in May 2015. But later, 21 excised tissue samples were too small to measure, and the samples from another 5 of them were considered unqualified. Another 11 patients withdrew from the study. Therefore only 28 patients completed the experiment. Finally, the ERCC1_202 expression level was only identified at the mRNA level but not at protein level.
Overall, ERCC1_202 showed a strong effect as a predictive biomarker in the advanced NSCLC patients receiving platinum based chemotherapy. The research in the future should be based on large multi-center prospective trials with the use of standard ERCC1_202 assessment methodology.
Supplementary Material
Supplementary figures and tables.
Abbreviations
ERCC-1, excision repair cross-complementation group 1; NSCLC, non-small cell lung cancer; cDNA, complementary DNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NER, nucleotide excision repair; bp, base pair(s). qPCR, quantitative polymerase chain reaction; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; RR, response rate; PFS, progression-free survival; HR, hazard ratio; TTP, time to progression; IEC, Independent Ethics Committee.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities and the ordinary university graduate student research innovation project of Jiangsu Province, China (no. KYLX15_0187). We thank the National Science Foundation of China (grant numbers 81271636) for the financial support to this work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V. et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. The New England journal of medicine. 2006;355:983-91
2. Hubner RA, Riley RD, Billingham LJ, Popat S. Excision repair cross-complementation group 1 (ERCC1) status and lung cancer outcomes: a meta-analysis of published studies and recommendations. PLoS One. 2011;6:e25164
3. Allingham-Hawkins D, Lea A, Levine S. ERCC1 Expression Analysis to Guide Therapy in Non-Small Cell Lung Cancer. PLoS Curr. 2010;2:RRN1202
4. Azuma K, Komohara Y, Sasada T, Terazaki Y, Ikeda J, Hoshino T. et al. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Sci. 2007;98:1336-43
5. Malottki K, Popat S, Deeks JJ, Riley RD, Nicholson AG, Billingham L. Problems of variable biomarker evaluation in stratified medicine research-A case study of ERCC1 in non-small-cell lung cancer. Lung cancer (Amsterdam, Netherlands). 2016;92:1-7
6. Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J. et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. The New England journal of medicine. 2002;346:92-8
7. Yu D, Shi J, Sun T, Du X, Liu L, Zhang X. et al. Pharmacogenetic role of ERCC1 genetic variants in treatment response of platinum-based chemotherapy among advanced non-small cell lung cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:877-84
8. Sodja E, Knez L, Kern I, Ovcaricek T, Sadikov A, Cufer T. Impact of ERCC1 expression on treatment outcome in small-cell lung cancer patients treated with platinum-based chemotherapy. European journal of cancer (Oxford, England: 1990). 2012;48:3378-85
9. Ota S, Ishii G, Goto K, Kubota K, Kim YH, Kojika M. et al. Immunohistochemical expression of BCRP and ERCC1 in biopsy specimen predicts survival in advanced non-small-cell lung cancer treated with cisplatin-based chemotherapy. Lung cancer (Amsterdam, Netherlands). 2009;64:98-104
10. Tantraworasin A, Saeteng S, Lertprasertsuke N, Arayawudhikul N, Kasemsarn C, Patumanond J. The prognostic value of ERCC1 and RRM1 gene expression in completely resected non-small cell lung cancer: tumor recurrence and overall survival. Cancer management and research. 2013;5:327-36
11. Li C, Liu M, Yan A, Liu W, Hou J, Cai L. et al. ERCC1 and the efficacy of cisplatin in patients with resected non-small cell lung cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:12707-12
12. Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S. et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. The New England journal of medicine. 2013;368:1101-10
13. Friboulet L, Postel-Vinay S, Sourisseau T, Adam J, Stoclin A, Ponsonnailles F. et al. ERCC1 function in nuclear excision and interstrand crosslink repair pathways is mediated exclusively by the ERCC1-202 isoform. Cell cycle (Georgetown, Tex). 2013;12:3298-306
14. Baba H, Baba Y, Uemoto S, Yoshida K, Saiura A, Watanabe M. et al. Changes in expression levels of ERCC1, DPYD, and VEGFA mRNA after first-line chemotherapy of metastatic colorectal cancer: results of a multicenter study. Oncotarget. 2015;6:34004-13
15. Friboulet L, Olaussen KA, Ponsonnailles F, Stoclin A, Dorvault N, Adam J. et al. ERCC1-202 isoform is responsible for all Known functions of ERCC1. CANCER RESEARCH. 2013:73
16. Zhao H, Zhang H, Du Y, Gu X. Prognostic significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with advanced non-small cell lung cancer receiving chemotherapy. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:12679-88
17. Xian-Jun F, Xiu-Guang Q, Li Z, Hui F, Wan-Ling W, Dong L. et al. ERCC1 and BRCA1 mRNA expression predicts the clinical outcome of non-small cell lung cancer receiving platinum-based chemotherapy. Pakistan journal of medical sciences. 2014;30:488-92
18. Qin X, Yao W, Li W, Feng X, Huo X, Yang S. et al. ERCC1 and BRCA1 mRNA expressions are associated with clinical outcome of non-small cell lung cancer treated with platinum-based chemotherapy. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:4697-704
19. Deng Q, Yang H, Lin Y, Qiu Y, Gu X, He P. et al. Prognostic value of ERCC1 mRNA expression in non-small cell lung cancer, breast cancer, and gastric cancer in patients from Southern China. Int J Clin Exp Pathol. 2014;7:8312-21
20. Jian-Wei B, Yi-Min M, Yu-Xia S, Shi-Qing L. Expression levels of ERCC1 and RRM1 mRNA and clinical outcome of advanced non-small cell lung cancer. Pakistan journal of medical sciences. 2013;29:1158-61
21. Zhang H, Li J, Zhang Y, Sun M, Zhao P, Zhang G. et al. ERCC1 mRNA expression is associated with the clinical outcome of non-small cell lung cancer treated with platinum-based chemotherapy. Genetics and molecular research: GMR. 2014;13:10215-22
22. Lafuente-Sanchis A, Zuniga A, Galbis JM, Cremades A, Estors M, Martinez-Hernandez NJ. et al. Prognostic value of ERCC1, RRM1, BRCA1 and SETDB1 in early stage of non-small cell lung cancer. Clin Transl Oncol. 2016;18:798-804
Author contact
![]() Corresponding author: Guoqiu Wu (+86-25-83272355, E-mail: nationballcom)
Corresponding author: Guoqiu Wu (+86-25-83272355, E-mail: nationballcom)

 Global reach, higher impact
Global reach, higher impact