3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(3):654-664. doi:10.7150/jca.28537 This issue Cite
Review
Prognostic and Clinicopathological Significance of MiR-155 in Hematologic Malignancies: A Systematic Review and Meta-analysis
1. Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
2. Hubei clinical medical center of cell therapy for neoplastic disease.
3. Collaborative Innovation Center of Hematology, Huazhong University of Science and Technology, Wuhan, Hubei 430022, China.
4. Institute of Orthopedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
Received 2018-7-15; Accepted 2018-11-5; Published 2019-1-1
Abstract
Background: Aberrant miR-155 expression has been reported in various types of hematologic malignancies. However, the prognostic and clinicopathological value of miR-155 remains unclear. Here, we performed this systemic review and meta-analysis to comprehensively evaluate the prognostic and clinicopathological significance of miR-155 expression in hematologic malignancies.
Methods: We systematically searched the PubMed, EMBASE, ISI Web of Science, Cochrane library databases and OVID to identify eligible studies published from Jan 1, 2008 to Aug 1, 2018. The pooled hazard ratios (HRs) and odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used to detect the prognostic and clinicopathological role of miR-155 in hematologic malignancies.
Results: A total of 18 studies including 2316 patients were enrolled in the present meta-analysis, indicating significant association between elevated miR-155 expression and poor overall survival (OS) in 2114 patients (pooled HR = 1.72, 95%CI [1.50-1.97], p<0.001). Elevated miR-155 expression level was related to shorter event free survival (EFS, pooled HR = 1.55, 95%CI [0.94-2.57], P=0.002), disease free survival (DFS, pooled HR = 1.38, 95%CI [1.13-1.68], P=0.001), progress free survival (PFS, pooled HR = 1.58, 95%CI [1.06-2.35], p<0.001) and treatment free survival (TFS, pooled HR = 1.67, 95%CI [1.16-2.39], P=0.006). Additionally, overexpression of miR-155 was found to be significantly related to FLT3/ITD presence (OR=4.751, 95%CI [3.229-6.990], P<0.001), more WT1 mutation (OR=2.090, 95%CI [1.240-3.522], P=0.006) and less CEBPA mutation (OR=0.477, 95%CI [0.286-0.794], P=0.004) in 552 AML patients.
Conclusion: MiR-155 expression was found to be associated with several leukemia-related phenotype and poor prognosis in hematologic malignancies. Therefore, miR-155 overexpression might be a convinced unfavorable prognostic indicator that helps the clinical decision-making process.
Keywords: MiR-155, Hematologic malignancies, Biomarker, Prognosis, Meta-analysis
Introduction
In recent years, “personalized medicine” is a new principle that aims at tailoring medical treatment of the individual patients, thereby ending the current “one-fits-all” strategy. At present stage, cancer diagnostics are mainly based on the clinical manifestation, morphology, histology, cytogenetic, immune-phenotyping, and molecular genetic data, but identification of the molecular pathway driving tumorigenesis often fails [1]. Therefore, exploring and identifying novel biomarkers for early diagnosis and prognosis is necessary for a better control of cancer. MicroRNAs (miRNAs) are non-coding, small (19-24 nucleotides in size) single-stranded RNAs that have emerged as potent regulators of the target genes messenger RNAs expression in the response of cell to both physiological and pathophysiological conditions [2]. MiRNAs are deregulated in many types of solid and hematologic malignancies, where they can function as oncogenes, favoring initiation and progression of cancers, or as tumor suppressors, preventing tumorigenesis. Reflecting pathological processes today, miRNAs are widely validated for their potential role in diagnostic, prognostic and novel therapeutic targeting for cancers.
MicroRNA-155 (miR-155) is a kind of microRNAs expressed in hematopoietic cells. Recent data indicate that miR-155 plays a very important role in the pathogenesis of hematological malignancies through regulating cell signal transduction pathways of cell proliferation, differentiation and apoptosis [3, 4]. The previous meta-analysis has shown that miR-155 has great potential as a novel noninvasive biomarker for human cancer detection, including several kinds of solid tumor and hematologic malignancies. [5]. Altered expression of miR-155 has been reported in several hematologic malignancies [2, 6-10], including diffuse large B-cell lymphoma (DLBCL), Hodgkin's lymphoma (HL), follicular lymphoma (FL), acute myeloid leukemia (AML), acute and chronic lymphoblastic leukemia (ALL, CLL), and myelodysplastic syndrome (MDS). Upregulated miR-155 expression was further associated with tumor progression and poor prognosis in several reports. Therefore, miR-155 may be an important indicator to assess the diagnosis, treatment and prognosis of patients with hematological malignancies. However, some studies reported its tumor-repressor role in several malignancies, such as Burkitt lymphoma. Obviously, there is a debate among miR-155's role in hematological malignancies. Meanwhile, most studies reported so far are limited in discrete outcome and sample size. To elaborate and assess the potential of miR-155 as a prognostic biomarker in hematologic malignancies, we conducted this systematic review and quantitative meta-analysis to clarify its prognostic and clinicopathological significance.
Materials and Methods
This systematic review and meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [11].
Literature search
Two authors (Tang and Mei) independently used the following tools: PubMed, EMBASE, ISI Web of Science, Cochrane library databases and OVID to obtain relevant articles (published from Jan 1, 2008 to Aug 1, 2018). We used the following combined text and Medical Subject Headings (MeSH) as follows: terms: "Lymphoma", "Leukemia", "Hematological Neoplasms" and "MIRNA155 microRNA, human". The complete search used for PubMed was: (((("Lymphoma" [Mesh] OR "Lymphomas" [Text Word] OR "Lymphoma, Malignant" [Text Word] OR "Lymphomas, Malignant" [Text Word] OR "Malignant Lymphoma" [Text Word] OR "Malignant Lymphomas" [Text Word]) OR ("Leukemia" [Mesh] OR "Leukemias" [Text Word] OR "Leucocythaemia" [Text Word] OR "Leucocythaemias" [Text Word] OR "Leucocythemia" [Text Word] OR "Leucocythaemias" [Text Word])) OR ("Hematological Neoplasms" [Mesh] OR "Hematologic Malignancy" [Text Word] OR "Neoplasms, Hematological" [Text Word] OR "Hematological Neoplasm" [Text Word] OR "Neoplasm, Hematological" [Text Word] OR "Hematologic Neoplasm" [Text Word] OR "Neoplasm, Hematologic" [Text Word] OR "Malignancies, Hematological" [Text Word] OR "Malignancy, Hematologic" [Text Word] OR "Hematological Malignancies" [Text Word] OR "Hematologic Malignancies" [Text Word] OR "Malignancies, Hematologic" [Text Word] OR "Malignancy, Hematologic" [Text Word] OR "Hematopoietic Neoplasms" [Text Word] OR "Neoplasms, Hematopoietic" [Text Word] OR "Hematopoietic Neoplasm" [Text Word] OR "Neoplasm, Hematopoietic" [Text Word] OR "Hematopoietic Malignancies" [Text Word] OR "Hematopoietic Malignancy" [Text Word] OR "Malignancies, Hematopoietic" [Text Word]))) AND ("MIRN155 microRNA, human" [Supplementary Concept] OR "miR-155, human" [Text Word] OR "miR-155 microRNA, human" [Text Word] OR "hsa-mir-155 microRNA" [Text Word] OR "miRNA-155, human" [Text Word] OR "microRNA-155, human" [Text Word] OR "miR-155-5p, human" [Text Word])). All eligible studies were considered for this review, and we also did a manual search, using the reference lists of key articles published.
Literature selection
The same two researchers (Tang and Mei) independently assessed all the included studies and extracted the data. Studies were considered eligible if they met the following inclusion criteria: (1) any type of hematologic malignancies was involved; (2) sufficient information about miR-155 expression and detection; (3) the expression levels of miR-155 were detected and divided into two groups (high expression VS low expression); (4) the survival curve or sufficient relevant data were provided to obtain HR for survival rates and their 95%CI; (5) published in English. Studies were excluded if they met the following criteria: (1) letters, case reports, reviews, conference abstracts and so on; (2) studies with insufficient data; (3) animal studies, cellular level studies or molecular level studies of miR-155; (4) multiple published reports. When there were several reports concerning the same study we included the high quality and most recent publication in our meta-analysis. Disagreement between the two reviewers was resolved by discussion with another reviewer (Hu).
Data extraction and Quality assessment
Data extraction was repeated independently by the two researchers (Tang and Peng), and any disagreement was resolved by a third researcher (Hu). For each study, the following characteristics were collected: the first author; publication year; country of the population enrolled; sample size; sample type, miR-155 expression level; number of patients and controls; cut off values; true and false positives and negatives; follow-up data; survival analysis methodology; HRs and 95%CIs and relevant clinicopathological parameters (gender, age, serum LDH level et al.). If HRs with 95%CIs that could not be directly obtained, we extracted necessary data from Kaplan-Meier Curves, and inputted the obtained survival rates at specified time points into the spreadsheet set up by Tierney JF et al. to calculate HRs and 95%CIs by using the Engauge Digitizer version 9.8 [12]. If possible, we asked for original data directly from the authors of the relevant studies. Newcastle-Ottawa Quality Assessment Scale (NOS) checklist [13] were used to assess the quality of the studies we selected for this meta-analysis. This scale uses a star system to evaluate a study in three domains: selection of participants, comparability of study groups, and the ascertainment of outcomes of interest.
Statistical analysis
Pooled HRs with corresponding 95%CIs were used to evaluate the association between miR-155 expression and prognosis, and an HR > 1 represented poor prognostic outcome for patients with elevated miR-155 expression. Pooled ORs and corresponding 95%CIs were used to analyze the association between miR-155 expression and clinicopathological parameters. The test for heterogeneity was carried out using a χ2-based Q test and I2 test. If no or moderate heterogeneity (I2 < 50% or P > 0.10), the fixed-effect model was applied; if not, the random-effects mode [14]. Subgroup analysis and meta-regression were further performed to explore the source of heterogeneity. Influence analysis was conducted to explore the influence of one individual study on the overall estimate. The Begg's funnel plot was used to visually evaluate the publication bias of all studies included in our meta-analysis. And then the Egger's bias indicator test was performed for each of the pooled study groups [15]. Analyses were carried out by using STATA statistical software package version 15.0 (Stata Corporation: College Station, TX, USA). A two-tailed P < 0.05 was considered to be statistically significant.
Results
Literature search information and study characteristics
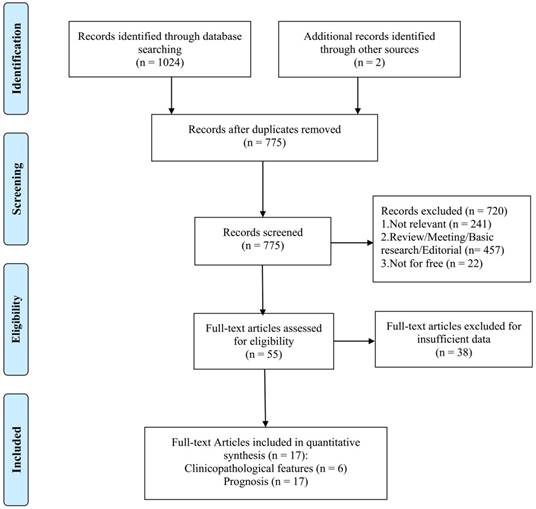
A total of 775 studies were retrieved from the databases according to the search strategy. As shown in Figure 1, 18 studies from 17 articles [6, 8, 16-30] were eligible for the final analysis on prognosis and clinicopathological features.
The comprehensive characteristics of studies included in this meta-analysis are shown in Table 1. A total of 2316 patients from 18 included eligible studies with relevant clinical data were included in this meta-analysis. The sample sizes of these studies ranged from 35 to 364. The year of publication ranged from 2012 to 2018. The miR-155 expression levels in these studies were mostly measured by quantitative real time PCR (qRT-PCR). As for cancer type, most kinds of hematologic malignancies were included. 12 studies were about leukemia (including ALL, ATL, AML and CLL), and 6 studies were about lymphoma. 5 studies were performed in Asia, and 13 studies in western countries. The cut-off value of most studies was based on the median. The follow-up time ranged from 800 days to more than 15 years. Most studies were of high quality with scores of NOS ≥ 6 except two with the score of NOS = 4 and 5, respectively.
Systematic qualitative summary of association between miR-155 and hematological malignancies
Altered expression of miR-155 has been reported in most kinds of hematologic malignancies. Here, we did a qualitative review of the association between aberrant expression of miR-155 and hematological malignancies. As shown in the Table 2, the expression of miR-155 was up-regulated in most kinds of hematologic malignancies, including DLBCL, B-NHL, HL, CLL, ALL, ATL and FLT3/ITD+ AML. Elevated expression of miR-155 correlated with poor prognosis, except that one study reported its tumor-repressor role in FLT3/ITD+ AML. In contrast to this, it has been found to be down-regulated in Burkitt lymphoma cell lines. The role of miR-155 in tumor mainly depends on the oncogenic function by its target genes and the involved underlying molecular pathways also presented in Table 2. Further exposition and analysis were shown in the discussion part.
Association between miR-155 and clinicopathological characteristics
We pooled all clinicopathological data from eligible studies to analyze their association with miR-155 expression. As shown in Table 3, increased expression level of miR-155 was significantly associated with FLT3/ITD presence (pooled OR = 4.751, 95%CI [3.229, 6.990], P<0.001), more WT1 mutation (pooled OR = 2.090, 95%CI [1.240, 3.522], P = 0.006) and less CEBPA mutation (pooled OR = 0.477, 95%CI [0.286, 0.794], P = 0.004) in 552 AML patients. However, no studies demonstrated significant association between miR-155 with patients' gender, age, serum LDH level, NPM mutation and remission (P = 0.657, 0.902, 0.177, 0.388 and 0.179, respectively).
Flow diagram of selecting relevant published works regarding miR-155 in hematologic malignancies.
Comprehensive characteristics of miR-155 test for prognosis in hematologic malignancies.
| Author | Year | Country | Tumor type | NO (high/low) | Specimen | Detection Method | Cut-off Value | Survival Analysis | Outcome | Source of HR | Follow-up | NOS Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhou et al.[30] | 2017 | China | ALL | 76(38/38) | BM | qRT-PCR | median | multivariate | OS | reported | NA | 6 |
| Ishihara et al.[20] | 2012 | Japan | ATL | 35(24/11) | plasma | qRT-PCR | median | K-M Curve | OS | K-M Curve | 1000 days | 4 |
| Marcucci et al.[21] | 2013 | USA | AML | 363(181/182) | BM/blood | nCounter | median | multivariate | OS, DFS | reported | 5 years | 9 |
| Metzeler et al.[22] | 2013 | USA | AML | 364(174/190) | BM/blood | Microarray | median | multivariate | OS, DFS | reported | 5 years | 8 |
| Ramamurthy et al.[8] | 2016 | USA | AML | 198(50/148) | BM | qRT-PCR | median | univariate | OS, EFS | reported | 7 years | 8 |
| Papaioannou et al.[26] | 2017 | USA | AML | 94(48/46) | blood | qRT-PCR | median | multivariate | DFS | reported | 10 years | 6 |
| Narayan et al.[28] | 2017 | Australia | AML | 59(10/49) | NA | qRT-PCR | 10-fold | K-M Curve | OS | K-M Curve | 800 days | 6 |
| Ferrajoli et al.[17] | 2013 | USA | CLL | 143(63/80) | plasma | qRT-PCR | 56th percentile | univariate, multivariate | OS | reported | 250 months | 7 |
| Cui et al.(1)[16] | 2014 | USA | CLL | 86(55/31) | PBMCs | qRT-PCR | median | multivariate | OS, TFS | reported | >15 years | 8 |
| Cui et al.(2)[16] | 2014 | USA | CLL | 181(95/86) | PBMCs | Microarray | median | multivariate | OS, TFS | reported | >15 years | 6 |
| Guinn et al.[18] | 2015 | USA | CLL | 109(53/56) | NA | nCounter | median | multivariate | OS, PFS | K-M Curve | 144 months | 7 |
| Papageorgiou et al.[19] | 2017 | Greece | CLL | 88(20/68) | PBMCs | qRT-PCR | median | multivariate | OS | reported | 120 months | 6 |
| Zhong et al.[23] | 2012 | China | DLBCL | 90(51/39) | FFPE | qRT-PCR | median | multivariate | OS | reported | 84 months | 7 |
| Iqbal et al.[24] | 2015 | USA | DLBCL | 54(27/27) | CS | qRT-PCR | 2-fold | K-M Curve | EFS | K-M Curve | 12 years | 8 |
| Go et al.[25] | 2015 | korea | DLBCL | 200(79/121) | FFPE | qRT-PCR | median | K-M Curve | OS, PFS | K-M Curve | 140 months | 7 |
| Ahmadvand et al.[27] | 2018 | Iran | DLBCL | 40(19/21) | plasma | qRT-PCR | median | univariate | OS | reported | 25 months | 5 |
| Wu et al.[29] | 2018 | China | DLBCL | 82(32/47) | FFPE | qRT-PCR | 12-fold | univariate, multivariate | OS, PFS | reported | 50 months | 8 |
| Bedewy et al.[6] | 2016 | Egypt | B-NHL | 54(30/24) | serum | qRT-PCR | median | multivariate | EFS | reported | 40 months | 6 |
Systematic qualitative summary of association between miR-155 and hematological malignancies
| Hematologic Malignancy | MiR-155 Expression | Prognosis | Association and Underlying Mechanism | Ref |
|---|---|---|---|---|
| Leukemia | ||||
| AML | Up-regulated, especially in FLT3/ITD+ AML | Poor | Inhibit SHIP1 and C/EBPβ, then promote PI3K-AKT and IL-6. PI3K-AKT reduced apoptosis and leukemogenesis, and IL-6 reduced cell differentiation and increased precursor cell proliferation. | [4, 8, 21, 22, 26, 28, 34, 35] |
| Up-regulated, especially in FLT3/ITD+ AML | Improved | Increase myelomonocytic markers (CD11b, CD14, and CD15) and the expression of cleaved caspase-3 with a enhanced apoptosis, reduced cell growth and clonogenic capacity. | [32] | |
| ALL | Up-regulated | Poor | NA | [30] |
| ATL | Up-regulated | Poor | NA | [20] |
| CLL | Up-regulated | Poor | Reduce SHIP1 expression and enhance sensitivity to BCR ligation. | [16-19] |
| Lymphoma | ||||
| DLBCL | Up-regulated, especially in ABC subtype | Poor | Downregulation of BCL6 and HDAC4. BCL6 mediates transcriptional repression in hematopoietic cell differentiation, leukemogenesis, and inflammation by recruiting HDAC4; Higher activity of NF-κB leads to decrease in PU.1 and consequent reduction in CD10 mRNA and protein; Inhibition of SMAD5 activity. SMAD5 plays a very important role in the signaling pathway by which TGF-β inhibits the proliferation of human hematopoietic progenitor cells. | [4, 23, 24, 27, 29, 38, 42, 43] |
| B-NHL | Up-regulated | Poor | Associated with non-germinal B-cell-like type, the presence of B symptoms, involvement of extranodal sites, and higher IPI and ECOG scores. | [6] |
| HL | Up-regulated | Poor | Down-regulation of PU.1 expression, thus inhibiting immunoglobulin diversification and class switch. | [44, 45] |
| BL | Down-regulated | NA | NA | [39] |
| Other | ||||
| MDS | Up-regulated | Poor | Promote aberrant hematopoietic stem cell self-renewal and progression to AML. MiR-155 targets SHIP-1 gene which was expressed in MDS progenitor cells. Loss of SHIP-1 protein expression may result in myeloid leukemia growth. | [9, 46] |
Association between miR-155 expression and clinicopathological characteristics in hematologic malignancies.
| Clinicopathological parameters | Studies/N | Patients/N | Pooled OR (95%CI, P) | Heterogeneity (P, I², Model) |
|---|---|---|---|---|
| Gender (male vs female) | 5 | 882 | 1.063(0.831,1.389), 0.657 | 0.209, 31.8%, Fixed |
| Age (≥60 vs <60 years) | 3 | 324 | 0.951(0.430, 2.104), 0.902 | 0.046, 62.5%, Random |
| Serum LDH level (elevated vs normal) | 3 | 592 | 1.424 (0.853-2.379), 0.177 | 0.949, 0.0%, Fixed |
| FLT3/ITD (present vs absent) | 2 | 552 | 4.751 (3.229-6.990), <0.001 | 0.916, 0.0%, Fixed |
| NPM mutation (yes vs no) | 2 | 552 | 1.179 (0.811-1.712), 0.388 | 0.583, 0.0%, Fixed |
| CEBPA mutation (yes vs no) | 2 | 552 | 0.477 (0.286-0.794), 0.004 | 0.283, 13.2%, Fixed |
| WT1 mutation (yes vs no) | 2 | 552 | 2.090 (1.240-3.522), 0.006 | 0.953, 0.0%, Fixed |
| Remission (complete vs not complete) | 3 | 299 | 0.329 (0.065-1.667), 0.179 | 0.001, 85.5%, Random |
Prognostic role of miR-155 in hematologic malignancies for OS
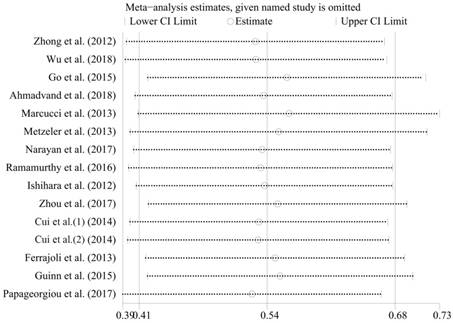
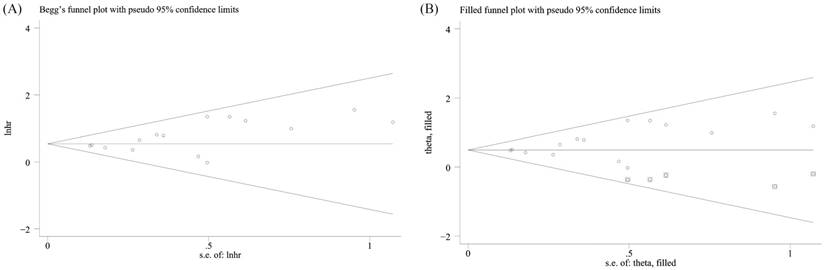
2316 patients in 18 studies (from 17 articles) [6, 8, 16-30] were investigated, and the result showed elevated miR-155 expression in poor survival in hematologic malignancies. A significant association was found between elevated miR-155 expression and poor OS in 2114 patients (pooled HR = 1.72, 95%CI [1.50-1.97], p < 0.001) (Figure 2A). Subsequently, five subgroup analyses of OS were performed by factors of the region, tumor type, B-cell malignancies or not, sample size and NOS score (Figure 2B-F, Table 4). There was no obvious heterogeneity (P = 0.593; I2 = 0.0%) across the selected datasets appraising the correlation between OS and miR-155. Moreover, significant heterogeneity was not found between groups of subgroup analyses. These results indicated that the included studies in the present meta-analysis were completely homogeneous and credible. In order to examine the stability of the pooled HRs in OS, influence analysis was carried out with the successive omission of each study (Figure 3). The leaving-one-out study revealed that there was no individual cohort influencing the results greatly. The Begg's funnel plot was demonstrated in Figure 4A, and no apparent asymmetry was found in Begg's funnel plot. The Begg's test linear regression test also proved that there was no significant publication bias (P = 0.166). However, the Egger's linear regression test indicated significant publication bias (P = 0.026). Using trim and fill analysis, we found that five studies evaluating the prognostic role of miR-155 expression in OS in hematologic malignancies remained unpublished (Figure 4B). The filled meta-analytic results for OS (pooled HR = 1.64, 95%CI [1.44-1.87], p < 0.001) supported our original results.
Prognostic role of miR-155 in hematologic malignancies for DFS, EFS, PFS, TFS
Meanwhile, we pooled several relevant studies to show the relationship between disease free survival (DFS), event free survival (EFS), progress free survival (PFS), treatment free survival (TFS) and miR-155 expression (Table 5). The results showed that elevated miR-155 expression level was significantly associated with shorter EFS (pooled HR = 1.55, 95%CI [0.94-2.57], P = 0.002), DFS (pooled HR = 1.38, 95%CI [1.13-1.68], P = 0.001), PFS (pooled HR = 1.58, 95%CI [1.06-2.35], p < 0.001) and TFS (pooled HR = 1.67, 95%CI [1.16-2.39], P = 0.006).
Forest plots of the association between miR-155 expression and OS. (A) overall. (B) tumor type. (C) B-cell malignancies or not. (D) region. (E) sample size. (F) NOS score.

Influence analysis of the studies enrolled in the present meta-analysis for OS.
Publication bias and influence analysis of the studies enrolled in the present meta-analysis for OS. (A) the Begg's funnel plot; (B) trim and fill analysis.
Overall and Subgroup meta-analysis of the relationship between miR-155 expression and OS.
| Subgroup | Studies/N | Patients/N | Pooled HR (95%CI, P) | Heterogeneity (P, I², Model) |
|---|---|---|---|---|
| Overall | 15 | 2114 | 1.72, (1.50-1.97), < 0.001 | 0.593, 0.0%, Fixed |
| Region | ||||
| west | 10 | 1631 | 1.72, (1.47-2.01), < 0.001 | 0.529, 0.0%, Fixed |
| east | 5 | 483 | 1.70, (1.28-2.25), < 0.001 | 0.543, 0.0%, Fixed |
| B-cell malignancies | ||||
| yes | 10 | 1095 | 1.77, (1.43-2.19), < 0.001 | 0.335, 11.7%, Fixed |
| no | 5 | 1019 | 1.69, (1.42-2.01), < 0.001 | 0.760, 0.0%, Fixed |
| Tumor type | ||||
| DLBCL | 4 | 412 | 1.80, (1.34-2.41), < 0.001 | 0.339, 10.7%, Fixed |
| CLL | 5 | 607 | 1.86, (1.33-2.61), < 0.001 | 0.257, 24.7%, Fixed |
| AML | 4 | 984 | 1.68, (1.41-2.00), < 0.001 | 0.543, 0.0%, Fixed |
| ALL | 1 | 76 | 0.98, (0.37-2.59), =0.967 | / |
| ATL | 1 | 35 | 3.27, (0.40-26.71), =0.269 | / |
| Sample size | ||||
| <100 | 8 | 516 | 2.53, (1.73-3.70), < 0.001 | 0.563, 0.0%, Fixed |
| ≥100 | 7 | 1598 | 1.63, (1.41-1.88), < 0.001 | 0.929, 0.0%, Fixed |
| NOS score | ||||
| <8 | 10 | 1350 | 1.67, (1.40-1.98), < 0.001 | 0.384, 6.2%, Fixed |
| ≥8 | 5 | 764 | 1.81, (1.45-2.27), < 0.001 | 0.696, 0.0%, Fixed |
Meta-analysis of the relationship between miR-155 expression and EFS, PFS, DFS and TFS.
| Outcome | Trials/N | Patients/N | Pooled HR (95%CI, P) | Heterogeneity (P, I2, Model) |
|---|---|---|---|---|
| EFS | 3 | 306 | 1.55, (0.94-2.57), 0.002 | 0.045, 67.7%, Random |
| PFS | 3 | 391 | 1.58, (1.06-2.35), <0.001 | 0.071, 62.2%, Random |
| DFS | 3 | 821 | 1.38, (1.13-1.68), 0.001 | 0.387, 0.0%, Fixed |
| TFS | 2 | 267 | 1.67, (1.16-2.39), 0.006 | 0.305, 4.9%, Fixed |
Discussion
Various miRNAs play critical roles in the causation of different hematological malignancies. MiR-155 is one of the important miRNAs that contributes greatly to the pathogenesis of diverse hematological malignancies and solid tumors with complex oncogenic as well as tumor repressor roles depending on the disease context and tissue type [2]. Overexpression of miR-155 has been found to be associated with many hematologic malignancies, suggesting its oncogenic role in their pathogenesis [4]. However, some studies reported its tumor-repressor role in several malignancies, such as Burkitt lymphoma. Therefore, there is a debate among miR-155's role in hematological malignancies. Meanwhile, most studies reported so far are limited in discrete outcome and sample size. To elaborate and assess the potential of miR-155 as a prognostic biomarker in hematologic malignancies, we conducted this systematic review and quantitative meta-analysis to clarify its prognostic and clinicopathological significance.
Our meta-analysis revealed that elevated expression level of miR-155 was significantly related to FLT3/ITD presence (pooled OR = 4.751, 95%CI [3.229-6.990]), more WT1 mutation (pooled OR=2.090, 95%CI [1.240-3.522]) and less CEBPA mutation (pooled OR=0.477, 95%CI [0.286-0.794]) in AML. However, no studies demonstrated significant association between miR-155 with patients' age, gender, serum LDH level, NPM mutation, WT1 mutation and remission. Notably, up-regulation of miR-155 was associated with FLT3/ITD mutated AML. Havelange et al. [31] found that miR-155 level in CN-AML was up-regulated, especially in the patients with FLT3/ITD mutation through the investigation of 48 cases of cytogenetically normal CN-AML. Thus, the role of miR-155 in the pathogenesis of AML may be related to the FLT3/ITD mutation in CN-AML. In a study by Palma et al. [32], knockdown of miR-155 in the FLT3-wildtype AML cells was shown to lead to resistance to cytarabine arabinoside induced apoptosis and to suppression of cell differentiation. Cammarata' group [33] showed that overexpression of miR-155 in AML with FLT3/ITD mutation was accompanied by up-regulation of corresponding transcription factors, such as PU.1 and C/EBP-β, suggesting that miR-155 maybe a target of AML. Additionally, we can find that WT1 and CEBPA mutation is also significantly related to miR-155 expression, and they have been considered as prognostic indicator for AML. However, the specific association between WT1, CEBPA mutation and miR-155 expression remains to be explored.
The present comprehensive meta-analysis pooled a total of 18 independent cohorts with 2316 patients for prognostic test of miR-155 in hematologic malignancies. The results indicated that miR-155 overexpression was a prognostic risk factor for OS (pooled HR=1.72, 95%CI [1.50-1.97]). There was no obvious heterogeneity (P=0.593; I2=0.0%) across the selected datasets appraising correlation between OS and miR-155. Subsequently, five subgroup analysis of OS were performed by region (west VS east), tumor type, B-cell malignancies (yes VS no), sample size (≥100 VS <100) and NOS score (≥8 VS <8), and no significant heterogeneity between groups of subgroup analysis. According to the results, we can find higher prognostic role of miR-155 in B-cell malignancies, especially in CLL and DLBCL. But we just used one dataset to evaluate the prognostic value of miR-155 in ALL and ATL patients because of the data limitation.
Meanwhile, we pooled several relevant studies to show the relationship between DFS, EFS, PFS, TFS and miR-155 expression. The results showed that elevated miR-155 expression level was significantly associated with shorter EFS, DFS, PFS and TFS. By combining HRs from Cox multivariate analysis, we found that elevated miR-155 expression level is an independent prognostic factor of DFS and TFS for hematologic malignancies. We used only limited studies to assess DFS, EFS, PFS and TFS, thus more and larger studies may be required.
Although our meta-analysis yielded an encouraging result of miR-155 for prognosis, several issues should be taken into consideration before suggesting any clinical conclusion. First, the cut-off values of high and low miR-155 expression were different among studies, although most of them were set to median. Second, the follow-up time were different among these studies from 800 days to more than 15 years. Third, differences of paper quality and sample size among these studies might cause bias in the meta-analysis although subgroup and meta-regression analysis did not show the paper quality or sample size as the resource of heterogeneity. Therefore, more and lager studied may be required. Fourth, HRs of five studies could not be directly obtained, thus calculating them through survival curves might not be precise enough. Lastly, most of the included studies reported positive results so our results might overestimate the prognostic significance of miR-155 in hematologic malignancies to some degree.
The obvious prognostic role of miR-155 in hematologic malignancies may be attributed to its underlying molecular mechanism and biological function. Different mechanisms have been postulated for the miR-155-mediated pathogenesis of hematological malignancies.
MiR-155-associated pathogenesis of acute myeloid leukemias has been proposed to be mediated through SHIP1 (Src homology 2 domain-containing inositol phosphatase) and C/EBP-β, two important regulators of B-cell maturation. MiR-155 is believed to promote the pathogenesis of AML by downregulating SHIP1, and thereby reversing SHIP1-mediated PI3K-Akt pathway suppression [4]. C/EBP-β is a transcription factor involved in negative regulation of the IL-6 signaling pathway in B-cells and also plays an important role in myeloid and lymphoid maturation [34]. Further, they found that knocking down SHIP1 or overexpressing miR-155 in HSPCs (Hematopoietic stem and progenitor cells) produced similar myeloproliferative phenotypes, with an increased number of CD11b+ myeloid cells in the bone marrow and spleen, decreased marrow erythropoiesis, and splenomegaly [35]. In contrast to this, Palma et al. [32] reported that overexpression of miR-155 was found to alter the expression of transcription factors and chromatin modifiers that regulate myeloid cell survival and differentiation, and upregulate mediators of the apoptotic cascade. Notably, our meta-analysis resolved this dispute, and we tend to support its indication for poor prognosis. Nevertheless, the further precise downstream pathways targeted by miR-155 remain to be elucidated.
Overexpression of miR-155 has also been found to be associated with the more aggressive and poorer prognosis type of CLL patients. Cui et al. [16] found that transfection of CLL cells with miR-155 reduced SHIP1 expression and enhanced responsiveness to B-cell receptor (BCR) ligation, whereas transfection with a miR-155 inhibitor had the opposite effect. Treatment of CLL or normal B cells with the CD40-ligand or B-cell-activating factor upregulated miR-155 and increased sensitivity to BCR ligation.
As for MDS patients, the relatively higher expressions of miR-155 had an adverse prognostic significance [9]. The dysregulation of miR-155 within the primitive bone marrow compartment may promote aberrant hematopoietic stem cell self-renewal and progression to AML. MiR-155 targets SHIP1 gene which was expressed in MDS progenitor cells, and loss of SHIP1 protein expression may result in myeloid leukemia growth. Because of its lack for sufficient data, we didn't include this study in our meta-analysis. Even so, this study supported our conclusion that miR-155 correlated with poor prognosis.
One of these proposed pathogenic mechanisms, especially for lymphomas such as DLBCL, is the down-regulation of B-cell lymphoma 6 protein (BCL6) and histone deacetylase 4 (HDAC4) by miR-155 [36]. BCL6 mediates transcriptional repression in processes such as hematopoietic cell differentiation, leukemogenesis, and inflammation by recruiting HDACs like HDAC4. Sandhu et al. [37] found that miR-155 expression inversely correlated with BCL6 and HDAC4 expression. BCL6 was indirectly targeted by miR-155 through Mxd1/Mad1 up-regulation, while its corepressor partner HDAC4 was found to be directly targeted. They also found that increased ectopic expression of HDAC4 in human activated B-cell (ABC)-type DLBCL cells reduces miR-155-induced proliferation and clonogenic potential and increases apoptosis. Up-regulation of miR-155 in DLBCL resulted in higher activity of NF-κB leading to decrease in PU.1 and consequent reduction in CD10 mRNA and protein, and this pathway also occurred in HL. Meanwhile, SMAD5 is another miR-155 target considered to be involved in the pathogenesis of DLBCL, which plays a very important role in the signaling pathway by which TGF-β inhibits the proliferation of human hematopoietic progenitor cells [38]. Although the decreased expression was reported in BL cell lines, the explicit expression in BL patients was unclear [39]. Therefore, this meta-analysis didn't include the BL patients. Further finds about miR-155' role in other kinds of lymphoma needs to be explored.
As miR-155 plays a very important role in oncogenesis, especially for the hematologic malignancies such as DLBCL, anti-miRNAs targeting it might prove to be of significant therapeutic benefit. Overexpression of miR-155 has been associated with drug resistance in several human cancers, including breast cancer, B-cell lymphoma and colon cancer. Roosbroeck et al. [40] demonstrated that overexpression of miR-155 induces resistance to chemotherapy, which can be reversed upon miR-155 inhibition. However, limitations to this treatment include the instability of free-floating anti-miRNAs in the plasma and their vulnerability to breakdown by nucleases, tissue uptake and renal clearance [4]. These limitations may be overcome by nanoparticle-based delivery of the anti-miRNAs to target tissues. In a study by Babar et al. [41], systemic delivery of antisense peptide nucleic acids encapsulated in unique polymer nanoparticles inhibits miR-155 and slows the growth of these “addicted” pre-B-cell tumors in vivo, suggesting a promising therapeutic option for lymphoma and leukemia. Nevertheless, more effective and reliable miR-155 targeted therapy remains to be discovered and explored.
Conclusion
In conclusion, our study suggested that miR-155 has strong potential to be considered as a novel prognostic biomarker in hematologic malignancies, and overexpression of miR-155 was associated with several clinicopathological features such as FLT3/ITD mutation in AML patients. This systematic review and meta-analysis is the first to demonstrate the prognostic and clinicopathological significance of miR-155 in hematologic malignancies, which may prove to be of great importance in the target treatment. In the future, more studies will be necessary to investigate the role of miR-155 in human hematological neoplasms. Further clear elucidation of the role and pathogenic mechanisms of miR-155 in hematological malignancies may provide useful clues on the development of effective treatment.
Abbreviations
MiR-155: micro RNA 155; DLBCL: diffused large B cell lymphoma; MM: multiple myeloma; ALL: acute lymphoblastic leukemia; AML: acute myelocytic leukemia; CLL: chronic lymphoblastic leukemia; ATL: acute T-cell lymphoblastic leukemia; B-NHL: B-cell non-Hodgkin lymphoma; HL: Hodgkin lymphoma; BL: Burkitt lymphoma; qRT-PCR: quantitative reverse transcription PCR; BM: bone marrow; PBMC: peripheral blood mononuclear cell; FFPE: formalin-fixed paraffin-embedded tissue sample; CS: clinical sample; OS: overall survival; PFS: progression free survival; DFS: disease free survival; EFS: event free survival; TFS: treatment free survival; HR: hazard ratio; OR: odds ratio; 95%CIs: 95% confidence intervals; NOS: Newcastle-Ottawa Scale; NA: not available; LDH: lactate dehydrogenase; BCR: B-cell receptor; BCL6: B-cell lymphoma 6 protein; HDAC4: histone deacetylase 4; SHIP1: Src homology 2 domain-containing inositol phosphatase; HSPCs: Hematopoietic stem and progenitor cells; IPI: International Prognostic Index; ECOG: Eastern Cooperative Oncology Group score; ABC: activated B-cell-like. Sample type; SMAD5: SMA and MAD-related protein 5.
Acknowledgements
We would like to thank the researchers and study participants for their contributions.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81770132 for Yu Hu and No. 81570116 for Heng Mei) and the Science and Technology Department of Hubei Province (No. 2018ACA141 for Yu Hu).
Authors' contributions
Lu Tang and Heng Mei collected and analyzed the data, wrote the paper; Yi-zhong Peng analyzed the data; Yu Hu conceived and designed this study, analyzed the data, wrote the paper; and all authors reviewed the paper, and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Due H, Svendsen P, Bødker JS, Schmitz A, Bøgsted M, Johnsen HE. et al. miR-155 as a Biomarker in B-Cell Malignancies. Biomed Res Int. 2016;2016:9513037
2. Jurkovicova D, Magyerkova M, Kulcsar L, Krivjanska M, Krivjansky V, Gibadulinova A. et al. miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma. 2014;61:241-51
3. Grasedieck S, Sorrentino A, Langer C, Buske C, Döhner H, Mertens D. et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121:4977
4. Ranganath P. MicroRNA-155 and Its Role in Malignant Hematopoiesis. Biomarker Insights. 2015;10:95-102
5. Wu C, Liu Q, Liu B. MicroRNA-155 hallmarks promising accuracy for the diagnosis of various carcinomas: results from a meta-analysis. Dis Markers. 2015;2015:327287
6. Bedewy AM, Elmaghraby SM, Shehata AA, Kandil NS. Prognostic value of miRNA-155 expression in B-cell Non-Hodgkin's lymphoma. Turk J Haematol. 2017;34:207
7. Jung I, Aguiar RC. MicroRNA-155 expression and outcome in diffuse large B-cell lymphoma. Br J Haematol. 2009;144:138
8. Ramamurthy R, Hughes M, Morris V, Bolouri H, Gerbing R, Wang Y. et al. miR-155 expression and correlation with clinical outcome in pediatric AML: A report from Children's Oncology Group. Pediatr Blood Cancer. 2016;63:2096-103
9. Choi Y, Hur EH, Moon JH, Goo BK, Choi DR, Lee JH. Expression and prognostic significance of microRNAs in Korean patients with myelodysplastic syndrome. Korean J Intern Med. 2017 [Epub ahead of print]
10. Narayan N, Bracken CP, Ekert PG. MicroRNA-155 expression and function in AML: An evolving paradigm. Experimental hematology. 2018;62:1-6
11. Knobloch K, Yoon U, Vogt PM. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. Journal of cranio-maxillo-facial surgery: official publication of the European Association for Cranio-Maxillo-Facial Surgery. 2011;39:91-2
12. Guyot P AA, Ouwens MJ, Welton NJ. Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC medical research methodology. 2012;12:9
13. Wells GA, Shea BJ, O'Connell D. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Non-Randomized Studies in Meta-Analysis. The Ottawa Hospital Research Institute. 2014 (3), 1-4
14. Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177
15. Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ (Clinical research ed). 1998;316:471
16. Cui B, Chen L, Zhang S, Mraz M, Fecteau J, Yu J. et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546-54
17. Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N. et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891
18. Guinn D, Ruppert AS, Maddocks K, Jaglowski S, Gordon A, Lin TS. et al. miR-155 expression is associated with chemoimmunotherapy outcome and is modulated by Bruton's tyrosine kinase inhibition with Ibrutinib. Leukemia. 2015;29:1210-3
19. Papageorgiou SG, Diamantopoulos MA, Scorilas A, Kontos CK, Bouchla A, Glezou E. et al. MicroRNA-155-5p Overexpression in Peripheral Blood Mononuclear Cells of Chronic Lymphocytic Leukemia Patients Is a Novel, Independent Molecular Biomarker of Poor Prognosis. Dis Markers. 2017;2017:1-10
20. Ishihara K SD, Tsuruda K, Inokuchi N, Nagai K, Hasegawa H, Yanagihara K, Kamihira S. Impact of miR-155 and miR-126 as novel biomarkers on the assessment of disease progression and prognosis in adult T-cell leukemia. Cancer Epidemiol. 2012;36:560-5
21. Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu Y, Mrózek K. et al. Clinical Role of microRNAs in Cytogenetically Normal Acute Myeloid Leukemia: miR-155 Upregulation Independently Identifies High-Risk Patients. J Clin Oncol. 2013;31:2086-93
22. Metzeler KH, Maharry K, Kohlschmidt J, Mrózek K, Becker H, Nicolet D. et al. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27:2023-31
23. Zhong H, Xu L, Zhong JH, Xiao F, Liu Q, Huang H. et al. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Exp Ther Med. 2012;3:763
24. Iqbal J, Shen Y, Huang X, Liu Y, Wake L, Liu C. et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggresive B-cell lymphoma. Blood. 2015;125:1137
25. Heounjeong G, Ji-Young J, Pil-Jong K, Young-Goo K, Soo Jeong N, Jin Ho P. et al. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035-49
26. Papaioannou D, Nicolet D, Volinia S, Nicolet D, Volinia S, Mrózek K. et al. Prognostic and biologic significance of long non-coding RNA profiling in younger adults with cytogenetically normal acute myeloid leukemia. Haematologica. 2017;102:1391-400
27. Ahmadvand M, Eskandari M, Pashaiefar H, Yaghmaie M, Manoochehrabadi S, Khakpour G. et al. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leukemia research. 2018;70:45-8
28. Narayan N, Morenos L, Phipson B, Willis SN, Brumatti G, Eggers S. et al. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31:808-20
29. Wu X, Wang F, Li Y, Wang X, Liu P, Zhang H. et al. Evaluation of latent membrane protein 1 and microRNA-155 for the prognostic prediction of diffuse large B cell lymphoma. Oncol Lett. 2018;15:9725-34
30. Zhou G, Cao Y, Dong W, Lin Y, Wang Q, Wu W. et al. The clinical characteristics and prognostic significance of AID, miR-181b, and miR-155 expression in adult patients with de novo B-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2017;58:1-9
31. Faraoni I LS, Ardiri D, Ciardi C, Fazi F, Lo-Coco F. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:26
32. Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V. et al. MicroRNA-155 as an inducer of apoptosis and cell differentiation in Acute Myeloid Leukaemia. Mol Cancer. 2014;13:79
33. Cammarata G AL, Salemi D, Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A, Bica MG, Cascio L, Floridia PM, Mineo AM, Russo M, Fabbiano F, Santoro A. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331-9
34. Scott L, Civin C, Rorth P, Friedman A. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725
35. O'Connell R, Chaudhuri A, Rao D, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113-8
36. Gadareth H, Frank S. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinforma. 2013;3:1-8
37. Sandhu SK VS, Costinean S, Galasso M, Neinast R, Santhanam R, Parthun MR, Perrotti D, Marcucci G, Garzon R, Croce CM. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eμ-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012;109:20047-52
38. Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111-6
39. Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ. et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes, chromosomes & cancer. 2006;45:147-53
40. Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez-Aguayo C, Fuentes-Mattei E. et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:2891-904
41. Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM. et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695-704
42. Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R. et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012;109:20047-52
43. Thompson RC, Herscovitch M, Zhao I, Ford TJ, Gilmore TD. NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem. 2011;286:1675-82
44. Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:253-64
45. Huskova H, Korecka K, Karban J, Vargova J, Vargova K, Dusilkova N. et al. Oncogenic microRNA-155 and its target PU.1: an integrative gene expression study in six of the most prevalent lymphomas. International journal of hematology. 2015;102:441-50
46. Lee DW, Futami M, Carroll M, Feng Y, Wang Z, Fernandez M. et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene. 2012;31:4085-94
Author contact
![]() Corresponding authors: Heng Mei, Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Dadao, Wuhan 430022, China, Tel.: +86-27-85726007, Fax: +86-27-85726387, E-mail: hmeiedu.cn. Yu Hu, Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Dadao, Wuhan 430022, China, Tel.: +86-27-85726007, Fax: +86-27-85726387, E-mail: dr_huyucom
Corresponding authors: Heng Mei, Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Dadao, Wuhan 430022, China, Tel.: +86-27-85726007, Fax: +86-27-85726387, E-mail: hmeiedu.cn. Yu Hu, Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Dadao, Wuhan 430022, China, Tel.: +86-27-85726007, Fax: +86-27-85726387, E-mail: dr_huyucom

 Global reach, higher impact
Global reach, higher impact