Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(18):3236-3246. doi:10.7150/jca.26051 This issue Cite
Research Paper
Establishment and Characterization of gc-006-03, a Novel Human Signet Ring Cell Gastric Cancer Cell Line Derived from Metastatic Ascites
1. Department of Oncology, Nanjing First Hospital, Nanjing Medical University, Nanjing, China
2. Department of Radiation Oncology, the Affiliated Huai'an Hospital of Xuzhou Medical College, Huai'an, China
*These authors contributed equally to this work
Received 2018-3-14; Accepted 2018-7-17; Published 2018-9-7
Abstract
Signet ring cell gastric cancer (SRCGC) is a special type of gastric cancer with rapid progression and poor prognosis. However, few available SRCGC cell lines from Chinese patients can be used for research, the molecular mechanism of its growth and metastasis is still incompletely understood. In this study, we established and characterized a novel SRCGC cell line, gc-006-03.The cells showed a tendency to pile up without contact inhibition. G-band karyotypes of gc-006-03 were revealed hypotriploid with a modal chromosome number of 51. Immunohistochemistry analysis showed that the cells were positive for CEA, CK7, CDX2 and Ki-67(45%), and negative for CK20, TTF1and Li-cadherin. Flow cytometry analysis showed that gc-006-3 had 25% of CD44+ cells. The cells possessed strong clonality and high plating efficiency, and the doubling time was 36h. The cells grew vigorously for more than 100 passages in serial culture. Meanwhile, the cells showed a high rate of tumor formation. Tumors were observed in all of the nude mice (5/5) given injections of the cells. The metastatic capability of the cell line was found in zebrafish injected the cells. The results of whole genome sequencing revealed the unique genomic characteristics of gc-006-03. In summary, this new stable cell line may be useful in basic and clinical research on gastric signet ring cell carcinoma.
Keywords: Signet ring cell gastric cancer, cell line, whole genome sequencing, TP53, CD44.
Introduction
Gastric cancer is the world's second most deadly cancer, with poor survival especially in the advanced patients [1-3]. Signet ring cell gastric cancer (SRCGC) is a special type of gastric cancer diagnosed by its microscopic features of signet ring cells [4]. It is reported that the incidence of SRCGC has grown to 99.8% in recent five years [5]. SRCGC has a specific epidemiology and oncogenesis and has two forms: early gastric cancer and advanced gastric cancer. The two forms have different prognosis to non-SRCGC. Patients with SRCGC had a better survival than patients with other gastric adenocarcinomas in 1520 early gastric cancer patients [6]. However, more than 3500 patients with advanced SRCGC showed a significantly worse 5-year survival rate than in non-SRCGC [7, 8]. Unfortunately, SRCGC is more frequent in stage4, T3/T4 and N2 gastric cancers than in early gastric cancer. Meanwhile, SRCGC is thought to be less chemosensitive than non-SRCGC, but recent reports suggest that it could have a specific sensitivity profile and be more sensitive to taxane-based chemotherapy or antiangiogenics [9]. However, basic research on SRCGC is still incomplete due to the lack of the suitable cell model. Therefore, it is absolutely necessary to establish a stable and available SRCGC cell line.
Cell lines established from human gastric cancers may provide useful tools to study the biology of the disease and to develop and test new therapeutic approaches. A large bank of well characterized cell lines should reflect the diversity of tumor phenotypes and provide adequate models for the study of tumor heterogeneity. Generally, well-characterized gastric carcinoma cell lines have proven to be more difficult to establish in long-term culture than colorectal carcinoma cell lines. The mostly number of gastric carcinoma lines have been described from Korea [10, 11] and Japan [12-15], A few lines have been established in other countries, including China [16-24]. We have established three cell lines of gastric carcinoma derived from primary tumors and metastatic sites in 32 Chinese patients since 2016. But only the gc-006-03 cell line, which an immortal human signet ring cell gastric cancer cell line derived from metastatic ascites, was cultured with high efficiency and well-characterized. This line would serve as a valuable tool for gastric carcinoma research.
Herein we describe the establishment and characterization of the gc-006-03 cell line. The cell line has been analyzed with respect to the growth property, cellular ultrastructure, neoplastic behavior in athymic BALB/c nude mice and zebrafish assay, karyotype, the expression of tumor-associated antigen, cell Line authentication by short tandem repeat (STR) profiling, excepted mycoplasma contaminated.
Massively sequencing technologies promise the capacity to paint a genome-wide portrait of mutation in gastric cancer. In order to comprehensively characterize the genome of this cell line and to serve as a model of SRCGC genome sequencing, we have generated 30× genomic sequence coverage using a PE150 sequence strategy. Here we counted results for point mutations, small insertions/deletions, copy number variation and structure variation in this cell. According to some well-known driver genes in gastric cancer (GC), status of these exon alterations in this cell line were presented for details, and organized mutated genes from the perspective of related biological pathways.
Materials and Methods
Establishment and cell culture conditions
The cell line was derived from metastatic ascites of a 55-year-old Chinese female patient with SRCGC, taken after 5-fluorouracil(5-FU) combined with oxaliplatin chemotherapy, and informed consent was signed by the patient before the study. First we extracted the patient's ascites, then separated the cells by partial solution of 1.077g density. Cancer cells were cultured in 1640 medium (Gibco) with 10% fetal bovine serum (FBS, Gibco), 10mM HEPES (Gibco), 100u/ml penicillin G (Gibco), 100µg/ml streptomycin sulfate (Gibco) and 0.25µg/ml amphotericin B (Gibco). After that, the cells were incubated in an atmosphere containing 5%C02 at 37℃.By repeating subculture of the cells in vitro, we established a signet ring cell gastric cancer cell line named gc-006. Furthermore, picked distinct monoclonal cells from gc-006 cell line were cultured into coated poly-L-sine in a 24-well plate containing 1ml of 1640 complete medium. And so gc-006-03 cell line (the subline of gc-006) was established.
Ultrastructural studies
Cell pellets were fixed in 2.5% glutaraldehyde, postfixed in 1% osmium tetroxide, stained with 1% uranyl acetate, and thin sections were examined by a Siemens 1A electron microscope.
Cell Line Authentication by Short Tandem Repeat (STR) profiling
The cell line, together with positive and negative control were amplified using GenePrint 10 System (Promega). Then, amplified products were processed using the ABI3730xl Genetic Analyzer. Data were analyzed using GeneMapper4.0 software and then compared with ATCC, DSMZ or JCRB databases for reference matching. STR assay at China Center for Type Culture Collection (Wuhan, China).
Chromosomal analysis
The logarithmic phase cells of 5, 10, 25 passages were taken for karyotype analysis,respectively. The cells were treated with colchicine for 2h,then with 0.075mol/L KCL solution, fixed with acetic acid glacial, dropped with ice wet glass, disposed with pancreatic enzyme after aging at room temperature, finally, utilized with Giemsa staining for banding analysis.
Immunohistochemical analysis
Morphology of the cells was confirmed using antibodies directed against target protein at the 30th passage. Cells were plated in 100-mm plates and grew to approximately 90% confluence. For immunohistochemical staining, tumor cells were collected by centrifuging as cell pellet. Slowly added neutral formalin into the tube and fixed the cell pellet overnight. Then processed the cell pellet to paraffin block for embedding which could be used for making histological sections.The sections were dewaxed with xylene and rehydrated with graded alcohol, then briefly immersed in water. Endogenous peroxidase activity was blocked by incubating the sections with 3% hydrogen peroxide for 10 min. For all biomarkers, heat-mediated antigen retrieval was performed by heating the sections (immersed in 0.01 mol/L citrate buffer, pH 6.0) in a microwave oven (450 W) for 10 min. The slides were then washed with phosphate-buffered saline (PBS) before incubation with respective primary antibody overnight at 4 °C. After being washed with PBS, the slides were incubated for 1 h with the secondary antibody, and then further washed for 3 × 5 min with PBS. Peroxidase reaction was developed in PBS using hydrogen peroxide as a substrate and DAB as a chromogen. Sections were counterstained with hematoxylin, dehydrated, and evaluated under a light microscope. Negative control sections were processed immunohistochemically after the primary antibody was replaced with PBS. Anti-vimentin (MA5-11883, Invitrogen), anti-CK7 (9021, abcom), anti-CK20 (854, abcom), anti-TTF1 (72876, abcom), anti-Li-cadherin (204960, abcom), anti-CDX2 (157524, abcom), anti-CEA (4451, abcom) and anti-Ki-67 (8191, abcom) were used in this study.
Flow cytometry analysis
Ninety percent confluent culture cells in a 100-mm cell plate were dissociated from plates using trypsin-EDTA, centrifuged, resuspended up to 107 nucleated cells per 100 µL of buffer, and then stained with 10 µL of human CD44-FITC (130-098-210, Miltenyi Biotec), isotype control antibody Mouse IgG1-FITC (130-098-847, Miltenyi Biotec) or human CD326 (EpCAM)-PE (130-098-115, Miltenyi Biotec), isotype control antibody Mouse IgG1-PE (130-098-845, Miltenyi Biotec) at 4℃ for 10 min, respectively. Analyses were performed using a BD FACSCanto Ⅱ (BD Biosciences).
Cell proliferation and colony formation assay
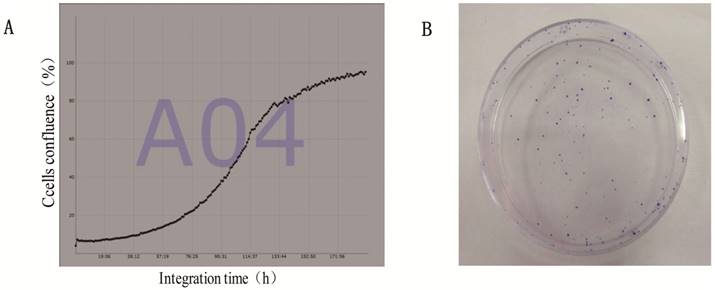
Suspensions of 1×103 logarithmic phase cells were placed on 24 well plates to ensure 1000ul per well, with 5 repeating wells each, respectively. Then, the cells were placed in JuLi Stage real-time cell count meter, counted every hour and recorded for total eight days. The population doubling time was calculated from the growth curve, which was drawn by cells confluence and integration time, according to the formula that (PDT)=(t-t0)lg2/(lgNt-lgN0). For studies of plating efficiency, 100 single suspension cells were placed onto three-60mm dishes each and cultured for 9 days. Plating efficiency was calculated by the ratio of the number of clones (more than 50 cells after crystal violet stain) to the total number of inoculated cells.
Nude mouse tumorigenicity assay
Tumorigenicity of gc-006-03 cell line was tested by inoculating 5×106 cells s.c. into one side shoulder and the contralateral hip of each of 5 male athymic nude mice, BALB/c background (Comparative Medicine Centre of Yangzhou University, Jiangsu, China) and observing one weekly for progressive growth. All animal studies were performed according to protocols approved by Animal Ethical and Welfare Committee of Nanjing Medical University (NJMU).
Zebrafish assay
Zebrafish zygotes were harvested and incubated for 24h in an incubator at 28.5°C. 1-Phenyl-3- (2-thiazolyl) -2-thiourea (PTU)(sigma) was added to water of the incubator to make its final concentration 0.2mM, and the zygotes were incubated for another 24h. Then, removed zebrafish embryonic membrane and injected tumor cells of gc-006-03 which had treated with Dil solutions (C7000, Invitrogen) before into zebrafish yolk sac. Four days after injection, Fluorescence microscopy (D-eclipse C1, Nikon) was used and the fluorescence intensity was analyzed using the image pro plus (6.0) software. All animal studies were performed according to protocols approved by Animal Ethical and Welfare Committee of NJMU.
DNA extraction
DNA was extracted with Qiagen extraction kits (Qiagen GmbH, Hilden, Germany) from the cell line.
Whole genome sequencing
Library preparation and sequencing were performed at Bionova Company.In brief, genomic DNAs were sheared using Covaris instrument to average size of 500 bp, fragment purified, ends repaired, A-tailed, ligated with appropriate adapters and amplified using Illumina sequencing primers. The libraries were applied to pair-end sequencing with read lengths of 150 nt on Illumina HiSeqXten platform.
Results
Cytomorphology
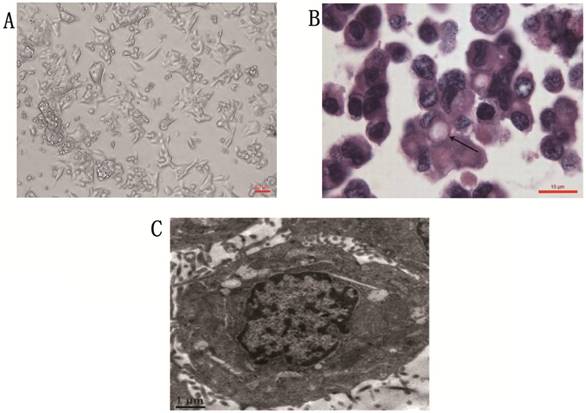
Phase contrast microscope revealed that the gc-006-03 cell line was strictly anchorage-independent, and formed multi-layered sheets with clusters upon confluence, was without obvious contact inhibition phenomenon (Figure 1A). Stained with hematoxylin and eosin, the cells presented morphology of malignant epithelial cells, and showed typical signet ring cell morphology (Figure 1B). Organelles were visible under the transmission electron microscopy, such as ribosome, rough endoparasitic reticulum and golgi complexes (Figure 1C).
STR matching analysis
By comparing the results of STR matching analysis with the information of cells in ATCC, DSMZ and JCRB, there had not found any gc-006-03 homologous cells. This proved the uniqueness of the cell line.
Chromosomal analysis
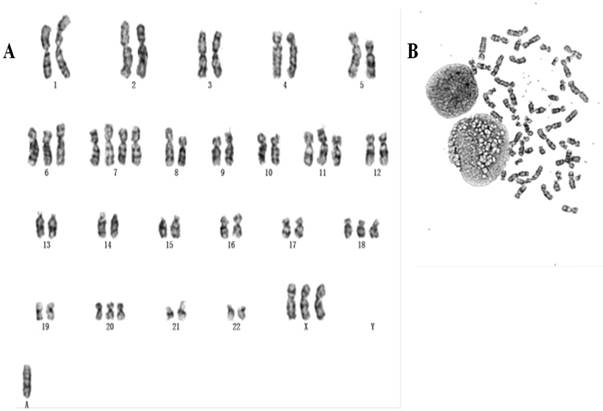
Chromosomes of 50 metaphase plates were analyzed for passage 5, 10, 25 respectively, indicating gc-006-03 relative stability. The karyotypes of over 58.5% of the cells fell within the modal chromosome number of 51±2, and showed a unimodal distribution. Representative karyotype of the clone with 54 chromosomes in gc-006-03 has been observed as 54, XX, +3, +7, +7, +8, +13, +18, +19, 20 (Figure 2A and 2B). And also they were founded double centromere, ectopic and derivative phenomenon.
Cytomorphology of cells from gc-006-03. (A) Cells were photographed under microscope. Scale bar, 50μm. (B) H&E staining of the gc-006-03 cells showed typical signet ring cell morphology (indicated by the arrows). Scale bar, 10μm. (C) Cell structure under the transmission electron microscopy. Scale bar, 1μm.
(A, B) Representative karyotype of the gc-006-03 cells. Approximately 58.5% of the gc-006-03 cells possessed the modal chromosomes number of 51±2.
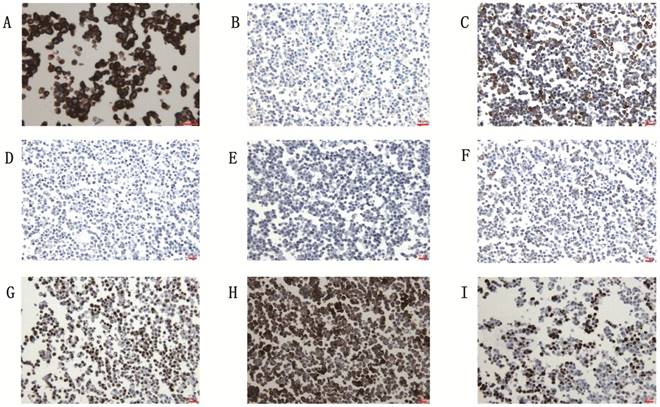
The cell line showed the characteristic of the epithelial tumor cells: keratin(+) (A), vimentin (-) (B) . Immunohistochemical staining with anti-ck7 (C), anti-ck20 (D), anti-ttf1 (E), anti-Li-cad (F), anti-adx2 (G), anti-CEA (H) and anti-Ki-67 (I). Scale bar, 10μm.
Immunohistochemistry
The gc-006-03 cell line was confirmed to the characteristic of epithelial tumor cells: cytokeratin (+), vimentin (-) (Figure 3A and 3B). Because of the cell line was source from the ascites of advanced patient with signet ring cell gastric cancer, so CK7, CK20 and TTF1 testing was needed to identify the source of the cells. Immunohistochemical analysis showed CK7 (2+), CK20 (-) and TTF1 (-) in gc-006-03 (Figure 3C, 3D and 3E), conformed to the characteristics of gastric carcinoma. Furthermore, the cell line presented Li-cadherin (+), CDX2 (2+), CEA (3+) and 45% positive of Ki-67(Figure 3F, 3G, 3H and 3I), which showed more biological characteristics of gastric carcinoma.
CD44 and CD326 expression assay
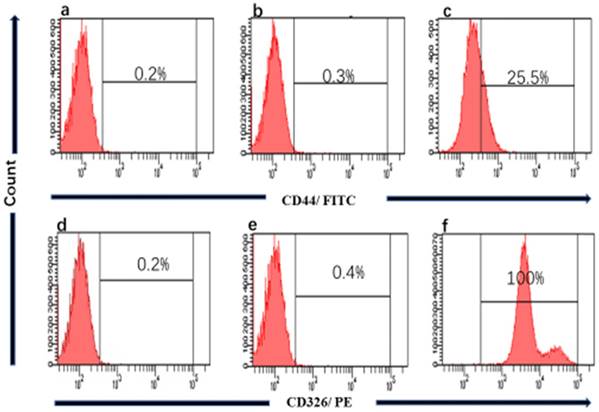
We performed the Flow cytometry analysis to confirm the source of the cell line and clarity the proportion of cancer stem cells (CSCs) in gc-006-03. The results showed that nearly 100% of CD326+ in gc-006-03 cells were compared to the control group (Figure 4), meaning that gc-006-03 derived from epithelial cells. Meanwhile, about 25.5% of CD44+ in gc-006-03 cells were compared to the control group (Figure 4). Therefore, this cell line could be used in the study of cancer stem cells theory.
Cell proliferation and clone formation ability
The average of doubling time for the gc-006-03 cells was 36h (Figure 5A).And the cells had a strong ability of clone formation, with approximately 70% cloning efficiency in soft agar (data was not shown here),while 88% in a 60mm cell plate (Figure 5B).
Tumorigenicity and metastatic capability in vivo
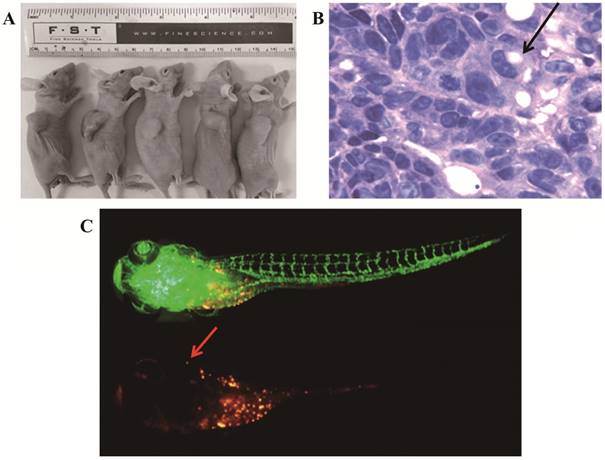
To confirm its tumorigenicity in vivo, we injected viable cells (5×106/site) into nude mice. Strikingly, xenograft tumors initiated with 100% incidence (5/5) after 2 weeks (Figure 6A). The hematoxylin- and eosin-stained tissue sections of the mice tumor xenografts showed histologic features of human signet ring cell gastric carcinoma, a few signet ring cells (Figure 6B). We used zebrafish to study the metastatic capability of gc-006-03 cells. After 7 days post injection (dpi), cancer cells showed a tendency to metastasise in zebrafish (Figure 6C).
Whole genome sequencing
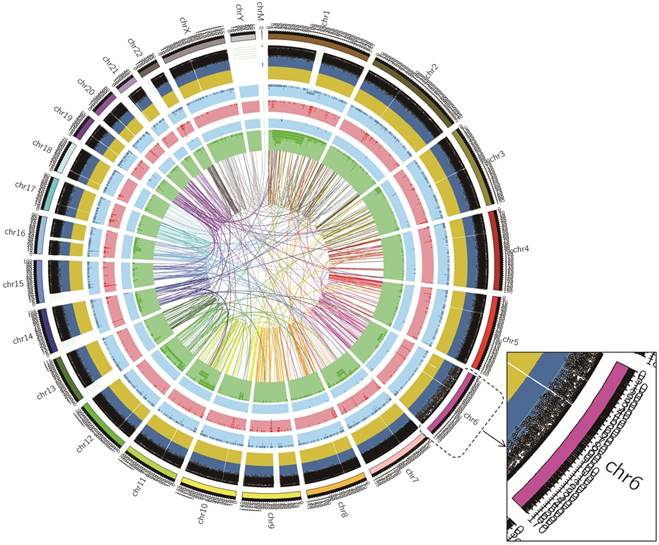
The DNA sample of the cell line was processed for the whole genome sequencing, the results showed 143,111.94Mb of the original bases. After the removal of low-qualitu reads, the sample got 922,451,234 clean reads (138,367.69Mb). The clean reads of the sample had high value of Q20 and Q30, indicating that sequencing data had good quality. The average content of GC was 40.91%.The statistics for the distribution of SNP, InDel, CNV and SV were shown in Table 1. Also, we showed the circos plot of whole genome sequencing (Figure 7). Meanwhile, we selected 23 important gastric cancer driven genes, and showed their corresponding SNP, InDel, CNV or SV variants in exon regions (Table 2). (We named this cell line gc-006-03 and the company labeled this cell line as gc_006_03).
Flow cytometry of CD44 and CD326 expression. a and d Histograms showing absence of CD44, CD326 marker respectively in gc-006-03 cells, when no antibody were used (technical control). b, e Histograms of mouse IgG1 stained with FITC-conjugated, with PE-conjugated mouse monoclonal antibody respectively, against Human CD44, CD326 respectively (isotype control). c and f Histograms of gc-006-03 cells stained with FITC-conjugated, PE-conjugated mouse monoclonal antibody respectively, against Human CD44 and CD326 showing.
Proliferation and clone formation ability of gc-006-03 cells. (A)Growth curves of gc-006-03 cells. Doubling time was 36h. (B)100 cells was used to clone formation assay in a 60mm cell plate.
Discussion
Despite the cell lines may only present a part of the characteristics of the tumor [25], it is still necessary to establish a stable model of advanced SRCGC to study the mechanism of its genesis and development, and then so as to guide the clinical treatment. Which cells of SRCGC established from primary tumors are relatively difficult to culture in vitro as permanent cell lines because of overgrowth of fibroblasts. The use of body fluids as a starting material for cell culture provides many technical advantages and has already been used for the establishment of signet ring cell carcinoma of the stomach [22, 26]. In this literature, for the first time, we isolated and established a new cancer cell line from metastatic ascites of the Chinese female patient with SRCGC, diagnosed by its microscopic characteristics as described by the World Health Organization, with a worse prognosis than other forms of gastric cancer in Asia and the United States [27, 28], named gc-006-03. And we confirmed that the cell line was not contaminated with mycoplasma, bacteria and fungi.
Tumorigenicity and metastatic capability in vivo. (A)The gc-006-03 cells formed tumors in 100% of mice (5 to 5 mice). (B)H&E staining of the tumor specimens in mice showed obvious signet ring cell charactistics (indicated by the arrows). (C)The gc-006-03 cells in zebrafish metastasized to its brain (indicated by the arrows).
Circos plot of whole genome sequencing. Chromosome ideograms are shown around the outer ring and are oriented pter-qter in a clockwise direction. Other tracks, which were divided into five parts, contain somatic alterations from outside to inside. SNP was indicated by the black spots according to the position. InDel results were represented in dark-blue and yellow layers, where dark-blue indicated deletion, yellow indicated insertion. Both light-blue and light-red layers represent these results of CNV, with blue rectangles representing genomic deletion of CNV, and red rectangles representing genomic duplication of CNV. Both light-blue and green layer and the lines of innermost represented the relative position of SV, blue rectangles indicated chromosomal deletion, dark-green rectangles indicated chromosomal inversion, and the line represented chromosomal translocations.
The results of the whole genome sequencing: statistics for the distribution of SNPs, InDels, CNVs and SVs.
| Term | Number |
|---|---|
| Total SNPs | 3,543,987 |
| Novel | 105,448 |
| Homozygous | 1,635,472 |
| Heterozygous | 1,908,515 |
| Intronic | 1,384,385 |
| 5' UTRs | 3,605 |
| 3' UTRs | 22,615 |
| SNPs in the coding region | |
| Synonymous | 10,551 |
| Missense | 10,153 |
| Stopgain | 97 |
| Stoploss | 26 |
| Startloss | 19 |
| Splicing | 182 |
| TI/Tv | 3.13 |
| Total InDels | 1,333,695 |
| Novel | 789,571 |
| Homozygous | 397,809 |
| Heterozygous | 935,886 |
| Intronic | 591,345 |
| 5' UTRs | 766 |
| 3' UTRs | 10,652 |
| InDels in the coding region | |
| Frames hift | 622 |
| Non-frameshift Insertion | 153 |
| Non-frameshift Deletion | 192 |
| Stoploss | 11 |
| Startloss | 10 |
| Splicing | 81 |
| Total CNVs | 2,628 |
| Exonic | 1,097 |
| Splicing | 2 |
| NcRNA | 0 |
| Intronic | 402 |
| 5' UTRs | 0 |
| 3' UTRs | 1 |
| Upstream | 48 |
| Downstream | 25 |
| Intergenic | 1,053 |
| Total SVs | 1,934 |
| Insertion | 0 |
| Deletion | 38 |
| Inversion | 582 |
| ITX | 992 |
| CTX | 322 |
| Exonic | 110 |
| Splicing | 6 |
| NcRNA | 0 |
| Intronic | 1,293 |
| 5' UTRs | 1 |
| 3' UTRs | 35 |
We used a series of immune histochenmical markers to confirm that the cell line has an epithelia nature of the stomach, instead of derived from the lungs or the thyroid gland. Furthermore, gc-006-03 expressed CDX2, Li-cadherin, and strongly expressed CEA. All of these three tumor markers are important to diagnosis, therapy control and monitoring of gastric cancer. However, CDX2 suggests a good prognosis of gastric cancer while Li-cadherin and CEA not. In general, different markers suggested different prognosis of gastric cancer. Therefore, in addition to gastric cancer, we still need to find more useful markers for signet ring cell gastric cancer. Moreover, gc-006-03 slightly expressed Ki-67. Ki-67 protein is widely used as a cellular marker for proliferation [29], and it is often studied in breast cancer, which is used as a marker of cellular proliferation and response to chemotherapy, and also is associated with patient prognosis [30-35]. However, the role of Ki-67 in gastric cancer is still not clear except that it can be used as a biomarker for predicting the prognosis of early gastric cancer [36]. Therefore, this cell line is helpful for the study of the mechanism of Ki-67 in cellular proliferation in gastric cancer.
Example of exon alterations in the gc-006-03 cell line.
| Classification | Gene symbol | Status of Genetic alterations (SNP,InDel,CNV and SV) |
|---|---|---|
| Cell cycle regulation | TP53 | p.Pro72Arg/c.215C>G |
| PTEN | Wild-type | |
| TTK | Wild-type | |
| CDKN2A | p.Glu33fs/c.97delG | |
| BRCA2 | p.Asn289His/c.865A>C, p.Asn991Asp/c.2971A>G, p.Val2466Ala/c.7397T>C, | |
| Chromatin remodeling | ARID1A | ARID1A-Del* |
| Cell adhesion | CDH1 | Wild-type |
| FAT4 | p.Arg172Cys/c.514C>T, p.Gln453Leu/c.1358A>T, p.Ala807Val/c.2420C>T, p.Gly833Glu/c.2498G>A, | |
| p.Gln1257Glu/c.3769C>G, p.Gly3526Asp/c.10577G>A, p.Ile3604Leu/c.10810A>C, | ||
| p.Ser3875Asn/c.11624G>A, p.Cys3928Tyr/c.11783G>A, | ||
| CTNNA1 | Wild-type | |
| CD44 | p.Lys417Arg/c.1250A>G, p.Ile479Thr/c.1436T>C, CD44...SLC1A2-ITX** | |
| Cytoskeleton and cell motility | RHOA | Wild-type |
| ALK pathway | CTNNB1 | Wild-type |
| APC | p.Val1822Asp, p.Ile169fs/c.505_508delATAG, p.Glu1554_Lys1555fs/c.4661_4662insA | |
| RNF43 | p.Leu418Met/c.1252C>A, p.Arg117His/c.350G>A, p.Ile47Val/c.139A>G, | |
| SMAD4 | Wild-type | |
| PIK pathway | PIK3CA | p.Glu545Gly/c.1634A>G |
| ERBB signaling | ERBB2 | p.Pro1140Ala/c.3418C>G, p.Pro1155Ala/c.3463C>G, p.Pro1170Ala/c.3508C>G |
| KRAS | Wild-type | |
| NRAS | Wild-type | |
| DNA replication | MSH6 | Wild-type |
| MSH3 | p.Ile79Val/c.235A>G, p.Gln949Arg/c.2846A>G, p.Ala1045Thr/c.3133G>A, p.Lys381fs/c.1141delA, | |
| MUTYH | p.Gln324His/c.972G>C, | |
| Fas signaling | MET | Wild-type |
Annotate: ITX: intrachromosomal translocation; fs: framshift; ins: insert; del: deletion.
* copy number losses of ARID1A at the chromosome 1 27022501-27024300
** structural variation of CD44 and SLC1A2 occurred intrachromosomal translocation
CD44 is a multi-structural and multifunctional cell surface molecule and the major cell surface receptor for hyaluronic acid [37, 38]. Many results show that CD44 plays an important role in Cancer stem cells (CSCs), including mediation of adhesion and indirect enhancement of the expression of antiapoptotic proteins [39-41]. Meanwhile, CD44 may be a new target for the treatment of gastric cancer [42]. We found that the positive rate of CD44 in gc-006-03 was 25%. So, this cell line may be a useful model for the study of CSCs and the targeted therapy of gastric cancer. Apart from this, structural variation in CD44-SLC1A2 gene was found in the data of structure variation (SV) and CD44-SLC1A2 may represent a class of gene fusions in gastric cancer that establish a pro-oncogenic metabolic milieu favoring tumor growth and survival [43].
We have sequenced the whole genome of the cell line and obtained the total results of SNP, InDel, CNV and SV in our study. Meanwhile, we listed the common mutation sites in the exon of 23 driver genes of gastric cancer, such as TP53, APC, ERBB2 and RNF43. Among them, TP53 Pro72Arg polymorphism (rs1042522) was the most widely studied. By encoding a protein called p53, TP53 gene involved in cell cycle control, apoptosis, senescence, and maintenance of DNA integrity [44]. Many studies have been performed to investigate the association between this TP53 polymorphism and the risk of cancer, including cervical cancer, colorectal cancer, breast cancer, lung cancer, ovarian cancer, and endometrial cancer [45]. FAT4 plays a crucial role in carcinogenesis as a key component of the Hippo signaling pathway [46, 47]. FAT4 Ala807Val polymorphism (rs1039808) is associated with the risk of esophageal cancer in non-drinkers in an Eastern Chinese population [48]. The function of MSH3 is to eliminate replication errors and maintain genomic stability and its gene polymorphisms have been studied in many kinds of tumors [49, 50]. Hirata et al. found evidence for an increased risk of sporadic prostate cancer with single nucleotide polymorphisms (SNPs) in the MSH3 variants rs1805355 and rs26279 [50]. MSH3 Ala1045Thr polymorphism (rs26279) was associated with the sensitivity to platinum-based chemotherapy in advanced non-small cell lung cancer patients [51]. MUTYH, as a base excision gene, plays an important role in many cancers and loss of MUTYH function in human cells could lead to accumulation of oxidative damage and genetic instability [52]. Many studies have revealed the relationship between MUTYH Gln338His polymorphisms (rs3219489) and the risk of many cancers, such as breast cancer [53], esophageal adenocarcinoma [54] and colorectal cancer [55]. Thus, these mutations in gc-006-03 also reveal its potential for further biological research.
In addition, many new mutations were found in these driven genes, which may reveal some new mechanism in cancer. CDKN2A, as an important tumor suppressor, prevents carcinogenesis via induction of cell growth arrest and senescence [56, 57]. The BRCA2 gene encodes a tumor suppressor protein which helps repair damaged DNA and ensures stability of cellular genetic material, and its mutations may increase or reduce the risk of cancer at various sites [58]. RNF43, as a tumor suppressor gene, restricts Wnt/β-catenin signaling pathway by removing Wnt receptors from the cell surface [59]. The ERBB2 gene promotes cell proliferation and suppresses apoptosis by encoding HER2 protein in gastric cancer [60-62]. Because some of these mutations are novel, they require further functional characterization. In our study, we only analyzed a very small fraction of the whole genome sequence, so we were looking for a platform to save the full results for your future research to download.
In summary, the cell line gc-006-03 was derived from metastatic ascites of an advanced patient with SRCGC. Gc-006-03 is a well-established cell line with its specific biological characteristics. Studying protein profile of this cell line may provide some new tumor-associated markers. Also this cell line might be a good model for the study of cancer stem cells and targeted therapy of SRCGC. At the same time, the whole genome sequencing of cell line also provides the direction for the study of the gene of SRCGC. However, according to the results of genome sequencing, we still need more studies to further explore its genetic characteristics.
Abbreviations
SRCGC: signet ring cell gastric cancer; CEA: carcinoembryonic antigen; CK7: cytokeratin 7; CK20: cytokeratin 20; CDX2: caudal type homeobox transcription factor 2; TTF1: human transcription termination factor 1; STR: short tandem repeat; GC: gastric cancer; PDT: population doubling time; CSCs: cancer stem cells; SNP: single nucleotide polymorphisms; InDel: insertion-deletion; CNV: copy number variations; SV: structure variation.
Acknowledgements
The work was partly supported by grants from the National Natural Science Foundation of China (Grant No. 81272469), the National 973 Basic Research Program of China (Grant No. 2013CB911300) and the clinical special project for Natural Science Foundation of Jiangsu Province (Grant No. BL2012016), and a grant from The Project of Plans for the Development of Science and Technology of Nanjing, China (Grant No. 201208020), and the grant from Nanjing 12th Five-Year Key Scientific Project of Medicine to Jinfei Chen.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:1893-907
2. Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M. et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. The New England journal of medicine. 2006;355:11-20
3. Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN. et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. The New England journal of medicine. 2001;345:725-30
4. Jass JR, Sobin LH, Watanabe H. The World Health Organization's histologic classification of gastrointestinal tumors. A commentary on the second edition. Cancer. 1990;66:2162-7
5. Henson DE, Dittus C, Younes M, Nguyen H, Albores-Saavedra J. Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973-2000: increase in the signet ring cell type. Archives of pathology & laboratory medicine. 2004;128:765-70
6. Ha TK, An JY, Youn HK, Noh JH, Sohn TS, Kim S. Indication for endoscopic mucosal resection in early signet ring cell gastric cancer. Annals of surgical oncology. 2008;15:508-13
7. Kim JP, Kim SC, Yang HK. Prognostic significance of signet ring cell carcinoma of the stomach. Surgical oncology. 1994;3:221-7
8. Li C, Kim S, Lai JF, Hyung WJ, Choi WH, Choi SH. et al. Advanced gastric carcinoma with signet ring cell histology. Oncology. 2007;72:64-8
9. Hultman B, Mahteme H, Sundbom M, Ljungman M, Larsson R, Nygren P. Benchmarking of gastric cancer sensitivity to anti-cancer drugs ex vivo as a basis for drug selection in systemic and intraperitoneal therapy. Journal of experimental & clinical cancer research: CR. 2014;33:110
10. Park JG, Frucht H, LaRocca RV, Bliss DP Jr, Kurita Y, Chen TR. et al. Characteristics of cell lines established from human gastric carcinoma. Cancer research. 1990;50:2773-80
11. Park JG, Yang HK, Kim WH, Chung JK, Kang MS, Lee JH. et al. Establishment and characterization of human gastric carcinoma cell lines. International journal of cancer. 1997;70:443-9
12. Hojo J. Establishment of cultured cell lines of human stomach cancer origin and their morphological characteristics. 1977.
13. Motoyama T, Hojo H, Watanabe H. Comparison of seven cell lines derived from human gastric carcinomas. Acta Pathologica Japonica. 1986;36:65
14. Kubota H, Harada T, Morikawa S, Nakamura T. In vivo selection of tumorigenic subline from non-tumorigenic human gastric carcinoma cells: In relation to proliferative properties In vivo and in nude mice. Gastroenterologia Japonica. 1988;23:371-83
15. Whelan R, Gibby E, Sheer D, Povey S, Hill BT. Characterization of a continuous cell line in culture established from a Krukenberg tumour of the ovary arising from a primary gastric adenocarcinoma. European Journal of Cancer & Clinical Oncology. 1988;24:1397-408
16. Dobrynin YV. ESTABLISHMENT AND CHARACTERISTICS OF CELL STRAINS FROM SOME EPITHELIAL TUMORS OF HUMAN ORIGIN. Jnci Journal of the National Cancer Institute. 1963;31:1173-95
17. Wolff E, Wolff E. Cultures organotypiques de longue durée de deux tumeurs humaines du tube digestif. European Journal of Cancer. 1966;2:93-102
18. Smith HS. In vitro properties of epithelial cell lines established from human carcinomas and nonmalignant tissue. J Natl Cancer Inst. 1979;62:225-30
19. CL L, C A, MH C-T, C G, AM C, F P. Characterization of a newly established human gastric cancer cell line HGT-1 bearing histamine H2-receptors.Cancer Res. 42:1541. Cancer research. 1982;42:1541-8
20. Barranco SC, Townsend CM Jr, Casartelli C, Macik BG, Burger NL, Boerwinkle WR. et al. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer research. 1983;43:1703-9
21. Lin CH, Fu ZM, Liu YL, Yang JL, Xu JF, Chen QS. et al. Investigation of SGC-7901 cell line established from human gastric carcinoma cells. Chinese Medical Journal. 1984;97:831
22. Dippold WG, Kron G, Boosfeld E, Dienes HP, Klingel R, Knuth A. et al. Signet ring stomach cancer: morphological characterization and antigenic profile of a newly established cell line (Mz-Sto-1). European Journal of Cancer & Clinical Oncology. 1987;23:697 703-1,6
23. Kai-hua W. A in vitro cell line (MGc80-3) of a poorly differentiated mucoid adenocarcinoma of human stomach (in chinese). Acta Biologiac Experimentalis Sinica. 1983:20-30
24. Yuhui C, Juezhi G, Jiaqing Z, Ping L, Qinghuan z. Establishment of a human gastric cancer cell line BGC-823 (in chinese). Journal of Peking University (Health Sciences). 1986
25. Elsasser HP, Lehr U, Agricola B, Kern HF. Structural analysis of a new highly metastatic cell line PaTu 8902 from a primary human pancreatic adenocarcinoma. Virchows Archiv B, Cell pathology including molecular pathology. 1993;64:201-7
26. Sekiguchi M, Sakakibara K, Fujii G. Establishment of cultured cell lines derived from a human gastric carcinoma. Japanese Journal of Experimental Medicine. 1978;48:61-8
27. Bamboat ZM, Tang LH, Vinuela E, Kuk D, Gonen M, Shah MA. et al. Stage-stratified prognosis of signet ring cell histology in patients undergoing curative resection for gastric adenocarcinoma. Annals of surgical oncology. 2014;21:1678-85
28. Taghavi S, Jayarajan SN, Davey A, Willis AI. Prognostic significance of signet ring gastric cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:3493-8
29. Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. Journal of cellular physiology. 2000;182:311-22
30. Niikura N, Sakatani T, Arima N, Ohi Y, Honma N, Kanomata N. et al. Assessment of the Ki67 labeling index: a Japanese validation ring study. Breast cancer (Tokyo, Japan). 2016;23:92-100
31. Ohno S, Chow LW, Sato N, Masuda N, Sasano H, Takahashi F. et al. Randomized trial of preoperative docetaxel with or without capecitabine after 4 cycles of 5-fluorouracil- epirubicin-cyclophosphamide (FEC) in early-stage breast cancer: exploratory analyses identify Ki67 as a predictive biomarker for response to neoadjuvant chemotherapy. Breast cancer research and treatment. 2013;142:69-80
32. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology: official journal of the European Society for Medical Oncology. 2011;22:1736-47
33. Niikura N, Iwamoto T, Masuda S, Kumaki N, Xiaoyan T, Shirane M. et al. Immunohistochemical Ki67 labeling index has similar proliferation predictive power to various gene signatures in breast cancer. Cancer science. 2012;103:1508-12
34. Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:7212-20
35. de Azambuja E, Cardoso F, de Castro G Jr, Colozza M, Mano MS, Durbecq V. et al. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. British journal of cancer. 2007;96:1504-13
36. Ko GH, Go SI, Lee WS, Lee JH, Jeong SH, Lee YJ. et al. Prognostic impact of Ki-67 in patients with gastric cancer-the importance of depth of invasion and histologic differentiation. Medicine. 2017;96:e7181
37. Gallatin WM, Weissman IL, Butcher EC. A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature. 1983;304:30-4
38. Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303-13
39. Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R. et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. The American journal of pathology. 1999;154:515-23
40. Hao J, Chen H, Madigan MC, Cozzi PJ, Beretov J, Xiao W. et al. Co-expression of CD147 (EMMPRIN), CD44v3-10, MDR1 and monocarboxylate transporters is associated with prostate cancer drug resistance and progression. British journal of cancer. 2010;103:1008-18
41. Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H. et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer cell. 2011;19:387-400
42. Skandalis SS, Gialeli C, Theocharis AD, Karamanos NK. Advances and advantages of nanomedicine in the pharmacological targeting of hyaluronan-CD44 interactions and signaling in cancer. Advances in cancer research. 2014;123:277-317
43. Tao J, Deng NT, Ramnarayanan K, Huang B, Oh HK, Leong SH. et al. CD44-SLC1A2 gene fusions in gastric cancer. Science translational medicine. 2011;3:77ra30
44. Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nature reviews Cancer. 2009;9:95-107
45. Dahabreh IJ, Schmid CH, Lau J, Varvarigou V, Murray S, Trikalinos TA. Genotype misclassification in genetic association studies of the rs1042522 TP53 (Arg72Pro) polymorphism: a systematic review of studies of breast, lung, colorectal, ovarian, and endometrial cancer. American journal of epidemiology. 2013;177:1317-25
46. Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nature reviews Cancer. 2007;7:182-91
47. Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer cell. 2008;13:188-92
48. Du J, Ji J, Gao Y, Xu L, Xu J, Zhu C. et al. Nonsynonymous polymorphisms in FAT4 gene are associated with the risk of esophageal cancer in an Eastern Chinese population. International journal of cancer. 2013;133:357-61
49. Song H, Ramus SJ, Quaye L, DiCioccio RA, Tyrer J, Lomas E. et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006;27:2235-42
50. Hirata H, Hinoda Y, Kawamoto K, Kikuno N, Suehiro Y, Okayama N. et al. Mismatch repair gene MSH3 polymorphism is associated with the risk of sporadic prostate cancer. The Journal of urology. 2008;179:2020-4
51. Xu XL, Yao YL, Xu WZ, Feng JG, Mao WM. Correlation of MSH3 polymorphisms with response and survival in advanced non-small cell lung cancer patients treated with first-line platinum-based chemotherapy. Genetics and molecular research: GMR. 2015;14:3525-33
52. Shinmura K, Yamaguchi S, Saitoh T, Takeuchi-Sasaki M, Kim SR, Nohmi T. et al. Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base pair in double-stranded DNA. Nucleic acids research. 2000;28:4912-8
53. Kappil M, Terry MB, Delgado-Cruzata L, Liao Y, Santella RM. Mismatch Repair Polymorphisms as Markers of Breast Cancer Prevalence in the Breast Cancer Family Registry. Anticancer research. 2016;36:4437-41
54. Kong F, Han XY, Luan Y, Qi TG, Sun C, Wang J. et al. MUTYH association with esophageal adenocarcinoma in a Han Chinese population. Asian Pacific journal of cancer prevention: APJCP. 2013;14:6411-3
55. Kuno T, Matsubara N, Tsuda S, Kobayashi M, Hamanaka M, Yamagishi D. et al. Alterations of the base excision repair gene MUTYH in sporadic colorectal cancer. Oncology reports. 2012;28:473-80
56. Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223-33
57. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. International journal of cancer. 2012;130:1715-25
58. Lawniczak M, Jakubowska A, Bialek A, Karpinska-Kaczmarczyk K, Lubinski J, Starzynska T. Possible association of the BRCA2 gene C5972T variant with gastric cancer: a study on Polish population. Polskie Archiwum Medycyny Wewnetrznej. 2015;125:39-45
59. Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665-9
60. Neve RM, Lane HA, Hynes NE. The role of overexpressed HER2 in transformation. Annals of oncology: official journal of the European Society for Medical Oncology. 2001;12(Suppl 1):S9-13
61. Rubin I, Yarden Y. The basic biology of HER2. Annals of oncology: official journal of the European Society for Medical Oncology. 2001;12(Suppl 1):S3-8
62. Boku N. HER2-positive gastric cancer. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2014;17:1-12
Author contact
![]() Corresponding author: Address: Department of Oncology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing, 210006, China; Fax: +86-25-87726234; Tel: +86-25-87726242; Email: jinfeichencom
Corresponding author: Address: Department of Oncology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing, 210006, China; Fax: +86-25-87726234; Tel: +86-25-87726242; Email: jinfeichencom

 Global reach, higher impact
Global reach, higher impact