Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(23):4363-4373. doi:10.7150/jca.27533 This issue Cite
Research Paper
Promotion of Cell Proliferation through Inhibition of Cell Autophagy Signalling Pathway by Rab3IP is Restrained by MicroRNA-532-3p in Gastric Cancer
Department of General Surgery, Nanfang Hospital, Southern Medical University, Guangdong Provincial Engineering Technology Research Center of Minimally Invasive Surgery, Guangzhou 510515, China
# co-first authors
Received 2018-5-29; Accepted 2018-8-28; Published 2018-10-22
Abstract
Background: RAB3A-interacting protein (Rab3IP) is known to be involved in cancer; however, its function during the proliferation of gastric cancer (GC) cells remains unknown. Therefore, this study aimed to explore the potential function of Rab3IP in GC.
Methods: The expression of Rab3IP and its clinical pathology value were determined by quantitative real-time PCR and immunohistochemistry. Rab3IP (knockdown and overexpression) and light chain 3 (LC3) lentiviruses were transfected into GC cells, and cell proliferation was measured using cell counting kit-8, plate clone formation, flow cytometry, and tumorigenesis assays. Cell autophagy was measured using a confocal laser scanning microscope and by western blotting. Luciferase reporter assay was performed to analyse the regulation of Rab3IP by microRNA-532-3p (miR-532-3p).
Results: Overexpression of Rab3IP in GC samples enhanced the cell proliferation ability, but decreased the number of autophagosomes and expression of LC3-II and sequestosome-1 (SQSTM1 or p62) markers. Furthermore, we found that miR-532-3p can bind to the 3ʹUTR region of RAB3IP and inhibit the proliferation ability of GC cells. Further, the expression of miR-532-3p negatively correlated with that of Rab3IP.
Conclusions: Our study elucidates the central role of Rab3IP in inducing proliferation of GC cells through its involvement in autophagy. miR-532-3p directly targets Rab3IP and represses its function, thereby demonstrating a novel regulatory link in GC.
Keywords: Rab3IP, miR-532-3p, cell proliferation, cell autophagy, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common malignant tumours and its mortality rate ranks third worldwide. As a result of national screening plans and improvements in chemotherapy, the mortality has significantly declined. However, the mortality rate of GC is relatively higher in developing countries than in developed countries1. With the accumulation of genetic mutations and epigenetic changes, the morbidity of GC could be higher2. In recent years, traditional chemotherapy and molecular target therapy have made great progress, but their efficacy in advanced stages of GC is still limited. Therefore, there is a pressing need to identify novel potential treatment targets.
Rab GTPases are members of the Ras GTPase superfamily, which regulate membrane identity and vesicle trafficking processes by switching cycles between the GDP- and GTP-bound states3. Previous studies have shown that RABs play an important role in epithelial cell transformation and motility of cancer cells. The guanine nucleotide exchange factors (GEFs) promote the expression of Rab proteins, which would boost GDP release by regulating specific residues in the switch regions of Rabs4. RAB3A-interacting protein (Rab3IP) is a Rab-specific GEF and a major activator of Rab proteins, which is directly regulated by Rab115. Previous studies by de Bruijn et al.6 have confirmed that Rab3IP interacts with SSX2, a cancer-related protein, in Hela cells. In addition, several studies have revealed that Rab3IP might play an important role in malignant cellular growth, as it was found to be upregulated, and is considered a tumour-specific marker in some malignant tumours such as colorectal cancer, major salivary carcinomas, and glioma7-9. In our previous study, we also found that, Rab3IP is upregulated in both gastric cancer cell lines and human tissues, and interacts with SSX2. Furthermore, we also revealed the involvement of Rab3IP and SSX2 in the epithelial-mesenchymal transition (EMT) and their potential implication in the invasion and metastasis of gastric cancer10. However, the potential function of Rab3IP during the proliferation of GC cells have not been totally revealed.
MicroRNAs (miRNAs) are a class of small-regulatory RNA molecules of ~22 nt in length that induce the inhibition of protein translation through binding to the 3ʹ untranslated regions (UTRs) of target mRNAs and causing their degradation 11, 12. Emerging evidences about the expression profile and function of miRNAs have proved their essential role in cellular differentiation, proliferation, and apoptosis in human cancer. Moreover, a series of miRNAs has been found to contribute to the cell cycle and apoptosis regulation in GC13. Nevertheless, the role and underlying mechanism of miRNAs in the regulation of Rab3IP-specific GEF associated with GC remains elusive and further investigation is required14.
In this study, we investigated the expression pattern and possible function of Rab3IP in proliferation and tumorigenicity of GC cells. Furthermore, we studied the involvement of Rab3IP in cell autophagy, which is supposed to be highly associated with the cell proliferation process of GC. Meanwhile, we also identified miR-532-3p as the miRNA that targets Rab3IP and illustrate its potential regulatory mechanism.
Materials and Methods
Tumour tissue specimens
Paired samples of GC and normal mucosal tissues were obtained from 150 patients who underwent standard gastrectomy at Nanfang Hospital of Southern Medical University from 2009 to 2014. Histological diagnosis, and pathological and clinical materials were evaluated and provided by the Department of Pathology and Department of General Surgery in Nanfang Hospital, according to the 8th version of American Joint Committee on Cancer (AJCC) stage guidelines. The Ethics Committee of Nanfang Hospital of Southern Medical University approved this study in compliance with the Declaration of Helsinki.
Cell culture and transfection
GC cell lines, including AGS, MKN45, BGC823, SGC7901, MGC803, and MKN28, and normal gastric mucosa cells (GES-1), were obtained from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 mixed with 10% foetal bovine serum under 5% CO2 at 37 ºC. Plasmids containing Rab3IP cDNA (the sequence results are shown in Supplementary materials) and Rab3IP-specific siRNAs (5'CCAGTGGGATTACAACCTA3') were purchased from OBIO Biotechnology Company (Shanghai, China). Rab3IP-knockdown lentiviruses were obtained from GeneChem Company (Shanghai, China) and were used to establish the Rab3IP-knockdown cell lines, namely AGS-Rab3IP (-) cells. Rab3IP overexpressing lentiviruses were also from GeneChem Company (Shanghai, China) and were used to generate the Rab3IP-overexpression cell lines, namely MKN45-Rab3IP (+) cells. Negative control cell lines were also established, called AGS-Rab3IP (-)-NC and MKN45-Rab3IP (+)-NC cells. The miR-450-5p, miR-217, miR-532-3p, miR-935, and miR-584-5p mimics as well as the negative control were purchased from OBIO Biotechnology Company (Shanghai, China) and transfected into GC cells, using the Lipofectamine 2000 Transfection Reagent (Thermo Fisher, USA).
Total RNA extraction, cDNA synthesis, and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cells or tissues, using RNAiso Plus reagent (Takara, Tokyo, Japan) according to the manufacturer's instructions. The concentration and quality of the RNA were measured by UV absorbance at 260 and 280 nm (260/280 nm) using the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). cDNA was then synthesised using the PrimScript TM RT Reagent Kit(Takara, Japan). qRT-PCR was performed using the SYBR Premix Ex Taq II Kit (Takara, Japan) to assess gene expression. Data were normalised to the geometric mean of the expression of the housekeeping gene GAPDH, and relative expression was calculated using the 2-ΔΔCT method.
Western blotting
Total proteins were collected from cells or tissues, using a PIPA lysis buffer (Biouniquer Technology, Nanjing, China) according to the manufacturer's instructions. Protein concentration was measured at 562 nm using the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). SDS-PAGE was used to separate equal amounts of protein lysates from fresh tissues or cells. Proteins were transferred onto PVDF membranes, which was followed with western blotting according to standard methods, using anti-Rab3IP (Cell Signaling Technology/CST, USA), anti-LC3 (CST, USA), anti-p62 (CST, USA), and anti-GAPDH (CST, USA) antibodies. GAPDH was used as the loading control.
Immunohistochemistry (IHC)
Standard two-step IHC was applied to investigate the expression of proteins in human GC tissue. Primary antibodies against Rab3IP (1:100) were incubated with the sections overnight at 4 ºC and Mayer's haematoxylin was used for nuclear counter staining. Slides were reviewed and evaluated by two blinded pathologists and the intensity of staining of the cancer cells was scored according to the standard stage as follows: 0 (negative, no staining), 1 (weak, light yellow), 2 (moderate, yellow brown), and 3 (intense, brown). High expression (or overexpression) was defined as an intensity score of ≥ 2, and a score of < 2 indicated low expression.
Cell counting kit 8 (CCK-8) assay
AGS and MKN45 cells were seeded into 96-well plates at a density of 3,000 cells per well and cultured for 24 h in RPMI Medium 1640 plus 10% FBS at 37 ºC and 5% CO2. RPMI 1640 in wells without cells served as blank control. Cells were quantified after 48 h using the CCK-8 assay (Donjindo, Kumamoto, Japan) and a microplate reader. The absorbance data (OD value) at 450 nm were collected for cell viability analysis.
Cell apoptosis assay
AGS and MKN45 were seeded into 6-well plates at a density of 1 × 106 cells per well and cultured for 24 h. Then the cells were washed with PBS three times, trypsinised, and harvested for staining using an annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis kit (Keygen, Nanjing, China). The staining of cells was assessed by flow cytometry and data analysed using the FlowJo software, with FITC-positive cells considered apoptotic.
Plate clone formation assay
For plate colony formation assays, AGS and MKN45 cells were seeded at a density of 500 cells/dish and incubated at 37 ºC, 5% CO2 for 1 week, and fresh medium was replenished as needed. The cell culture was terminated once visible clones appeared. The cell clones were washed with PBS, fixed for 15 minutes and stained with Giemsa. The number of stained clones was then counted and analysed.
Confocal laser scanning microscopy (CLSM)
AGS and MKN45 cells were transfected with LC3 double-fluorescent autophagy virus, and positive transformants selected in the presence of puromycin. Following this, the cells were cultured in RPMI 1640 with or without treatment with 3-methyladenine (3-MA, MedChemExpress, Shanghai China), rapamycin (RAPA, MedChemExpress, Shanghai China), or miR-532-3p. After cell adhesion was achieved, the culture medium was replaced as described above. At the time indicated, the cells were fixed with 4% paraformaldehyde, washed, then the cell nuclei were mounted with DAPI at 37 ºC for 20 min. Fluorescence measurements were performed using an FV10i confocal microscope (Olympus, Tokyo, Japan).
Dual-luciferase reporter assay
MKN45 cells were seeded into a 96-well plate. Forty-eight hours after being co-transfected with pmirGLO-WT-Rab3IP-3ʹUTR (wild type) or pmirGLO-MT-Rab3IP-3ʹUTR (mutant type) plasmid along with gga-miR-532-3p mimic, firefly and Renilla luciferase activities were measured using a Dual-GLO Luciferase Assay System Kit (Promega, USA) according to the manufacturer's instructions. Luminescence was measured using a Fluorescence/ Multi-Detection Microplate Reader (BioTek, USA) and firefly luciferase activities were normalised to Renilla luminescence in each well.
In vivo cell proliferation assay
Male Sprague-Dawley rats aged 6 to 8 weeks were purchased from the Experimental Animal Center of Southern Medical University, which is certified by the Guangdong Provincial Bureau of Science. The rats were raised in a clean environment, and all animal experiments were performed according to ethical practices. To establish the xenograft model, 5 × 106 cells in 0.2 mL of serum-free DMEM were injected subcutaneously into the right flank of nude mice. miR-532-3p agomir (GenePharma Company, Shanghai, China) was dissolved in PBS and injected into the tumour after the injection of cancer cell and the mice were killed 2 weeks later. The tumours were surgically removed from the mice and weighed.
Statistical analyses
We used SPSS statistical package version 19.0 for Windows (SPSS, Chicago, IL, USA) and GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA, USA) for statistical analysis. We used Mann-Whitney test to compare the mRNA levels of RAB3IP and miR-532-3p between GC tissues and normal tissues. The relationship between Rab3IP and miR-532-3p was assessed by linear regression analysis. Comparison among multiple groups was made using one-way ANOVA. P <0.05 was considered to indicate a statistically significant difference.
Results
Rab3IP is upregulated in GC
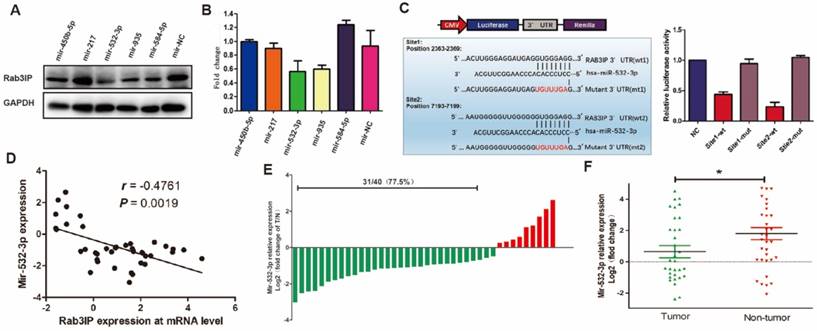
qRT-PCR, western blotting, and IHC were utilised to clarify the expression pattern of Rab3IP in GC. From the results of qRT-PCR, RAB3IP transcript levels were found to increase in GC samples compared with those in normal gastric mucosa samples (Fig 1A). Consistently, Rab3IP showed a higher expression in GC samples at the protein level (Fig 1B). To further characterise Rab3IP expression in GC, tissue microarray (TMA) containing 150 samples of GC was used for IHC analysis; here, 67.3% (101 / 150) of the samples were classified as Rab3IP-positive (Fig 1D). Furthermore, we analysed the relationship between the expression of Rab3IP with the relative clinical parameters. Rab3IP's clinical pathology value was further studied using Rab3IP expression in 150 cases of GC (Table 1). The chi-square test results showed that the expression level of Rab3IP significantly correlated with tumour size, differentiation, tumour (T) stage, lymph node (N) stage, and serum CEA and CA-724 levels. However, analysis of CA-199 serum levels, lymphatic invasion, perineural infiltration, vascular invasion, and M staging were excluded. Among all the gastric cell lines, GES-1 cells showed least expression of Rab3IP. Furthermore, AGS cells showed the highest expression compared to other GC cell lines, while the expression in MKN 45 cells was relatively lower (Fig 1C).
Rab3IP is overexpressed in gastric cancer (GC). A. Expression of Rab3IP transcripts in paired GC tissues and peritumoral normal tissues. B. Western blot analysis of Rab3IP in paired GC tissues and peritumoral normal tissues. C. Western blot analysis of Rab3IP in GC cell lines. D. Tissue microarray (TMA) analysis of Rab3IP in primary human GC tissues. ** means P<0.001. Scar bar, 100 nm.
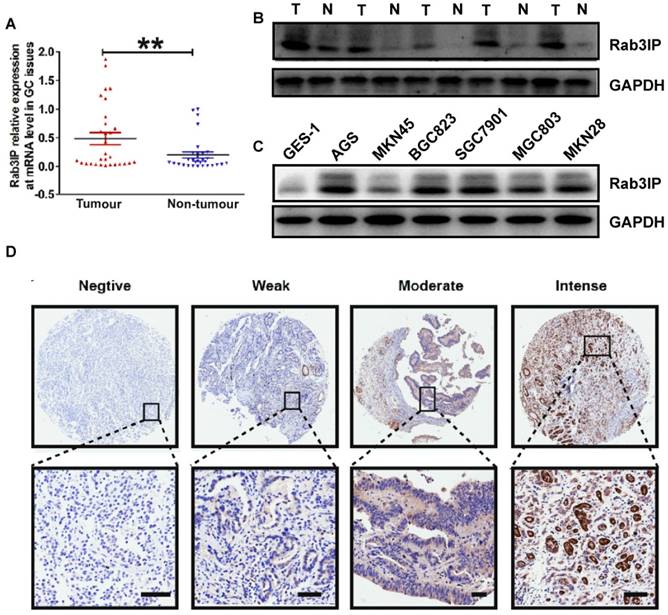
Depletion of Rab3IP suppresses cell proliferation in gastric cancer. A. Quantitative real-time-PCR analysis to confirm knock-down efficiency of Rab3IP transcripts in AGS cells. B. CCK-8 assay using AGS-Rab3IP (-) and AGS- Rab3IP (-)-NC cells. C. Clone formation assay using AGS-Rab3IP (-) and AGS- Rab3IP (-)-NC cells. D. Apoptosis rates of AGS-Rab3IP (-) and AGS- Rab3IP (-)-NC cells using Annexin V-FITC/PI kit. E. Tumour xenograft model established by subcutaneous injection in nude mice (n = 3). Data points are presented as mean tumour weight (gram) and tumour volume (mm3). *means P<0.05, ** means P<0.01, *** means P<0.001.
Depletion of Rab3IP reduces proliferation of GC cells
The observation that overexpression of Rab3IP was correlated with tumour size and T stage further inspired us to examine the functional role of Rab3IP in GC cell lines. To investigate the effects of Rab3IP on the proliferation of GC cells, lentivirus-mediated knockdown of Rab3IP was employed in the AGS cells, as the expression was relatively higher in AGS cells based on western blotting results (Fig 1C). The qRT-PCR results showed that lentivirus targeting Rab3IP efficiently reduced the mRNA expression of RAB3IP in AGS cells (Fig 2A). Owing to the reduction in Rab3IP levels, AGS-Rab3IP (-) cells showed lower OD value in the CCK-8 assay (Fig 2B), fewer cell colonies in the plate clone formation assay (Fig 2C), and a 2-fold higher apoptotic rate in flow cytometry (Fig 2D), compared to the AGS-Rab3IP (-)-NC control group. In addition, elimination of Rab3IP significantly suppressed the tumorigenicity of AGS cells in vivo, as AGS-Rab3IP (-) cells exhibited less tumour mass and volume compared to the AGS-Rab3IP (-)-NC control group (Fig 2E).
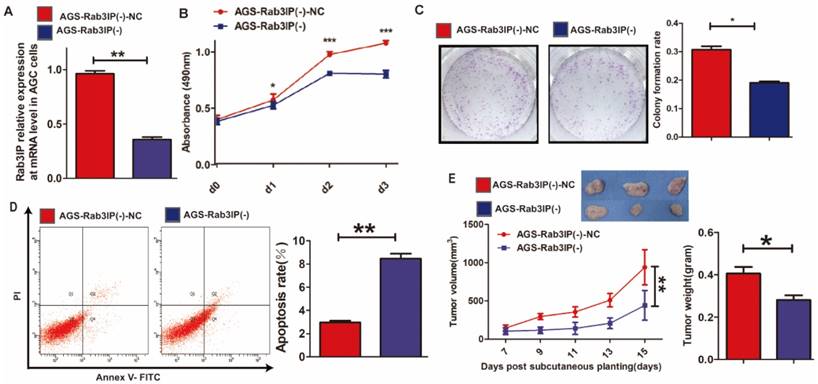
Upregulation of Rab3IP enhances proliferation of GC cells
Rab3IP was overexpressed in MKN45 cells using a lentivirus system, since the expression was relatively lower in MKN45 cells based on western blotting results (Fig 1D). After the transfection of Rab3IP plasmid, the MKN45-Rab3IP (+) group showed higher expression of RAB3IP transcripts (60-fold higher) compared to the control group (Fig 3A). Owing to the upregulation of Rab3IP, MKN45-Rab3IP (+) cells showed higher OD value in the CCK-8 assay (Fig 3B), more cell colonies in the plate clone formation assay (Fig 3C), and a lower apoptotic rate in flow cytometry (Fig 3D), compared to the MKN45-Rab3IP (+)-NC control group. Furthermore, overexpression of Rab3IP dramatically promoted cell proliferation in vivo, as MKN45-Rab3IP (+) cells exhibited greater tumour mass and volume (Fig 3E).
Rab3IP inhibits autophagy of GC cells
Cell autophagy has been increasingly recognised to be associated with cell proliferation in GC15. To identify whether Rab3IP regulates cell proliferation in GC owing to its involvement in the process of cell autophagy, we investigated the effect of Rab3IP on autophagy-related markers. The CCK-8 results revealed that depletion of Rab3IP significantly reduced the proliferation ability of AGS cells, but this was reversed in the presence of 3-MA, a classic cell autophagy inhibitor (Fig 4A). On the contrary, enhanced cell proliferation induced by overexpression of Rab3IP was suppressed in the presence of RAPA, a classic cell autophagy inducer (Fig 4B). Confocal laser scanning microscopy (CLSM) showed that depletion of Rab3IP led to more autophagosomes, while upregulation of Rab3IP led to fewer autophagosomes; however, these phenomena could be reversed by the addition of 3-MA and RAPA, respectively (Fig 4C and Fig 4D). To ascertain whether expression of Rab3IP is associated with autophagy, western blotting was used to detect autophagy-related markers (Fig 4E). We observed an increase in the intensity of both the LC3-II and p62 bands in the AGS-Rab3IP (-) group having lower expression of Rab3IP compared with that in the negative control group. However, this increase was again restrained by the addition of 3-MA. Furthermore, MKN45-Rab3IP (+) cells that overexpressed Rab3IP exhibited a decrease in the intensity of both LC3-II and p62 bands, which was reversed by the addition of RAPA.
Rab3IP is a direct target of miR-532-3p in GC
To further explore the upstream regulatory pathway of Rab3IP in GC, we utilised publicly available bioinformatic algorithms (TargetScan and miRMALK) to predict its upstream microRNAs. The results predicted five miRNAs (miR-450-5p, miR-217, miR-532-3p, miR-935, and miR-584-5p) as potential regulators targeting Rab3IP in GC. Of these, the transfection of miR-532-3p mimic showed the most significant reduction in Rab3IP expression, both at the protein (Fig 5A) and mRNA levels (Fig 5B), compared to the control and other four miRNAs. Luciferase reporter assay was performed to further corroborate this finding, A significant decrease in luciferase activity of the wild type vector was observed, whereas mutation in the putative binding site in the 3ʹUTR region of RAB3IP abrogated the suppressive ability of miR-532-3p, verifying that miR-532-3p regulates endogenous RAB3IP by directly binding to its 3ʹUTR region in GC cells (Fig 5C). To further measure the original expression level of miR-532-3p in GC, qRT-PCR analysis was performed using 40 paired GC samples. Simultaneously, the mRNA levels of RAB3IP were determined to verify its correlation with miR-532-3p. Consistent with the bioinformatics prediction, the expression of miR-532-3p was negatively correlated with Rab3IP expression (r=-0.4761, p=0.0019) (Fig 5D). Moreover, miR-532-3p was downregulated in 77.5% (31/40) of the GC samples (Fig 5E), while Rab3IP was upregulated in 75% (30/40) of the samples (Fig 1A).
Rab3IP upregulation enhances proliferation of GC cells, which is reversed by miR-532-3p
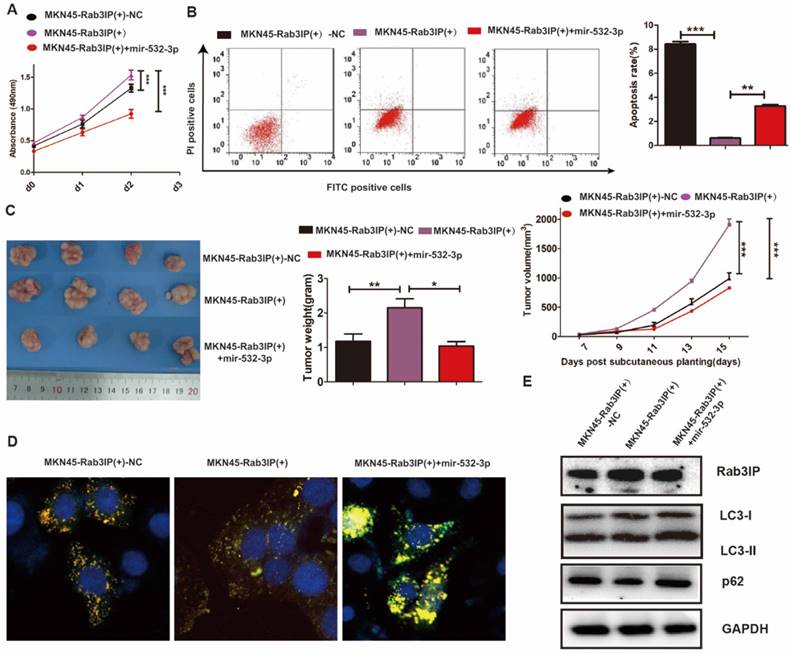
Based on the observation that the expression levels of miR-532-3p and Rab3IP are negatively correlated, their functional link was further explored. Since Rab3IP was previously demonstrated to enhance the proliferation of GC cells through inhibition of cell autophagy, we determined whether miR-532-3p could reverse this enhancing capacity. As expected, the enhanced cell proliferation induced by overexpression of Rab3IP was suppressed by the addition of miR-532-3p in the CCK-8 assay (Fig 6A). Apoptosis assay was used to determine the function of miR-532-3p, as significant alteration induced by miR-532-3p was observed compared to that in the MKN45-Rab3IP (+) group (Fig 6B). To verify the effect of miR-532-3p on tumorigenesis in vivo, GC cells were implanted subcutaneously into nude mice and miR-532-3p showed its potential in decreasing the tumour burden and slowing down the tumour growth rate (Fig 6C). Furthermore, we used CLSM to observe the indicated cells and found that miR-532-3p could induce more autophagosomes (Fig 6D). Consistent with the CLSM results, decrease in the intensity of both the LC3-II and p62 bands was observed in the MKN45-Rab3IP (+) group having higher expression of Rab3IP compared with the negative control group, and this decreased expression could be reversed by miR-532-3p (Fig 6E).
Discussion
Several studies have shown that the expression of GTPase proteins is abnormal in many tumours, such as breast cancer, lung cancer, GC, and prostate cancer16-19. A novel molecular classification of GC has been put forward, which includes four subtypes: 1. EB virus positive, 2. Microsatellite instability (MSI) type, 3. Genome-stabilised (GS) type, 4. Chromosomal instability (CIN) type. The RHOA gene mutation or the RHO family GTPase activating protein gene fusion phenomenon is more common in the GS type20. Moreover, GTPases are widely involved in the signal pathway of cell proliferation and differentiation, which might have a huge impact on the development of cancer.
Rab3IP is one of the Rab family GTPase-activating proteins, which is known to be involved in intracellular vesicular trafficking and cytoskeleton regulation. Recent research concerning about the biological function of Rah3IP(Rabin8), has illustrated that the guanine nucleotide exchange reaction of Rab8 Protein was catalyzed by guanine nucleotide exchange factor Rabin8. According to the biophysical and structural details shown, mechanism of GDP displacement in the Rab8/Rabin8 system could be described as a disturbance of the structure of the nucleotide-binding regions of Rab8, in particular switch I and switch II. In addition, Rabin8 was identified as the specific GEF for Rab8 and affects the GTP/GDP exchange21, 22.Rab3IP has been reported to be closely related to the occurrence, development, and metastasis of colorectal cancer, salivary gland cancer, and cervical cancer9, 23. In our previous study, we also found that, Rab3IP is upregulated in both gastric cancer cell lines and human tissues, and interacts with SSX2. Furthermore, we also revealed the involvement of Rab3IP and SSX2 in the epithelial-mesenchymal transition (EMT) and their potential implication in the invasion and metastasis of gastric cancer10. However, the specific mechanism of action of Rab3IP during the proliferation of GC cells have not been fully elucidated. Our study demonstrated that Rab3IP is upregulated in GC tissues. The expression level of Rab3IP was found to be highly correlated with tumour size, differentiation degree, tumour (T) stage, lymph node (N) stage, and serum CEA and CA-724 levels. To explore the biological function of Rab3IP in GC, we depleted Rab3IP in AGS cells. This caused the in vitro proliferative capacity of AGS cells to be significantly inhibited, while apoptosis increased. Consistently, the tumour volume and quality in AGS-transfected nude mice were significantly reduced. On the other hand, the overexpression of Rab3IP in MKN45 cells caused enhanced cell growth rate both in vitro and in vivo. As observed in GC, Rab3IP was found to be overexpressed in gliomas7. The authors used a 12q13-21 amplicon-specific genomic microarray and a bioinformatics amplification prediction tool, which showed that Rab3IP was one of the overexpressed genes in the amplified region, thereby confirming that Rab3IP is related to the proliferation of gliomas at the gene level. These results suggested that Rab3IP could promote tumour cell proliferation in GC. According to recent advancement in the molecular function of Rab3IP, it was proved to interact with SSX2, Rab8 and other proteins as an activator of GTP/GDP exchange. Thus, we hypothesis that it plays an important role in progress of gastric cancer. In addition, Rab proteins may affects the actions of Rab3IP. However, due to the limitation of the current study, we have not found the direct relation between Rab3IP biological function and the Rab3IP-Rab8 interaction21, 22.
Autophagy is a highly conserved lysosome-dependent catabolic pathway in eukaryotes. It is closely related to various physiological and pathological processes and plays an important role in the development of tumours. The mTOR signalling pathway is a classical autophagy signalling pathway. Studies have shown that GTPases are involved in the mTOR signalling pathway and regulate autophagy in cancer cells24. Many Rab proteins, such as Rab1, Rab5, Rab7, Rab9A, Rab11, Rab23, Rab32, Rab33B, and Rab37, are involved in autophagy25-27. Rabl3 has been shown to be involved in the inhibition of autophagosome formation in lung cancer cells28. Furthermore, downregulation of Rab25 and Rab7 can promote autophagy of ovarian cancer cells29, 30. Therefore, we speculated that Rab3IP might promote the proliferation and differentiation of GC cells by affecting autophagy. We thus transfected AGS-Rab3IP (-) cells, MKN45-Rab3IP (+) cells, and their negative controls with LC3 double-fluorescing autophagic virus and observed them using CLSM.
Overexpression of Rab3IP promotes cell proliferation in gastric cancer. A. Quantitative real-time-PCR analysis to show overexpression of Rab3IP transcripts in MKN45 cells. B. CCK-8 assay using MKN45-Rab3IP (+) and MKN45- Rab3IP (+)-NC cells. C. Clone formation assay using MKN45-Rab3IP (+) and MKN45- Rab3IP (+)-NC cells. D. Apoptosis rates of MKN45-Rab3IP (+) and MKN45- Rab3IP (+)-NC cells using Annexin V-FITC/PI kit. E. Tumour xenograft model established by subcutaneous injection in nude mice (n = 3). Data points are presented as mean tumour weight (gram) and tumour volume (mm3). *means P<0.05, ** means P<0.01, *** means P<0.001.
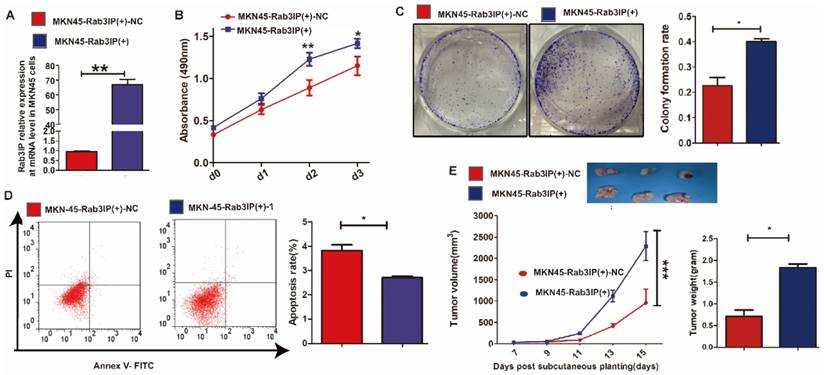
Rab3IP promotes cell autophagy in gastric cancer. A. Effect of downregulation of Rab3IP and addition of 3-methyladenine (3-MA) on cell proliferation using CCK-8 assay. B Effect of overexpression of Rab3IP and addition of RAPA on cell proliferation using CCK-8 assay. C LC3 fluorescence in AGS cells after downregulation of Rab3IP and addition of 3-MA. D LC-3 fluorescence in MKN45 cells after overexpression of Rab3IP and addition of RAPA. E Western blot analysis of Rab3IP, LC3-I, LC3-II, p62, and GAPDH in AGS and MKN45 cells. *** means P<0.001. Scale bar, 50 nm.
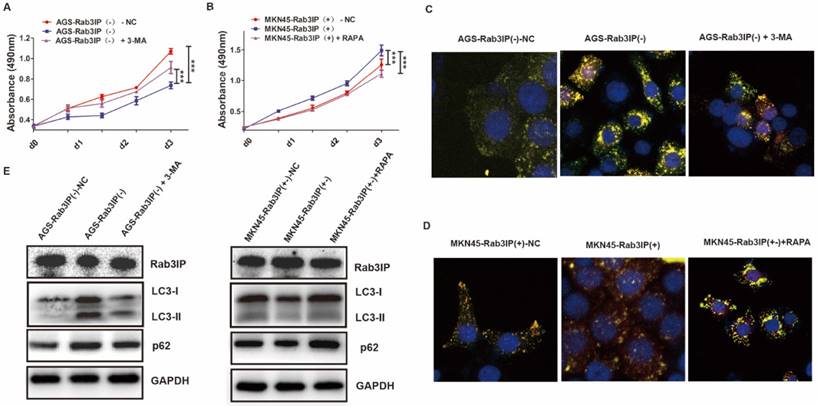
Rab3IP is a direct target of miR-532-3p in gastric cancer (GC). A. Western blot analysis of Rab3IP in the indicated cells treated with different mimics. B. Expression analysis of Rab3IP in the indicated cells treated with different mimics. C. Luciferase assay revealed that miR-532-3p directly binds to the 3ʹUTR region of Rab3IP. D. Correlation between miR-532-3p and Rab3IP at mRNA level in 40 paired human gastric cancer tissues and adjacent normal mucosa tissues. E. Upregulation of miR-532-3p was found in (31/40) 77.5% of GC cases. Data represents the log2 ratio of miR-532-3p level in GC tissues compared to the corresponding non-tumour tissues. F. The expression of miR-532-3p in GC tumour (T) versus non-tumour (NT) tissues, as detected by qRT-PCR. *means P<0.05.
The results showed that depletion of Rab3IP led to more autophagosomes, while upregulation of Rab3IP led to fewer autophagosomes; these effects were reversed by the addition of an autophagy inhibitor (3-MA) and enhancer (RAPA), respectively. To further corroborate the correlation of Rab3IP expression with autophagy, we used western blotting to detect autophagy-related markers and observed a similar trend. Autophagy can promote cell proliferation as a physiological process in normal cells. However, autophagy has been shown to inhibit proliferation in cancer cells like bladder cancer cells and breast cancer cells31, 32. In our study, upregulation of Rab3IP significantly promoted cell proliferation. Considering that it also led to the reduction of the number of autophagosomes, we concluded that the effect of Rab3IP on the proliferation of GC cells is mediated through regulation of autophagy.
Enhanced cell proliferation induced by Rab3IP is restrained by miR-532-3p in gastric cancer. A. Effect of overexpression of miR-532-3p and Rab3IP on cell proliferation using CCK-8 assay. B. Effect of overexpression of Rab3IP and addition of miR-532-3p on cell apoptosis using Annexin V-FITC/PI assay. C. Tumour xenograft model established by injecting Rab3IP-overexpressed MKN45 cells, while miR-532-3p agomir, a small molecule inhibitor, was injected into the tumour (n = 3). Data points are presented as mean tumour weight (gram) and tumour volume (mm3). D. LC-3 fluorescence in MKN45 after introduction of Rab3IP and addition of miR-532-3p. E. Western blot analysis of Rab3IP, LC3-I, LC3-II, p62, and GAPDH in MKN45 cells after introduction of Rab3IP and addition of miR-532-3p. *means P<0.05, ** means P<0.01, *** means P<0.001. Scar bar, 50 nm.
There is increasing evidence that miRNAs play an important role in several key biological processes of GC and its pathogenesis. miRNAs are important inhibitors of oncogenes, such as Rab40b, Rab40c, Rab18, Rab22A, and other Rab family members, in various cancers33-36. Here, we found that miR-532-3p can directly target Rab3IP and that its expression is closely related to the expression level of Rab3IP in GC tissues. Earlier studies have found that miR-532-3p, a tumour suppressor, targets cancer cells by inducing new oncogenes, such as KRAS, MKL2, WNT9A, CSNK2A2, CHD4, and SH3PXD2A, in ovarian and lung adenocarcinoma37-40. Here, we demonstrate that miR-532-3p is downregulated in GC and exerts its biological function by targeting Rab3IP. We verified the low expression levels of miR-532-3p in GC tissues and demonstrated that miR-532-3p could reverse Rab3IP's promotion of proliferation in GC cells. Subsequently, we studied the biological function of miR-532-3p in GC cells and nude mice. The results showed that miR-532-3p can induce the production of more autophagosomes and reduce the tumour burden in nude mice. In addition, the expression of Rab3IP and miR-532-3p was linearly correlated in GC cells, confirming that the regulation of Rab3IP by miR-532-3p plays an important role in the development of GC.
Conclusion
In summary, this study clarifies the essential link between miR-532-3p and Rab3IP in regulating the proliferation ability and their involvement in the autophagy process in human GC cells. Taking this into consideration, Rab3IP could serve as a potential target for the treatment of GC. However, the present study has some limitations. Firstly, the mechanism of how Rab3IP regulates cell proliferation through autophagy remains unclear. In addition, how miR-532-5p regulates cell proliferation in GC through the Rab3IP/autophagy pathway requires further exploration.
Correlation between Rab3IP protein expression and the clinicopathological parameters.
| Variables | Cases, n | Rab3IP protein expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | ||||
| Male | 105 | 36 | 69 | 0.573 |
| Female | 45 | 13 | 32 | |
| Age at diagnosis (years) | ||||
| <60 | 92 | 25 | 67 | 0.077 |
| ≥60 | 58 | 24 | 34 | |
| BMI | ||||
| <25 | 116 | 42 | 74 | 0.100 |
| ≥25 | 34 | 7 | 27 | |
| Abdominal surgery | ||||
| Yes | 130 | 42 | 88 | 0.802 |
| No | 20 | 7 | 13 | |
| Comorbidity | ||||
| Yes | 102 | 35 | 67 | 0.580 |
| No | 48 | 14 | 34 | |
| Complication | ||||
| Yes | 59 | 17 | 42 | 0.478 |
| No | 91 | 32 | 59 | |
| CA-199 | ||||
| ≤37 | 120 | 42 | 78 | 0.279 |
| >37 | 30 | 7 | 23 | |
| CEA | ||||
| ≤5 | 116 | 43 | 73 | 0.038 |
| >5 | 34 | 6 | 28 | |
| CA-125 | ||||
| ≤6 | 99 | 38 | 61 | 0.044 |
| >6 | 51 | 11 | 40 | |
| Size (diameter, cm) | ||||
| <3 | 38 | 19 | 19 | 0.015 |
| ≥3 | 112 | 30 | 82 | |
| Differentiation Degree | ||||
| Well and medium | 43 | 20 | 23 | 0.033 |
| Poorly and undifferentiated | 107 | 29 | 78 | |
| Lymphatic invasion | ||||
| Yes | 81 | 30 | 51 | 0.227 |
| No | 69 | 19 | 50 | |
| Nerve invasion | ||||
| Yes | 47 | 18 | 29 | 0.351 |
| No | 103 | 31 | 72 | |
| Vascular invasion | ||||
| Yes | 91 | 33 | 58 | 0.287 |
| No | 58 | 16 | 43 | |
| T stage | ||||
| 1 and 2 | 38 | 19 | 19 | 0.015 |
| 3 and 4 | 112 | 30 | 82 | |
| N stage | ||||
| 0 | 47 | 24 | 23 | 0.002 |
| 1 and 2 | 103 | 25 | 78 | |
| M stage | ||||
| M0 | 128 | 43 | 85 | 0.631 |
| M1 | 22 | 6 | 16 | |
Abbreviations
Rab3IP: RAB3A interacting protein; GC: gastric cancer; T: tumour tissues; N: normal tissues; AGS- Rab3IP (-)-NC cells: Negative control AGS cells; CCK-8: cell counting kit-8; AGS-Rab3IP (-) cells: Rab3IP-knockdown cell lines; MKN45-Rab3IP (+) cells: Rab3IP overexpression MKN45 cell lines: MKN45- Rab3IP (+)-NC cells: Negative control MKN45 cell; CCK-8: cell counting kit-8; FITC: fluorescein isothiocyanate; PI: propidium iodide; 3-MA: 3-Methyladenine; RAPA: Rapamycin; LC3: Light chain 3; p62: Sequestosome 1; miR-450b-5p: microRNA-450b-5p; miR-27: microRNA-27; miR-532-3p: microRNA-532-3P; miR-584-5p: microRNA-584-5p; miR-NC: microRNA- negative control; UTR: Untranslated region; r: correlation coefficient; BMI: Body Mass Index; CA-199: Carbohydrate Antigen 199; CEA: Carcinoembryonic antigen; CA-125: Carbohydrate Antigen 125; T: Tumour; N: Lymph node; M: Metastasis.
Supplementary Material
Supplementary information.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81500399; 81672446), the Guangzhou City Science and Technology Project-Zhujiang Technology New Star Project (201710010017), the Natural Science Foundation of Guangdong Province, China (2017A030306023), the Special Funds for the Cultivation of Guangdong College Students' Scientific and Technological Innovation (pdjhb0111), the State's Key Project of Research and Development Plan (2017YFC0108300, 2017YFC0108301), the Southern Medical University Clinical Research Start-Up Project (LC2016ZD003), the Key Clinical Specialty Discipline Construction Program ([2011]170), and the Outstanding Youth Development Program Fund (2017J006).
Ethical considerations
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. All institutional and national guidelines for the care and use of laboratory animals were followed.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: Potential diagnostic and therapeutic applications. Surg Today. 2011;41:24-38
3. Kelly EE, Horgan CP, Goud B, McCaffrey MW. The Rab family of proteins: 25 years on. Biochem Soc Trans. 2012;40:1337-1347
4. Blumer J, Rey J, Dehmelt L. et al. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200:287-300
5. Wang J, Ren J, Wu B. et al. Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc Natl Acad Sci U S A. 2015;112:148-153
6. de Bruijn DR, Dos SN, Kater-Baats E. et al. The cancer-related protein SSX2 interacts with the human homologue of a Ras-like GTPase interactor, RAB3IP, and a novel nuclear protein, SSX2IP. Genes Chromosomes Cancer. 2002;34:285-298
7. Fischer U, Keller A, Leidinger P. et al. A different view on DNA amplifications indicates frequent, highly complex, and stable amplicons on 12q13-21 in glioma. Mol Cancer Res. 2008;6:576-584
8. Zhang L, Mitani Y, Caulin C. et al. Detailed genome-wide SNP analysis of major salivary carcinomas localizes subtype-specific chromosome sites and oncogenes of potential clinical significance. Am J Pathol. 2013;182:2048-2057
9. Hur K, Cejas P, Feliu J. et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635-646
10. Ren H, Xu Z, Guo W, Deng Z, Yu X. Rab3IP interacts with SSX2 and enhances the invasiveness of gastric cancer cells. Biochem Bioph Res Co. 2018
11. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704-714
12. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-469
13. Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35-47
14. Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849-864
15. Zhou H, Yuan M, Yu Q, Zhou X, Min W, Gao D. Autophagy regulation and its role in gastric cancer and colorectal cancer. Cancer Biomark. 2016;17:1-10
16. Wu J, Sang M, Cao W, Zheng J, Pei D. [Identification analysis of eukaryotic expression plasmid Rap2a and its effect on the migration of lung cancer cells]. Zhongguo Fei Ai Za Zhi. 2014;17:643-648
17. McSherry EA, Brennan K, Hudson L, Hill AD, Hopkins AM. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res. 2011;13:R31
18. Saigusa S, Tanaka K, Mohri Y. et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer. 2015;18:84-92
19. Yang C, He H, Zhang T, Chen Y, Kong Z. Decreased DAB2IP gene expression, which could be induced by fractionated irradiation, is associated with resistance to gammarays and alphaparticles in prostate cancer cells. Mol Med Rep. 2016;14:567-573
20. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209.
21. Wang J, Ren J, Wu B. et al. Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc Natl Acad Sci U S A. 2015;112:148-153
22. Guo Z, Hou X, Goody RS, Itzen A. Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J Biol Chem. 2013;288:32466-32474
23. Zhang L, Mitani Y, Caulin C. et al. Detailed Genome-Wide SNP Analysis of Major Salivary Carcinomas Localizes Subtype-Specific Chromosome Sites and Oncogenes of Potential Clinical Significance. The American Journal of Pathology. 2013;182:2048-2057
24. Martin TD, Chen X, Kaplan REW. et al. Ral and Rheb GTPase Activating Proteins Integrate mTOR and GTPase Signaling in Aging, Autophagy, and Tumor Cell Invasion. Mol Cell. 2014;53:209-220
25. Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348-358
26. Feng ZZ, Jiang AJ, Mao AW. et al. The <em>Salmonella</em> effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J Biol Chem. 2018
27. Song Y, Shang D, Cheng H, Zhou R. The small GTPase RAB37 functions as an organizer for autophagosome biogenesis. Autophagy. 2018:1-3
28. Zhang W, Sun J, Luo J. High Expression of Rab-like 3 (Rabl3) is Associated with Poor Survival of Patients with Non-Small Cell Lung Cancer via Repression of MAPK8/9/10-Mediated Autophagy. Med Sci Monitor. 2016;22:1582-1588
29. Lu Z, Yang H, Sutton MN. et al. ARHI (DIRAS3) induces autophagy in ovarian cancer cells by downregulating the epidermal growth factor receptor, inhibiting PI3K and Ras/MAP signaling and activating the FOXo3a-mediated induction of Rab7. Cell Death Differ. 2014;21:1275-1289
30. Liu Y, Tao X, Jia L. et al. Knockdown of RAB25 promotes autophagy and inhibits cell growth in ovarian cancer cells. Mol Med Rep. 2012;6:1006-1012
31. Zhao C, She T, Wang L. et al. Daucosterol inhibits cancer cell proliferation by inducing autophagy through reactive oxygen species-dependent manner. Life Sci. 2015;137:37-43
32. Ying L, Huang Y, Chen H. et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407-411
33. Yang Q, Jie Z, Cao H. et al. Low-level expression of let-7a in gastric cancer and its involvement in tumorigenesis by targeting RAB40C. Carcinogenesis. 2011;32:713-722
34. Zhang B, Yin Y, Hu Y. et al. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32:331
35. Liu J, Zhang J, Li Y, Wang L, Sui B, Dai D. MiR-455-5p acts as a novel tumor suppressor in gastric cancer by down-regulating RAB18. Gene. 2016;592:308-315
36. Li Y, Jia Q, Wang Y, Li F, Jia Z, Wan Y. Rab40b upregulation correlates with the prognosis of gastric cancer by promoting migration, invasion, and metastasis. Med Oncol. 2015;32:126
37. Wang F, Chang JT, Kao CJ, Huang RS. High Expression of miR-532-5p, a Tumor Suppressor, Leads to Better Prognosis in Ovarian Cancer Both In Vivo and In Vitro. Mol Cancer Ther. 2016;15:1123-1131
38. Bai L, Wang H, Wang AH, Zhang LY, Bai J. MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. Plos One. 2017;12:e173912
39. Hu S, Zheng Q, Wu H, Wang C, Liu T, Zhou W. miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017;177:15-19
40. Griesing S, Kajino T, Tai MC. et al. Thyroid transcription factor-1-regulated microRNA-532-5p targets KRAS and MKL2 oncogenes and induces apoptosis in lung adenocarcinoma cells. Cancer Sci. 2017;108:1394-1404
Author contact
![]() Corresponding authors: Yanfeng Hu, Guangzhou 510515, China, yfenghucom, +86-020-84112354 and Guoxin Li, Guangzhou 510515, China, gzliguoxincom, +86-020-61641682
Corresponding authors: Yanfeng Hu, Guangzhou 510515, China, yfenghucom, +86-020-84112354 and Guoxin Li, Guangzhou 510515, China, gzliguoxincom, +86-020-61641682

 Global reach, higher impact
Global reach, higher impact