Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(18):3326-3333. doi:10.7150/jca.25666 This issue Cite
Research Paper
Overexpression of Translocation Associated Membrane Protein 2 Leading to Cancer-Associated Matrix Metalloproteinase Activation as a Putative Metastatic Factor for Human Oral Cancer
1. Department of Oral Science, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan
2. Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University Hospital, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan
3. Department of Dentistry and Oral-Maxillofacial Surgery, Eastern Chiba Medical Center, 3-6-2 Okayamadai, Togane, Chiba 283-8686, Japan
4. Department of Clinical Oncology, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan
Received 2018-2-22; Accepted 2018-8-17; Published 2018-9-7
Abstract
Translocation associated membrane protein 2 (TRAM2) has been characterized as a component of the translocon that is a gated channel at the endoplasmic reticulum (ER) membrane. TRAM2 is expressed in a wide variety of human organs. To date, no information is available regarding TRAM2 function in the genesis of human cancer. The purpose of this study was to investigate the status of the TRAM2 gene in oral squamous cell carcinoma (OSCC) cells and clinical OSCC samples. Using real-time quantitative reverse transcriptase-polymerase chain reaction, Western blotting analysis, and immunohistochemistry, we detected accelerated TRAM2 mRNA and protein expression levels both in OSCC-derived cell lines and primary tumors. Moreover, TRAM2-positive OSCC tissues were correlated closely (P<0.05) with metastasis to regional lymph nodes and vascular invasiveness. Of note, knockdown of TRAM2 inhibited metastatic phenotypes, including siTRAM2 cellular migration, invasiveness, and transendothelial migration activities with a significant (P<0.05) decrease in protein kinase RNA(PKR) - like ER kinase (PERK) and matrix metalloproteinases (MMPs) (MT1-MMP, MMP2, and MMP9). Taken together, our results suggested that TRAM2 might play a pivotal role in OSCC cellular metastasis by controlling major MMPs. This molecule might be a putative therapeutic target for OSCC.
Keywords: Oral squamous cell carcinoma, Translocation associated membrane protein 2, Tumor metastasis, Tumor invasion, Matrix metalloproteinase
Introduction
Translocation associated membrane protein 2 (TRAM2) is a component of the translocon, which not only transports proteins but also is a channel at the endoplasmic reticulum (ER) [1-8]. Through the translocon, the proteins synthesized by ribosome are transported into the ER. The folded proteins in the ER then are secreted to the Golgi apparatus. In addition, the C-terminal domain of TRAM2 interacts with the main calcium pump of the ER to regulate the calcium concentration in the ER [9]. The translocon also is an ER calcium leak channel that maintains calcium homeostasis [3-5].
TRAM2 expression is regulated by the runt-related transcription factor 2 gene (RUNX2) and participates in a RUNX2-associated osteogenic reaction [10]. TRAM2 is expressed ubiquitously in several organs, such as the heart, liver, and kidney [11]. However, the expression status and function of TRAM2 in cancers, especially oral squamous cell carcinoma (OSCC), have not been fully characterized.
In the current study, we report the status of TRAM2 expression in OSCC cells and 106 clinical samples. We also describe the detailed mechanism of tumoral metastasis using TRAM2 knockdown models in vitro.
Materials and Methods
Ethics statement
The ethics committee of Chiba University approved the study protocol (approval number, 680), which was conducted according to the tenets of the Declaration of Helsinki. All patients and their families provided written informed consent before inclusion in the study.
Cells and clinical OSCC tissue samples
Nine OSCC-derived cell lines (HSC-2, HSC-3, HSC-4, KOSC-2, Sa3, Ca9-22, SAS, HO-1-u-1, and HO-1-N-1) [12,13]were obtained from the Japanese Collection of Research Bioresources Cell Bank (Ibaraki, Osaka, Japan) and the RIKEN BioResource Center (Tsukuba, Ibaraki, Japan). Human normal oral keratinocytes (HNOKs) were obtained from young patients at Chiba University Hospital. Primary HNOKs were cultured in oral keratinocyte medium (ScienCell Research Laboratories, Carlsbad, CA, USA) supplemented with penicillin and streptomycin (ScienCell Research Laboratories) and oral keratinocyte growth supplement (ScienCell Research Laboratories). Human umbilical vein endothelial cells (HUVEC) (Lonza Group Ltd., Basel, Switzerland) were cultured in endothelial cell growth medium (Lonza Group Ltd.). OSCC cell lines were cultured in Dulbecco's modified Eagle medium (DMEM) (Sigma-Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum (FBS) (HyClone, Logan, UT, USA) and 50 units/ml of penicillin and streptomycin (Sigma-Aldrich).
One hundred six clinical OSCC samples were obtained at Chiba University Hospital. All tissue samples were fixed in 20% formalin and embedded in wax. Four-micron sections were prepared for immunohistochemistry (IHC) analysis.
mRNA expression analysis
Total RNA was isolated from cells using Trizol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan). Gene expression was analyzed by real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) using FastStart Essential DNA Probes Master (Roche Diagnostics, Mannheim, Germany) and the LightCycler 480 Instrument (Roche Diagnostics). Primers were designed using the Universal ProbeLibrary Assay Design Center (http://lifescience.roche.com/). The primer sequences used for qRT-PCR were: TRAM2, forward, 5′- CTC ATC AAG AGG GAA TCT GGT T -3′; reverse, 5′- TTA GTC CGT GGG GAG GTT C -3′; universal probe #8 (Roche Diagnostics), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward, 5′-AGC CAC ATC GCT CAG ACA C-3′; reverse, 5′-GCC CAA TAC GAC CAA ATC C-3′; universal probe #60 (Roche Diagnostics).
Western blotting analysis
Western blotting analysis was conducted with 20 μg of protein as described previously [14-16]. Primary antibodies were rabbit anti-TRAM2 antibody (dilute concentration, 1:1000, Thermo Fisher Scientific), mouse anti-GAPDH antibody (dilute concentration, 1:1000, Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti-protein kinase RNA(PKR) - like ER kinase (PERK) antibody (dilute concentration, 1:1000, Santa Cruz Biotechnology), rabbit anti-MT1-MMP antibody (dilute concentration, 1:1000, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-MMP2 antibody (dilute concentration, 1:500, Merck KGaA, Darmstadt, Germany), and rabbit anti-MMP9 antibody (dilute concentration, 1:3000, GeneTex Inc., Irvine, CA, USA). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Promega, Madison, WI, USA) was used as secondary antibodies.
IHC
IHC was performed as described previously to evaluate the intensity of the TRAM2 immunoreaction [16-19]. IHC scores of OSCC and normal tissues were calculated using a semi-quantitative scoring system [20-25] ranging from 0 to 3, in which 0 indicates absent, 1 weak or focal, 2 moderate, and 3 strong immunoreactivity. The standard deviation score of +3 for normal tissue was determined as the cutoff point for the TRAM2 IHC scores. The IHC scores from the 106 patients were classified as TRAM2-positive and -negative.
Transfection of TRAM2 siRNA
TRAM2 siRNA (siTRAM2) (Santa Cruz Biotechnology) and control siRNA (siControl) (Santa Cruz Biotechnology) were transfected into the OSCC-derived cells, SAS and HSC4, using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions. After transfection, qRT-PCR and Western blotting analyses were conducted to analyze the knockdown efficiency of TRAM2.
Cellular proliferation assay
To examine the effectiveness of TRAM2 knockdown on cellular growth, we conducted a cellular proliferation assay in which the SiTRAM2 and siControl cells were seeded in 60-mm dishes at a density of 1 × 104 viable cells/dish. The cells were counted using the LunaTM Automated Cell Counter (Logos Biosystems, Annandale, VA, USA) every 24 h from 24 to 120 h.
Cellular migration assay
To examine the effectiveness of TRAM2 knockdown on cellular migration, we conducted the cellular migration assay. The siTRAM2 and siControl cells were seeded on 6-well plates and cultured in DMEM containing 10% FBS until a confluent monolayer formed. A wound was created using a micropipette tip in the middle of each well, and the cells then were cultured in serum-free medium. The wounded area was visualized at 12 and 24 h and measured using the Lenaraf220b free software (available at http://www.vector.co.jp/soft/dl/win95/art/se312811.html).
Cellular invasiveness assay
To examine the effectiveness of TRAM2 knockdown on cellular invasion, we conducted a cellular invasion assay in which 2.0×105 cells of siTRAM2 and siControl cells were seeded on 6-well plates with an 8-μm pore size cell culture insert (Corning, Corning, NY, USA). One milliliter of serum-free DMEM and 2 ml of DMEM containing 10% FBS were added in the upper side and lower side of the insert respectively. After 48 h, the insert was washed with phosphate buffered saline, and the cells on the upper side were removed with cotton swabs. Cells adhering to the lower surface of the insert membrane were fixed with methanol and stained with crystal violet. The numbers of stained invaded cells were counted in five random fields using a light microscope at ×100 magnification.
Transendothelial migration assay
To examine the effectiveness of TRAM2 knockdown on transendothelial migration, we conducted a transendothelial migration assay using a CytoSelectTM tumor transendothelial migration assay kit (Cell Biolabs, Inc., San Diego, CA, USA) according to the manufacturer's instructions. Briefly, HUVEC cells (5.0×104) diluted in 300 μl of culture medium were seeded on 24-well plates with an 8-μm pore size cell culture insert (Corning), and 500 μl of culture medium was added to the lower side of the insert. The cells were incubated 72 h until a monolayer formed. The siTRAM2 and siControl cells (1.0×106 cells) were diluted in 1 ml of serum-free medium; 2 μl of CytoTrackerTM solution (Cell Biolabs, Inc.) was added and incubated for 1 h. Suspensions of siTRAM2 and siControl cells were seeded on 24-well plates with an 8-μm pore size cell culture insert above the HUVEC monolayer. After a 24 h incubation, the non-migratory cells were removed with cotton swabs. In contrast, the migratory cells were resolved in a cell lysis buffer (Cell Biolabs, Inc.) and incubated for 5 min at room temperature with shaking. The cell lysate (100 μl) was transferred to 96-well plates and measured using the microplate reader TECAN Infinite 200 PRO (Tecan, Männedorf, Switzerland) at 480 nm / 520 nm.
Zymography
To examine the secreted MMPs from siTRAM2, we conducted gelatin zymography assay using a gelatin-zymography kit (Primary Cell, Sapporo, Japan) according to the manufacturer's instructions. Briefly, after culturing the siTRAM2 and siControl cells in the serum-free DMEM for 48 h, the cell culture medium were concentrated using Centrifugal Filter Units (Merck KGaA, Darmstadt, Germany). The condition medium were mixed with an SDS sample buffer and separated on gelatin gels by electrophoresis. After washing, the gels were and incubated for 48 h at 37℃ in the enzymatic reaction buffer and stained with coomassie brilliant blue. The gels were destained in methanol/acetic solution.
Statistical analysis
The Student's t-test was used to analyze the significance of the TRAM2 mRNA expression, cellular proliferation assay, cellular invasion assay, and transendothelial migration assay. The χ2 test was used to evaluate the correlations between the TRAM2 IHC scores and each clinicopathological parameter.
Results
Up-regulation of TRAM2 in OSCC-derived cell lines
To analyze the expression status of TRAM2, the nine OSCC-derived cell lines and HNOKs were subjected to qRT-PCR and Western blotting analyses. TRAM2 mRNA expression was up-regulated significantly (P<0.05) in all OSCC-derived cell lines compared with the HNOKs (Fig. 1A). TRAM2 protein expression also was significantly up-regulated in all the OSCC-derived cell lines compared with the counterpart (Fig. 1B).
TRAM2 mRNA expression in OSCC-derived cell lines and primary OSCC tissues. (A) Quantification of TRAM2 mRNA expression in OSCC-derived cell lines by qRT-PCR. The TRAM2 mRNA expression is significantly (*P<0.05, Student's t-test) up-regulated in nine OSCC-derived cell lines compared with the HNOKs. (B) Western blotting analysis of TRAM2 in the OSCC-derived cell lines and HNOKs. TRAM2 protein expression is up-regulated in the nine OSCC-derived cell lines compared with the HNOKs. TRAM2 protein densitometric data are normalized to the GAPDH protein levels. The values are expressed as a percentage of the HNOKs. (C, D) Representative IHC results of TRAM2 protein in (C) normal oral tissue and (D) primary OSCC tissue. Original magnification, ×200. Scale bars, 100 µm. (E) The status of the TRAM2 protein expression levels in primary OSCCs and normal counterparts (n=106) using a semi-quantitative scoring system. The TRAM2 protein expression levels in the OSCCs are markedly (*P<0.05, Student's t-test) higher than in the normal oral tissues.
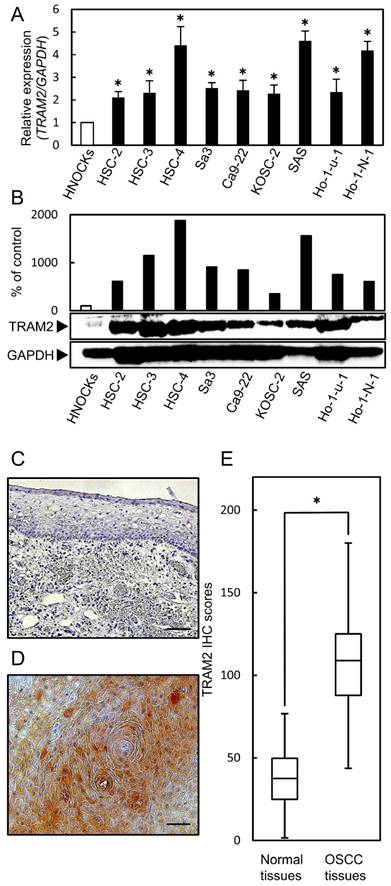
Evaluation of TRAM2 expression in primary OSCC tissues
A semi-quantitative IHC scoring system was used to evaluate the intensity of the TRAM2 immunoreactivity. Fig. 1C and D show representative IHC data of TRAM2 in the normal tissue and primary OSCC tissue. TRAM2 expression in the cytosol of the primary OSCC tissues was higher than that in normal tissues (Fig. 1C, D). The TRAM2 IHC scores of the primary normal oral tissues and OSCC tissues ranged from 1.6 to 76.8 (median, 37.55) and 43.7 to 180.1 (median, 108.95), respectively (Fig. 1E). The IHC scores in the primary OSCC tissues were significantly higher than in the normal oral tissues. We classified that IHC scores over 91.5 as TRAM2-positive. In the clinical classifications, TRAM2-positive OSCCs were associated significantly (P<0.05) with lymph node metastasis, stage, and vascular invasion (Table 1).
Correlation between TRAM2 expression and clinical classification in OSCCs
| Clinical classification | Results of IHC number of patients(%) | P value | ||
|---|---|---|---|---|
| Total | TRAM2(-) | TRAM2(+) | ||
| Age at surgery (years) | ||||
| <60 | 25 | 8 (32) | 17 (68) | 0.7291 |
| ≧60 | 81 | 23 (28) | 58 (72) | |
| T-primary tumor | ||||
| T1 | 23 | 11 (48) | 12 (52) | 0.054 |
| T2 | 36 | 12 (33) | 24 (67) | |
| T3 | 8 | 1 (13) | 7 (88) | |
| T4 | 39 | 7 (18) | 32 (82) | |
| N-regional lymph node | ||||
| Positive | 43 | 8 (19) | 35 (81) | 0.0466* |
| Negative | 63 | 23 (37) | 40 (63) | |
| Stage | ||||
| I | 19 | 9 (47) | 10 (53) | 0.0318* |
| II | 25 | 10 (40) | 15 (60) | |
| III | 16 | 5 (31) | 11 (69) | |
| IV | 46 | 7 (15) | 39 (85) | |
| Histopathologic type | ||||
| Poorly | 4 | 2 (50) | 2 (50) | 0.5538 |
| Moderately | 36 | 9 (25) | 27(75) | |
| Well | 66 | 20 (30) | 46 (70) | |
| Vascular invasion | ||||
| Positive | 40 | 6 (25) | 34 (85) | 0.0121* |
| Negative | 66 | 25 (38) | 41 (62) | |
*P<0.05
Transfection of TRAM2 siRNA
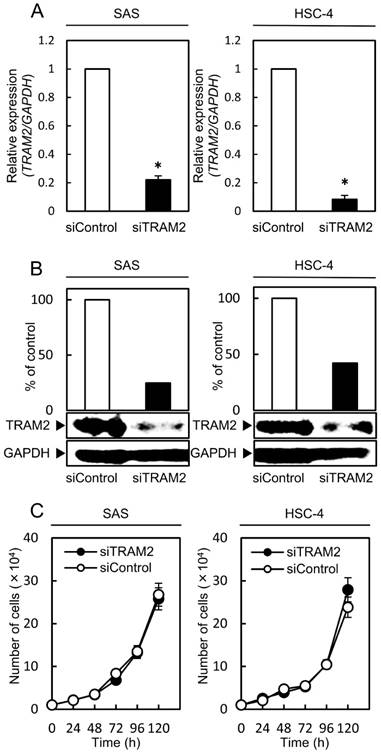
To investigate the function of TRAM2 in OSCC-derived cells, siTRAM2 or siControl was transfected into SAS and HSC4 cells. qRT-PCR and Western blotting analysis evaluated the efficacy of the TRAM2 siRNA transfection. TRAM2 mRNA expression decreased markedly (P<0.05) in the siTRAM2 cells (Fig. 2A). The expression level of TRAM2 protein in the siTRAM2 cells also was lower than in the siControl cells (Fig. 2B).
A cellular proliferation assay of the TRAM2 siRNA transfected cells. (A) The expression levels of TRAM2 mRNA in siTRAM2 cells (SAS and HSC-4). The TRAM2 mRNA expression in the siTRAM2 cells is decreased markedly (*P<0.05) compared with the siControl cells. (B) The expression level of TRAM2 protein in the siTRAM2 cells is also lower than in the siControl cells. (C) A cellular proliferation assay of the siTRAM2 cells shows no significant difference between the siTRAM2 and siControl cells.
Functional analyses of TRAM2 siRNA transfected cells
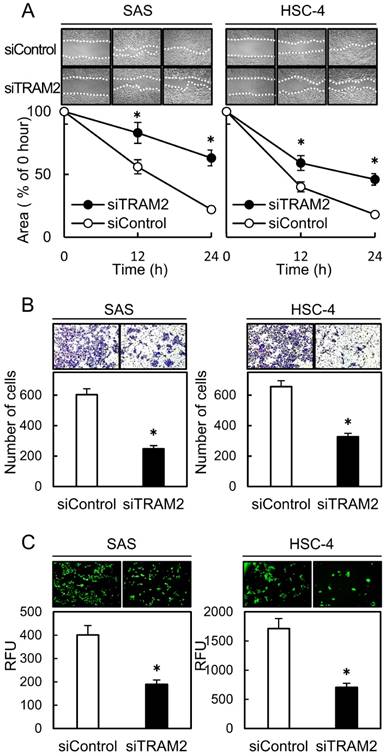
A cellular proliferation assay, cellular migration assay, cellular invasion assay, and transendothelial migration assay were performed to evaluate the effect of TRAM2 knockdown. The cellular proliferation assay showed no significant difference between the siTRAM2 cells and siControl cells (Fig. 2C). The cellular migration assay showed that the wounded areas of the siControl cells decreased significantly (P<0.05) compared with those of the siTRAM2 cells (Fig. 3A). The cellular invasion assay showed that the number of cells invading the siTRAM2 cells decreased significantly (P<0.05) compared with the siControl cells (Fig. 3B). The transendothelial migration assay showed that the relative fluorescence units (RFUs) of the siTRAM2 cells decreased significantly (P<0.05) compared with siControl cells (Fig. 3C).
A cellular migration assay, invasiveness assay, and transendothelial migration assay of TRAM2 siRNA transfected cells. (A) A cellular migration assay of the siTRAM2 cells shows that the wounded areas in the siControl cells are decreased significantly (*P<0.05) compared with those of the siTRAM2 cells. (B) A cellular invasion assay of the siTRAM2 cells shows that the number of cells invading the siTRAM2 cells is significantly (*P<0.05) decreased compared with siControl cells. (C) A transendothelial migration assay of the siTRAM2 cells shows that the RFUs of siTRAM2 are decreased significantly (*P<0.05) compared with the siControl cells.
PERK expression level in TRAM2 siRNA transfected cells
PERK is a marker of ER stress which activates some transcription factors. We investigated the expression of PERK by Western blotting analysis. The expressions of PERK in the siTRAM2 cells were significantly lower than those in the siControl cells (Fig. 4A).
PERK and MMP family expressions of the siTRAM2 transfected cells. (A) Western blotting analysis shows the expression levels of PERK protein in the siTRAM2 cells. The expression level of PERK in the siTRAM2 cells is significantly lower than in the siControl cells. (B) Western blotting analysis shows the expression levels of the MMP family protein in the siTRAM2 cells. The expression levels of MTI-MMP, MMP2, and MMP9 in the siTRAM2 cells are significantly lower than in the siControl cells. (C, D) Gelatin zymography assay shows secretion of MMPs. Secretion of proMMP-9, MMP-9, proMMP-2, and MMP-2 of the siTRAM2 cells was decreased compared with siControl cells.
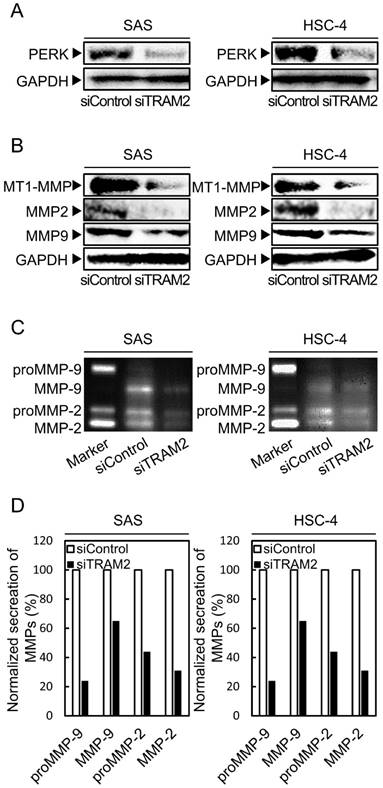
MMP family expression levels in TRAM2 siRNA transfected cells
The MMP family is involved in breakdown of the extracellular matrix; thus, MMP family overexpression is related to cellular invasion by cancer cells [26-30]. We investigated the expressions of several types of MMPs (MT1-MMP, MMP2, and MMP9) by Western blotting analyses. The expressions of MTI-MMP, MMP2, and MMP9 in the siTRAM2 cells were significantly lower than those in the siControl cells (Fig. 4B). Furthermore, we investigated the secretion levels of MMPs by gelatin zymography assay. Secretion of proMMP-9, MMP-9, proMMP-2, and MMP-2 of the siTRAM2 cells was decreased compared with siControl cells (Fig. 4C, D).
Discussion
Since the 5-year survival rate is less than 20% in patients with OSCC with metastasis to the regional lymph nodes [31-34], tumoral metastasis is a significant prognostic factor. However, there currently is no critical marker for diagnostic/therapeutic targets for patients with OSCC with metastatic lesion(s).
The current study found for the first time that TRAM2 was up-regulated significantly in OSCC-derived cell lines and primary OSCCs compared with the normal counterparts. Cellular migration, cellular invasion, and expression of the MMP family (MT1-MMP, MMP2, and MMP9) were inhibited in the TRAM2 knockdown cells. Furthermore, highly expressed TRAM2 OSCCs were correlated significantly (P<0.05) with metastasis to the regional lymph nodes and vascular invasion in the tumoral tissues examined (Table 1).
MT1-MMP, MMP2, and MMP9 are believed to promote tumoral metastasis and invasiveness of the cancer cells [28,35,36]. Several studies have reported that MTI-MMP activates MMP2 and MMP9, suggesting that cancer cells with high expressions of MMP2 and MMP9 are likely to metastasize to other organs. The current result, i.e., significant overexpression of MMPs in OSCC cell lines, confirmed a possible role of these MMPs in the accelerated metastasis of OSCCs.
The function of TRAM protein is still unknown including cancer progression. We assumed that abnormal expression of TRAM2 possibly induced ER stress, leading to highly tumoral metastasis [37-44]. Consistent with our hypothesis, PERK and MMPs expressions were dramatically decreased in the TRAM2 knockdown cells (Fig. 4). Moreover, the current study showed that cellular migration, cellular invasiveness, and transendothelial migration decreased significantly with inhibition of MMP expression in TRAM2 knockdown cells (Fig. 3, 4B, C, D). Thus, we suggested that abnormal expression of TRAM2 induced ER stress, an activated PERK, and up-regulation of MMPs, leading to highly tumoral metastasis.
In summary, TRAM2 expression is increased significantly in OSCC and plays a pivotal role in OSCC cellular metastasis by controlling major MMPs. This molecule might be a putative therapeutic target for OSCC.
Abbreviations
TRAM2: Translocation associated membrane protein 2; ER: endoplasmic reticulum; OSCC: oral squamous cell carcinoma; MMPs: matrix metalloproteinases.
Acknowledgements
We thank Lynda C. Charters for editing this manuscript. The authors received no financial support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Walter P. Protein translocation. Travelling by TRAM. Nature. 1992;357:22-3
2. Johnson AE, Haigh NG. The ER translocon and retrotranslocation: is the shift into reverse manual or automatic? Cell. 2000;102:709-12
3. Coppenolle F Van, Abeele F Vanden, Slomianny C, Flourakis M, Hesketh J, Dewailly E. et al. Ribosome-translocon complex mediates calcium leakage from endoplasmic reticulum stores. J Cell Sci. 2004;117:4135-42
4. Flourakis M, Coppenolle F Van, Lehen V, Beck B, Skryma R, Prevarskaya N. Passive calcium leak via translocon is a first step for iPLA 2 -pathway regulated store operated channels activation. FASEB J. 2006;20:1215-7
5. Lomax RB, Camello C, Coppenolle F Van, Petersen OH, Tepikin A V. Basal and Physiological Ca2+ leak from the endoplasmic reticulum of pancreatic acinar cells. Second messenger-activated channels and translocons. J Biol Chem. 2002;277:26479-85
6. Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25-35
7. Agarraberes FA, Dice JF. Protein translocation across membranes. Biochim Biophys Acta. 2001;1513:1-24
8. Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615-30
9. Stefanovic B, Stefanovic L, Schnabl B, Bataller R, Brenner DA. TRAM2 protein interacts with endoplasmic reticulum Ca2+ pump Serca2b and is necessary for collagen type I synthesis. Mol Cell Biol. 2004;24:1758-68
10. Pregizer S, Barski A, Gersbach CA. Identification of novel Runx2 targets in osteoblasts : cell type-specific BMP-dependent regulation of Tram2. J Cell Biochem. 2007;102:1458-71
11. Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S. et al. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:37-43
12. Tanzawa H, Uzawa K, Kasamatsu A, Endo-Sakamoto Y, Saito K, Ogawara K. et al. Targeting gene therapies enhance sensitivity to chemo- and radiotherapy of human oral squamous cell carcinoma. Oral Sci Int. 2015;12:43-52
13. Kasamatsu A, Uzawa K, Usukura K, Koike K, Nakashima D, Ishigami T. et al. Loss of heterozygosity in oral cancer. Oral Sci Int. 2011;8:37-43
14. Endo Y, Uzawa K, Mochida Y, Shiiba M, Bukawa H, Yokoe H. et al. Sarcoendoplasmic reticulum Ca2+ ATPase type 2 downregulated in human oral squamous cell carcinoma. Int J Cancer. 2004;110:225-31
15. Kasamatsu A, Uzawa K, Nakashima D, Koike H, Shiiba M, Bukawa H. et al. Galectin-9 as a regulator of cellular adhesion in human oral squamous cell carcinoma cell lines. Int J Mol Med. 2005;16:269-73
16. Saito T, Kasamatsu A, Ogawara K, Miyamoto I, Saito K, Iyoda M. et al. Semaphorin7A Promotion of Tumoral Growth and Metastasis in Human Oral Cancer by Regulation of G1 Cell Cycle and Matrix Metalloproteases: Possible Contribution to Tumoral Angiogenesis. PLoS One. 2015;10:e0137923
17. Uzawa K, Ono K, Suzuki H, Tanaka C, Yakushiji T, Yamamoto N. et al. High prevalence of decreased expression of KAI1 metastasis suppressor in human oral carcinogenesis. Clin Cancer Res. 2002;8:828-35
18. Kitajima D, Kasamatsu A, Nakashima D, Miyamoto I, Kimura Y, Saito T. et al. Tie2 Regulates Tumor Metastasis of Oral Squamous Cell Carcinomas. J Cancer. 2016;7:600-7
19. Uzawa K, Kasamatsu A, Saito T, Takahara T, Minakawa Y, Koike K. et al. Long-term culture of human odontoma-derived cells with a Rho kinase inhibitor. Exp Cell Res. 2016;347:232-40
20. Shiiba M, Ishige S, Saito Y, Shimizu T, Minakawa Y, Kasamatsu A. et al. Down-regulated expression of family with sequence similarity 3, member B (FAM3B), in oral squamous cell carcinoma. Oral Sci Int. 2012;9:9-16
21. Elnaggar A, Farag AH, Gaber ME, Hafeez MA, Ali MS, Atef AM. AlphaVBeta3 Integrin expression within uterine endometrium in unexplained infertility: a prospective cohort study. BMC Womens Health. 2017;17:90
22. Creus M, Ordi J, Fábregues F, Casamitjana R, Ferrer B, Coll E. et al. alphavbeta3 integrin expression and pinopod formation in normal and out-of-phase endometria of fertile and infertile women. Hum Reprod. 2002;17:2279-86
23. Koide N, Kasamatsu A, Endo-Sakamoto Y, Ishida S, Shimizu T, Kimura Y. et al. Evidence for Critical Role of Lymphocyte Cytosolic Protein 1 in Oral Cancer. Sci Rep. 2017;7:43379
24. Kita A, Kasamatsu A, Nakashima D, Endo-Sakamoto Y, Ishida S, Shimizu T. et al. Activin B Regulates Adhesion, Invasiveness, and Migratory Activities in Oral Cancer: a Potential Biomarker for Metastasis. J Cancer. 2017;8:2033-41
25. Kimura Y, Kasamatsu A, Nakashima D, Yamatoji M, Minakawa Y, Koike K. et al. ARNT2 Regulates Tumoral Growth in Oral Squamous Cell Carcinoma. J Cancer. 2016;7:702-10
26. Rundhaug JE. Matrix metalloproteinases and angiogenesis. J Cell Mol Med. 2005;9:267-85
27. Coussens LM, Fingleton B, Matrisian LM. Matrix Metalloproteinase Inhibitors and Cancer : trials and tribulations. Science. 2002;295:2387-92
28. Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9-34
29. Jimenez L, Jayakar SK, Ow TJ, Segall JE. Mechanisms of invasion in head and neck cancer. Arch Pathol Lab Med. 2015;139:1334-48
30. Kim HJ, Park C, Park BW, Lee H, Jung WH. Expression of MT-1 MMP, MMP2, MMP9 and TIMP2 mRNAs in ductal carcinoma in situ and invasive ductal carcinoma of the breast. Yonsei Med J. 2006;47:333-42
31. Sankaranarayanan R, Ramadas K, Amarasinghe H, Subramanian S, Johnson N. Oral Cancer: Prevention, Early Detection, and Treatment. The International Bank for Reconstruction and Development / The World Bank. 2015 Chapter 5
32. Noguti J, Foot C, Moura GDE, Protasio G, Jesus PDE, Hugo V. et al. Metastasis from oral cancer : an overview. Cancer Genomics Proteomics. 2012;9:329-35
33. Kapoor C, Vaidya S, Wadhwan V, Malik S. Lymph node metastasis: A bearing on prognosis in squamous cell carcinoma. Indian J Cancer. 2015;52:417-24
34. Goodman M, Liu L, Ward K, Zhang J, Almon L, Su G. et al. Invasion characteristics of oral tongue cancer: frequency of reporting and effect on survival in a population-based study. Cancer. 2009;115:4010-20
35. Al-raawi D, Abu-el-zahab H, El-shinawi M, Mohamed MM. Membrane type-1 matrix metalloproteinase ( MT1-MMP ) correlates with the expression and activation of matrix metalloproteinase-2 ( MMP-2 ) in inflammatory breast cancer. Int J Clin Exp Med. 2011;4:265-75
36. Arvelo F, Sojo F, Cotte C. Tumour progression and metastasis. Ecancermedicalscience. 2016;10:617
37. Hammadi M, Oulidi A, Gackière F, Katsogiannou M, Slomianny C, Roudbaraki M. et al. Modulation of ER stress and apoptosis by endoplasmic reticulum calcium leak via translocon during unfolded protein response : involvement of GRP78. FASEB J. 2013;27:1600-9
38. Ryan AJ, Larson-Casey JL, He C, Murthy S, Brent Carter A. Asbestos-induced disruption of calcium homeostasis induces endoplasmic reticulum stress in macrophages. J Biol Chem. 2014;289:33391-403
39. Liu J, Du L. PERK pathway is involved in oxygen-glucose-serum deprivation-induced NF-kB activation via ROS generation in spinal cord astrocytes. Biochem Biophys Res Commun. 2015;467:197-203
40. Kitamura M. Control of NF-κB and Inflammation by the Unfolded Protein Response. Int Rev Immunol. 2011;30:4-15
41. Willy JA, Young SK, Stevens JL, Masuoka HC, Wek RC. CHOP links endoplasmic reticulum stress to NF- B activation in the pathogenesis of nonalcoholic steatohepatitis. Mol Biol Cell. 2015;26:2190-204
42. Park JM, Kim A, Oh JH, Chung AS. Methylseleninic acid inhibits PMA-stimulated pro-MMP-2 activation mediated by MT1-MMP expression and further tumor invasion through suppression of NF-κB activation. Carcinogenesis. 2007;28:837-47
43. Lee YH, Park JH, Cheon DH, Kim T, Park YE, Oh E-S. et al. Processing of syndecan-2 by matrix metalloproteinase-14 and effect of its cleavage on VEGF-induced tube formation of HUVECs. Biochem J. 2017;474:3719-32
44. Fanjul-Fernández M, Folgueras AR, Cabrera S, López-Otín C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim Biophys Acta. 2010;1803:3-19
Author contact
![]() Corresponding authors: K. Uzawa, Department of Oral Science, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone: +81-43-226-2300; Fax: +81-43-226-2151; E-mails: uzawakchiba-u.jp and A. Kasamatsu, Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone: +81-43-226-2300; Fax: +81-43-226-2151; E-mails: kasamatsuachiba-u.jp
Corresponding authors: K. Uzawa, Department of Oral Science, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone: +81-43-226-2300; Fax: +81-43-226-2151; E-mails: uzawakchiba-u.jp and A. Kasamatsu, Department of Dentistry and Oral-Maxillofacial Surgery, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan. Phone: +81-43-226-2300; Fax: +81-43-226-2151; E-mails: kasamatsuachiba-u.jp

 Global reach, higher impact
Global reach, higher impact