3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(3):721-729. doi:10.7150/jca.28254 This issue Cite
Review
The Oncogenic Potential of SUV39H2: A Comprehensive and Perspective View
1. Department of Immunology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China;
2. National Clinical Research Center for Cancer, Tianjin, China;
3. Key Laboratory of Cancer Prevention and Therapy, Tianjin, China;
4. Tianjin's Clinical Research Center for Cancer, Tianjin, China;
5. Key Laboratory of Cancer Immunology and Biotherapy, Tianjin, China.
Received 2018-7-2; Accepted 2018-11-9; Published 2019-1-1
Abstract
Epigenetic modifications at the histone level have attracted significant attention because of their roles in tumorigenesis. Suppressor of variegation 3-9 homolog 2 (SUV39H2, also known as KMT1B) is a member of the SUV39 subfamily of lysine methyltransferases (KMTs) that plays a significant role in histone H3-K9 di-/tri-methylation, transcriptional regulation and cell cycle. Overexpressions of SUV39H2 at gene, mRNA and protein levels are known to be associated with a range of cancers: leukemia, lymphomas, lung cancer, breast cancer, colorectal cancer, gastric cancer, hepatocellular cancer and so on. Accumulating evidence indicates that SUV39H2 acts as an oncogene and contributes to the initiation and progression of cancers. It could, therefore, be a promising target for anti-cancer treatment. In this review, we focus on the dysregulation of SUV39H2 in cancers, including its clinical prognostic predictor role, molecular mechanism involved in cancer occurrence and development, relevant inhibitors against cancer, and its epigenetic modification interaction with immunotherapy. A better understanding of the SUV39H2 will be beneficial to the development of molecular-targeted therapies in cancer.
Keywords: SUV39H2, Histone methyltransferase, epigenetic modification, cancer
Introduction
Suppressor of variegation 3-9 homolog 2 (SUV39H2, also known as KMT1B) is a member of the SUV39 subfamily of lysine methyltransferases (KMTs) and is known to be localized in nucleus; it is the second H3K9 selective methyltransferases recognized after its homolog SUV39H1 [1, 2]. In humans, the SUV39 family proteins, namely SUV39H1 (KMT1A), SUV39H2 (KMT1B), SETDB1 (KMT1E), SETDB2 (KMT1F), G9A (EHMT2) and G9a-like protein (GLP1), contain pre-SET (N-SET) and post-SET (C-SET) domains besides the SET domain and mediate H3K9 methylation [3-6]. SUV39H2 is primarily responsible for di- and tri-methylation of H3K9; H3K9me3 is associated with heterochromatin organization at pericentric and telomeric repeats, transcriptional repression, and epigenetic silencing of the domains in euchromatin [1, 7-12]. SUV39H2 possesses three protein isoforms and its exon 3 could affect its histone methyltransferases (HMTases) activity, stability, and sub-nuclear localization [13]. SUV39H2 forms a complex along with Rb-related pocket proteins to silence E2F-responsive genes; tri-methylation of H3K9 in E2F-responsive genes by SUV39H2 contributes to regulating cell cycle and cell differentiation [14, 15]. H3K9me3 repressive heterochromatin conformation is accumulated in p53 pro-apoptotic and anti-proliferation target promoters [16]. Suv39h maintain normal biological function in mammal [17-23]. Suv39h-mediated H3K9me3 serves a function in directing DNA methylation to major satellite repeats at pericentric heterochromatin [24-26]. Suv39h-deficient mice showed growth restriction [27, 28]. Suv39h have been shown to regulate telomere length in mammalian and Suv39h-deficient mice exhibit genome instability and severely impaired viability [28-31]. SUV39H2 prefers H3 as substrate, especially unmethylated H3 peptide; SUV39H2 also methylates other non-histone candidate substrates weakly in human via specificity profile [7, 32]. Alternative splicing and automethylation have an effect on its function and specificity [13, 33].SUV39H2 dimethylates K134 on H2AX protein, which plays a critical role in DNA repair, and trimethylates LSD1 at Lys 322 for LSD1 stabilization in human [3, 34-36].
A summary of the roles of SUV39H2 in cancers.
| Cancer type | Function | Reference |
|---|---|---|
| Hematopathy | ||
| CLL | Associated with cytogenetic aberrations and hTERT | [24] |
| ALL | Maintains cell viability; enhances chemoresistance | [25] |
| AML | HSPA14-SUV39H2 fusion in normal karyotype AML | [46] |
| CML | ARC decreases the SUV39H2 expression | [34] |
| Lymphomas | Loss of Suv39h1/h2 leads to tumorigenesis | [23] |
| Lung cancer | ||
| YF454A, and erlotinib decrease its expression | [33] | |
| SNPs predict the susceptibility to lung cancer | [47] | |
| Methylates LSD1 and maintains LSD1 stabilization | [31] | |
| Its automethylation regulation | [28] | |
| Promotes the conformation of γ-H2AX; enchances the radio- and chemo- resistance | [29,50] | |
| Transcription inhibition of OPTN | [47] | |
| Breast cancer | ||
| Correlated with metastatic biology, poor survival | [36];[52];[54] | |
| Mutations | [53] | |
| Recruited by PR to methylate histone H3K9 for stabilization of HP1γ binding | [58] | |
| ERβ represses the expression of SUV39H1/2 to promote transcription activated by p53 | [59] | |
| Digestive system cancers | ||
| CRC | Promotes proliferation, progression and metastasis via tri-methylation of the SLIT1 promoter | [37] |
| The RF8 epitope triggers CTL responses | [32] | |
| GC | Suv39h1/2 knockdown results in Twist1 expression | [64] |
| Methylates H3K9 at the promoter of P16 locus to decrease P16 expression | [62] | |
| HCC | H3K9me3-HP1α complex promote C-myc expression | [65] |
| Urinary & Reproductive system cancers | ||
| PCa | Interacts with AR and MAGE-A11 to enhance AR transcription | [41] |
| Ovarian cancer | Exon 3 could maintain its stability, localization and catalytic activity | [11] |
| Other cancers | ||
| Bowen's disease | Arsenic reprograms E2F1 promoter by deregulating SUV39H2 | [71] |
Although identified as an embryonic-specific protein and restricted to the testis in adult tissues of healthy individuals, SUV39H2 is ubiquitously overexpressed in cancer tissues, such as leukemia, lymphomas, lung cancer, breast cancer, colorectal cancer, gastric cancer, and hepatocellular cancer [1, 13, 28, 30, 34, 37-47]. Increasing evidence has confirmed that dysregulation of SUV39H2 contributes to carcinogenesis and is involved in invasion and metastasis of malignancy [33, 36, 38, 41, 42, 44]. Recently, researches have also clarified that Suv39h2 knockout decrease the incidence of nonalcoholic steatohepatitis and myocardial infarction [48-50]. In this review, we focus on the prognostic value of SUV39H2 in cancers, describe the off-target effects of SUV39H2 on non-histone proteins in epigenetic modification, discuss its potential mechanisms in tumorigenesis and immunotherapy, and, summarize SUV39H2 inhibition as a potential target for anti-cancer therapies (Table 1).
1.0 SUV39H2 in hematopathy
1.1 Leukemia
In chronic lymphocytic leukemia (CLL), the cytogenetic analyses have shown that high SUV39H2 transcript levels are associated with cytogenetic aberrations. On the contrary, low SUV39H1 expression levels have been shown to be correlated with complex and altered karyotypes in CLL. The differential expression of SUV39H1/2 is related to genomic instability. The human telomerase reverse transcriptase (hTERT) positively correlated with expression level of SUV39H2 in CLL patients, not with SUV39H1, SUV420H1, and SUV420H2 [29]. In the CD19+B cells, histone H3K9 methylation level is significantly increased. However, SUV39H2 mRNA level has no difference between CLL and controls [51]. In acute lymphocytic leukemia (ALL), Mutonga et al. [30] has demonstrated that SUV39H2 protein and mRNA is expressed at dramatically high levels in patients with newly diagnosed ALL, in comparison with those healthy or obtained at remission. It is worth noting that SUV39H2 and H3K9me3 were not detected in normal peripheral blood mononuclear cells (nPBMNCs). SUV39H2 knockdown could impair the viability likely by inducing apoptosis in ALL cell lines. What's more, SUV39H2 overexpression enhances chemoresistance to doxorubicin, cytarabine , and dexamethasone in Jurkat cells [30]. It can contribute to the diagnosis of ALL or definition of the cancer recurrence by detecting the overexpression of SUV39H2.
A study focused on fusion transcripts in acute myeloid leukemia (AML) indicated that HSPA14-SUV39H2 fusion and the other seven fusions are exclusively detected by RNAseq method in normal karyotype AML. The fusion gets rid of the last 3 exons in HSPA14 and connects the rest of the HSPA14 parts with the first exon in SUV39H2 [52]. Considering difficulties in diagnosis and treatment of normal karyotype AML, HSPA14-SUV39H2 fusion may serve as a novel diagnostic genetic marker and an ideal therapy target. Lin et al. [39] revealed decreased expression of Suv39h2 when K-562 cells were subjected to exposure to arecoline (ARC), indicating that ARC may be carcinogenic through regulation of post-translational modifications.
1.2 Lymphomas
As described above, it implies that SUV39H2 overexpression is participated in leukemogenesis as an oncogene. However, a previous study indicated Suv39h1/h2 may play a tumor suppressor role in Lymphomas. Suv39h-deficient mice were shown to have increased incidence of slow-progressing B-cell lymphomas. This study showed that combined disruption of Suv39h1/h2 results in loss of H3K9 trimethylation at pericentric chromatin, severely impaired heterochromatin structures, and genome instabilities, all of which represents tendency to cancer [28].
SUV39H2 methylates non-histone candidate substrates. (A) SUV39H2 trimethylates LSD1 on lysine 322. SUV39H2-caused LSD1 methylation maintains LSD1 stabilization by suppressing LSD1 polyubiquitination; (B) SUV39H2 dimethylates histone H2AX on lysine 134 and then improve the formation of phosphorylated histone H2AX (γ-H2AX).
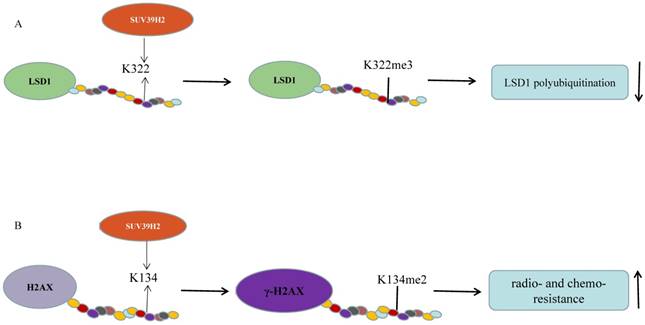
In summary, the role of SUV39H2 in hematological malignancy may depend on the cancer model. SUV39H2 causes resistance to chemotherapy, but the mechanism of its role of promoting to resistance to chemotherapy is poorly studied. However, SUV39H2 inhibitor may act as a chemotherapeutic sensitizer to improve the efficacy of chemotherapy in anti-cancer therapy.
2.0 SUV39H2 in lung cancer
Recently a novel therapy which combines the histone deacetylase inhibitor, YF454A and erlotinib, has been introduced to restore EGFR-TKI resistance in non-small cell lung cancer (NSCLC). Several essential genes involved in cell-cycle progression: SUV39H2, MCM7, MSH6, TIPIN, and E2F3A, are up-regulated in lung adenocarcinoma in contrast to normal tissues obtained from the Cancer Genome Atlas database (TCGA). The combined therapy has decreased the expression levels of SUV39H2 and other up-regulated genes [38]. According to the results of the whole genome direct sequencing in Korean, eight novel single nucleotide polymorphisms (SNPs) in the 3'-UTR of SUV39H2 have been identified. SUV39H2 genotypes with 1624C allele (G/C + C/C) is significantly related to an increased susceptibility to squamous cell carcinoma (SCC), in older age group. In contrast to non-smokers, smokers carrying the variant genotypes SUV39H2(1624 G→C) have a significantly higher risk of lung cancer in older age groups [53]. Abnormal expression level of SUV39H2 and SUV39H2 SNPs make it possible for SUV39H2 to become a molecular diagnostic marker clinically.
Piao et al. validated that SUV39H2 could trimethylate LSD1 on lysine 322 and SUV39H2 knockdown causes a decreased LSD1 expression without change in the mRNA levels (Figure 1A). SUV39H2-caused LSD1 methylation maintains LSD1 stabilization by suppressing LSD1 polyubiquitination. When the methylation site, lysine 322, is mutant, LSD1 showed decreased binding affinities to CoREST complex. SUV39H2 is confirmed to be important in regulating LSD1-target genes, such as SOX2 and CDKN1A (p21/CIP1) [36]. This team further investigated that SUV39H2 have the ability of automethylation at lysine 392 located in the post-SET domain. The automethylation decreases binding affinity to substrate proteins, including LSD1 and histone H3. With the treatment of chaetocin, the level of SUV39H2 automethylation declines [33]. Recently, the same phenomenon occurs in the SUV39H2 homologue, Clr4 in the fission yeast Schizosaccharomyces pomb [54, 55]. Previously, they reported that SUV39H2 could dimethylate histone H2AX on lysine 134 and then improve the formation of phosphorylated histone H2AX (γ-H2AX), which is involved in DNA-damage repair pathway and could enhance radio-/chemo- resistance of cancer as a prognostic biomarker of various human cancers (Figure 1B) [34, 56]. SUV39H2 knockdown makes HeLa cells and RERF-LC-AI cells more sensitive to doxorubicin and radiation respectively, by inhibiting H2AX methylation and subsequently inhibiting the production of γ-H2AX. Sone et al. suggested that SUV39H2 has oncogenic activity and deregulation of γ-H2AX by inhibiting SUV39H2 may be an ideal strategy for cancer treatment [34]. However, SUV39H2 is controversial as a function of K134 methylation [57]. Recently, two synthesized compounds have been found to inhibit SUV39H2 methyltransferase activity. OTS193320 could suppress SUV39H2 to attenuate γ-H2AX levels combined with doxorubicin which could significantly reduce cell viability in MDA-MB-231 cell line. Compared to a single agent, combination therapy has more significant effects. In mouse xenograft models using MDA-MB-231and A549 cell line, OTS186935 shows an obvious growth inhibition effect [56]. We have further determined the carcinogenic ability of SUV39H2 in lung adenocarcinoma cancer. RNA-seq has been performed to find differential genes and tumor pathway. SUV39H2 trimethylates H3K9 at the promoter of OPTN and inhibits its expression, which is associated with autophagy-related apoptosis [47, 58].
In conclusion, the SUV39H2 polymorphism is worth elucidating to predict the outcomes in lung cancer patients. SUV39H2 may act as a cofactor to modify oncogene in carcinogenic process through automethylation. SUV39H2 may cooperate with LSD1 to participate in tumorigenesis by regulating non-histone proteins, such as p53, E2F, MYPT1, and DNMT1. Anti-SUV39H2 treatment may reverse drug resistance and simultaneously enhance the sensitivity to conventional anti-cancer treatments via decreasing the γ-H2AX level.
3.0 SUV39H2 in breast cancer
In the MMTV-PyMT breast cancer (BC) model, the enrichment of SUV39H2 as well as Smyd3, Setd3/5, Prmt3/6, and Nsd1/2 in lung metastases compared to disseminated or primary tumor cells [41]. Moreover, the expression level of SUV39H2 increases considerably in the basal-like BC, and the mRNA level of SUV39H2 correlated with bad prognosis in BC [59]. According to the assessment of the chromatin modification enzyme profiles in human solid cancers, there is rare occurrence of mutations in SUV39H2 identified in epithelial cancers. But SUV39H2 mutations was identified in BC, which probably means a polymorphism in BC [60]. Bioinformatics analysis based on TCGA was conducted to reveal SUV39H2 and other novel genes (SUV39H1, DNMT3B, EZH2 and AURKB) expressions significantly increase in triple negative (TN) disease and positively associate with high Ki67 levels, TN status and tumor grade. The higher expression of SUV39H2 correlated with shorter survival time [61]. SUV39H2 appears to be associated with more aggressive phenotypes and metastatic biology. SUV39H2 and other epigenetic regulatory enzymes could be used to in the division of cancer stage and identification of tumor recurrence. And these indicators may have better prognostic value than TN status.
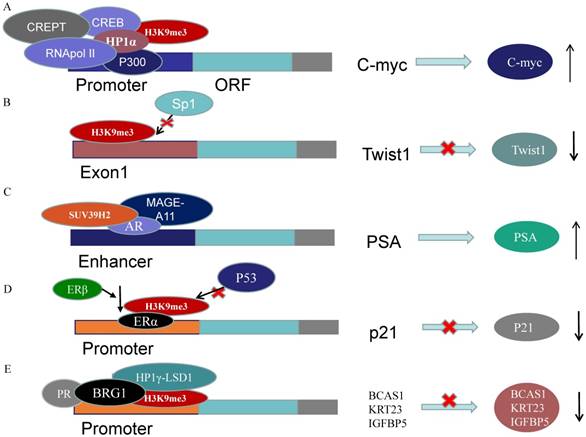
A recent study has provided evidence that ERα recruits SUV39H1/H2 and then form H3K9me3 repressive heterochromatin marks at estrogen-repressed genes, like p21, to silence the transcription activated by p53. In ERα-positive cells, ERβ abrogates the repressive heterochromatin by down-regulating SUV39H1 and SUV39H2 and reduces ERα-p53 binding. Ultimately, ERβ releases the p53-ERα transcriptional block and promotes anti-proliferative and pro-apoptotic capabilities (Figure 3D) [62]. Strikingly, Nacht et al. demonstrated that progesterone receptor (PR) recruits BRG1 and the HP1γ-LSD1 complex, which is anchored by the HP1γ binding to the H3K9me3 signal, to form a repressive complex on genes promoters. On repressive PR binding sites (PRBs), SUV39H2 is responsible for H3K9me3 conformation which stabilizes HP1γ binding to H3K9me3. The ligand-activated PR recruits the repressive complex to repress target genes concerning cell proliferation along with MSK1 and ERK (Figure 3E) [63].
Emerging data have confirmed that dysregulation of HMTs, including SUV39H2, triggers aberrant histone methylation patterns and subsequently leads to BC pathogenesis [64]. The metastatic process is reprogrammed and new epigenetic biomarkers are acquired during metastatic formation. SUV39H2 is implicated in hormone regulation in tumorigenesis and related to aggressive and metastatic biology. As a consequence, SUV39H2 may play a key role in cancer invasion and metastasis, as a potential independent predictor of survival in TNBC or advanced BC.
4.0 SUV39H2 in digestive system cancers
4.1 Colorectal carcinoma
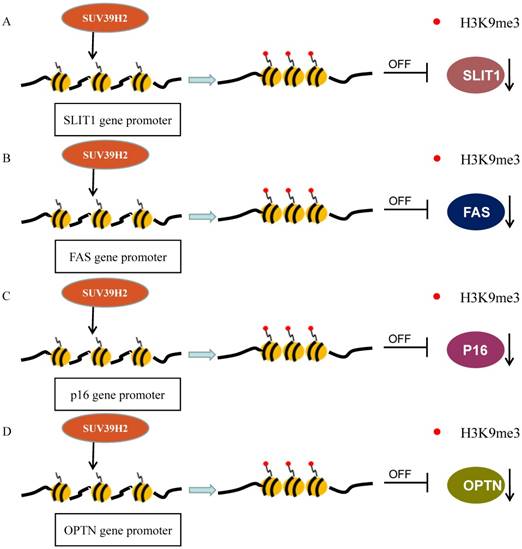
A data analysis from GEO dataset indicated that SUV39H2 is generally overexpressed and its overexpression is significantly associated with the TNM stage and metastasis in colorectal carcinoma (CRC). SUV39H2 overexpression relates to poorer prognosis in comparison with low levels of SUV39H2. Then, rescue experiments have verified that SUV39H2 could promote CRC proliferation and metastasis via silencing SLIT1 expression (Figure 2A). SUV39H2 catalyzes the H3K9me on the promoter of SLIT guidance ligand 1 (SLIT1) to silence SLIT1 gene. In addition, the expression of KCTD12 and GUCA2A, were also inversely modulated by SUV39H2 [42].
Another significant research has revealed that a peptide encoded in SUV39H2 has immunogenicity and can lead to the response of cytotoxic CD8+T-cell (CTL) to cancer cells. It is a novel cancer-testis antigen restricted HLA-A24. The RF8 peptide derived from SUV39H2 is displayed by HLA-A24 in colon cancer cells. This epitope triggers CTL responses specifically to cancer cells which express HLA-A24 along with SUV39H2 [37]. The FAS gene has been reported to be silenced by its promoter H3K9me3 to evade the immune surveillance and enhance chemoresistance. The level of H3K9me3 in the FAS promoter region is significantly higher in metastatic CRC cells than in primary CRC cells. Particularly, Verticillin A could decrease H3K9me3 level on promoter to recover its expression, by the way of repressing histone methyltransferases SUV39H1/2 and G9a/GLP, in metastatic CRC (Figure 2B). Verticillin A makes tumors more sensitive to FasL+ tumor-infiltrating CTLs by deregulating SUV39H1/2 and G9a to restore Fas expression [65].
SUV39H2 methylates H3 to suppress transcriptions. (A) SUV39H2 trimethylates the SLIT1 promoter to inhibit its transcription; (B) SUV39H2 methylates H3K9 at the promoter of FAS to inhibit its expression. (C)SUV39H2 methylates H3K9 at the promoter of p16 to decrease its transcription. (D) SUV39H2 methylates H3K9 at the promoter of OPTN to suppress its expression.
4.2 Gastric carcinoma
Previous studies suggested that CBX7, the core component of PcG repressor complex-1 (PRC1), could recruit and up-regulate SUV39H2 in gastric cancer (GC) cells and subsequently SUV39H2 trimethylate H3K9 at the promoter and Regulatory Domain (RD) of p16 gene to induce the downregulation of P16 expression (Figure 2C). The complexes of CBX7-SUV39H2 are detected in the nucleus. The transcription of p16 gene could be rescued by siRNA-knockdown of Suv39h2 [66]. This discovery links PRC1 binding and H3K9me3 formation, the known repressive epigenetic landmarks, to establish a reliable pathway for the epigenetic inactivation of the p16 gene. However, Hepatitis B virus X protein (HBx) represses p16 gene expression to promote the HCC progression via the increase of H3K9me3 on the p16 promoter. HBx decreases JMJd2B expression but not increases SUV39H2 or other methyltransferase [67]. The DNA methylation in the CpG-rich region within Twist1 coding exon 1, rather than its promoter, is closely related to Twist1 gene silence in GC cells. The level of H3K9me3 in the exon is high, which is induced by Suv39h1/2, in Twist1 expression-negative GC cells. Suv39h1/2 expression levels are inversely related with the Twist1 expression and Suv39h1/2 knockdown increase Twist1 expression. Suv39h1/2 may disturb the binding of the Sp1 to inhibit Twist1 expression, through the DNA methylation and the heterochromatin state (Figure 3B) [68].
4.3 Hepatocellular carcinoma
Recently, Xin Xet al. observed that the tri-methylation activity of SUV39h2 could be enhanced by miR675 and PKM2. Furthermore, the increased H3K9me3-HP1α complex recruit RNApol II, CREB and P300 on the C-myc promoter to promote C-myc expression, which contributes to the malignant transformation (Figure 3A) [69]. The mRNA and protein expression levels of SUV39H2, G9a and EZH2 are up-regulated in hepatocellular carcinoma (HCC) tissues and SUV39H2 is highly associated with the HCV infection and tumor stage [43]. In hepatocytes, Suv39H2 increases the incidence of nonalcoholic steatohepatitis and steatosis likely via increasing H3K9me3 on Sirt1 gene promoter and promoting SIRT1 trans-repression in mice [48, 50]. These all imply a potential role of SUV39H2 in HCC pathogenesis and progression.
SUV39H2 regulates genes expressions acting as cofactor. (A) H3K9me3-HP1α complex recruit RNApol II, CREB and P300 on the C-myc promoter to promote C-myc expression. (B) Suv39h1/2 may disturb the binding of the Sp1 to inhibit Twist1 expression. (C) SUV39H2 interacts with AR and MAGE-A11 to enhance the PSA expression. (D) ERβ abrogates H3K9me3 on p21 promoter by decreasing SUV39H1/2 to promote anti-proliferative and pro-apoptotic capabilities. (E) PR recruits BRG1 and HP1γ-LSD1 complex, which is anchored by the HP1γ binding to the H3K9me3 signal, to form a repressive complex on genes promoters.
4.4 Esophageal squamous cell carcinomas
In esophageal squamous cell carcinomas (ESCC), SUV39H2 and the other 33 genes are related with lymph node metastasis by gene expression profile analysis [44].
In a word, SUV39H2 overexpression relates with TNM stage and metastasis in most digestive system cancers. What's more, SUV39H2 overexpression relates to negative prognosis. It raises a possibility that SUV39H2 could be a novel biomarker in cancer stage and predicting prognosis.
Collectively, SUV39H2 plays a pivotal role in pathogenesis and progression of digestive system tumors via several pathways and immune regulation. There may be SUV39H2-SLIT1 axis and SUV39H2-FAS axis in CRC, CBX7/SUV39H2-P16 axis in GC and SUV39H2-C-myc axis in HCC. SLITs functions through cooperation with roundabout (ROBO) receptors [70]. We speculate that ROBO receptors may function as the downstream factors of SUV39H2-SLIT1 axis. SUV39H2 causes chemoresistance by silencing FAS expression in CRC. Considering the SUV39H2 peptide as a novel tumor-associated antigens (TAAs) triggered cytotoxic CTL, and the importance of H3K9me3 in Fas involved in FasL+ CTLs response, we speculate that down-regulation of SUV39H2 could improve immunotherapy efficacy and may be an ideal candidate for future vaccination therapies based on CTL. ATRA inhibits the production of cytokine IL6, TNF, via up-regulating SUV39H2 to increase the H3K9me3 levels on the promoters [71]. Recently, verticillin A has been reported to be likely to improve the efficacy of anti-PD-1/PD-L1 immunotherapy [72]. Through these studies, we have a hypothesis that SUV39H2 may be involved in the epigenetic regulation of immunotherapy.
5.0 SUV39H2 in reproductive and urinary system cancers
A report has indicated that exon 3 in SUV39H2 could modulate the gene expression via exon 3 alteration in ovarian cancer cell lines. The alternative inclusion of exon 3 in SUV39H2 regulates the expression of target genes: H19, KRT15, C5orf46 and so on [13]. The mRNA levels of SUV39H2 is not affected by the knockdown of NRF-1, which works as a transcription factor in neurite outgrowth in neuroblastoma cells [73, 74].
SUV39H2 has been reported to be upregulated in prostate cancer (PCa) compared to normal prostate tissues [45]. SUV39H2 has been verified acts as an AR coactivator to enhance the AR transcriptional activity in PCa cells. SUV39H2 co-localizes with AR and melanoma antigen-A11 (MAGE-A11), the AR coregulator, in the cytoplasm or nucleus. SUV39H2 interacts with the NH2-terminal region of AR and MAGE-A11. They are recruited to the PSA enhancer region to enhance AR transcriptional activity in the existence of androgen (Figure 3C) [46]. EPI-001 which interacts with AR NH2-terminal region, is the experimental drug in the treatment of prostate cancer[75, 76]. This indicates that SUV39H2 may have played a role in the molecular mechanism of EPI-001.
6.0 SUV39H2 in other cancers
Arsenic was found to downregulate SUV39H2 to decrease the binding of me-H3K9 on the E2F1 promoter which could increase the activation and expression of E2F1. Ultimately Arsenic promotes E2F1 to bind to the Aurora-A promoter which increases the expression of Aurora-A. SUV39H2 could increase centrosome amplification by epigenetic regulation of E2F1 and Aurora-A. The centrosome amplification results in apoptosis. These events interact with anti-apoptotic pathway triggered by TNF-α to conform the pathognomonic features in arsenic-induced Bowen's disease [77].
SUV39H2 overexpression has no significant association with prognosis of oral squamous cell carcinoma (OSCC) in Taiwan, unlike other histone modification enzymes: ARK2, G9a, EZH2, and SUV39H1 [78]. Evidence showed that the NOR1 expression could alter H3K9me3 levels, but mRNA expression levels of SUV39H1/2 and G9a are not affected by NOR1 in nasopharyngeal carcinoma (NPC) [79].
As mentioned before, SUV39H2 is not only involved in arsenic-induced Bowen's disease, but also associated with HCV affection in HCC patients. Some researchers have proved that high-carbohydrate/fat input and ischemia could promote its expression [48, 49]. Therefore, we hypothesized that the carcinogen and bad stimuli may be the main cause which brings about the increased expression of SUV39H2. Though the expression levels of SUV39H2 remains normal for a while, the enzyme activities might be affected.
Conclusions and perspective
In this review, we summarize the dysregulation of SUV39H2 in cancers and reveal two important issues of SUV39H2: the cellular and molecular mechanisms of SUV39H2 in cancer, and the significance in cancer detection and prevention. On one hand, cancer is characterized by its multi-step and complex progression with genetic mutations and epigenetic modification. Different from gene mutations, histone methylation is a reversible process. As mentioned earlier, SUV39H2 plays a role in three aspects: translational, post-translational and post- translational modification. It's reasonable to make the assumption that SUV39H2 have a negative regulation on TSGs via promoter trimethylation and cooperates with oncogenes to promote cancer formation. Therefore, elucidating the cellular and molecular mechanisms underlying SUV39H2-mediated suppression on TSG and the use of SUV39H2 inhibitors appears to be an attractive method in anti-cancer therapy. What's more, SUV39H2 enhances resistance to chemotherapy via increasing γ-H2AX and silencing FAS. SUV39H2 inhibitors can also combine with conventional treatment including radiotherapy, chemotherapy and immunotherapy, to reduce drug resistance and improve its efficacy. On the other hand, cancer is heterogeneous and the metastatic process is reprogrammed and new epigenetic biomarkers are acquired during metastatic formation. SUV39H2 appears to be associated with more aggressive phenotypes and metastatic biology. More importantly, SUV39H2 is hardly expressed in normal tissues. SNPs and mutations of SUV39H2 make it have the opportunity to be an ideal independent predictor in cancer detection and prevention.
In conclusion, SUV39H2 maintains normal biological function, loss of which leads to tumorigenesis, and its overexpression contributes to cancer initiation and progression as an oncogene in most instances. Elucidating the epigenetics molecular mechanisms of SUV39H2 and the clinical data statistics for identifying SUV39H2 to be a novel target in cancer therapies and a novel prognostic or diagnostic indicator in cancer.
Abbreviations
Suppressor of variegation 3-9 homolog 2: SUV39H2; lysine methyltransferases: KMTs; histone methyltransferases: HMTases; chronic lymphocytic leukemia: CLL; acute lymphocytic leukemia: ALL; acute myeloid leukemia: AML; non-small cell lung cancer: NSCLC; single nucleotide polymorphisms: SNPs; phosphorylated histone H2AX: γ-H2AX; Optineurin: OPTN; breast cancer: BC; triple negative: TN; colorectal carcinoma: CRC; cytotoxic CD8+T-cell: CTL; tumor suppressor gene: TSG; gastric cancer: GC; Hepatocellular carcinoma: HCC; esophageal squamous cell carcinomas: ESCC; prostate cancer: Pca.
Acknowledgements
This work was supported by a grant from National Key Technology R&D Program (No.2015BAI12B12) and national natural science foundation of china (81572265).
Competing Interests
The authors have declared that no competing interest exists.
References
1. O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G. et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423-33
2. Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T. et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633-6
3. Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227
4. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838-49
5. Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H. et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570
6. Rao VK, Pal A, Taneja R. A drive in SUVs: From development to disease. Epigenetics. 2017;12:177-86
7. Schuhmacher MK, Kudithipudi S, Kusevic D, Weirich S, Jeltsch A. Activity and specificity of the human SUV39H2 protein lysine methyltransferase. Biochim Biophys Acta. 2015;1849:55-63
8. Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94-9
9. Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF. et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591-8
10. Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116-20
11. Kudithipudi S, Schuhmacher MK, Kebede AF, Jeltsch A. The SUV39H1 Protein Lysine Methyltransferase Methylates Chromatin Proteins Involved in Heterochromatin Formation and VDJ Recombination. ACS Chem Biol. 2017;12:958-68
12. Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U. et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411-20
13. Mauger O, Klinck R, Chabot B, Muchardt C, Allemand E, Batsche E. Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions. Nucleic Acids Res. 2015;43:1869-82
14. Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796-809
15. Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I. et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. Embo j. 2004;23:605-15
16. Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol. 2012;19:478-84 S1
17. Yoshioka H, McCarrey JR, Yamazaki Y. Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice. Biol Reprod. 2009;80:804-12
18. Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55:277-90
19. Khani F, Thaler R, Paradise CR, Deyle DR, Kruijthof-de Julio M, Galindo M. et al. Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation. J Cell Biochem. 2017;118:1262-72
20. Zhang S, Wang F, Fan C, Tang B, Zhang X, Li Z. Dynamic changes of histone H3 lysine 9 following trimethylation in bovine oocytes and pre-implantation embryos. Biotechnol Lett. 2016;38:395-402
21. Ideno H, Shimada A, Imaizumi K, Kimura H, Abe M, Nakashima K. et al. Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr Patterns. 2013;13:84-90
22. Luzzani C, Solari C, Losino N, Ariel W, Romorini L, Bluguermann C. et al. Modulation of chromatin modifying factors' gene expression in embryonic and induced pluripotent stem cells. Biochem Biophys Res Commun. 2011;410:816-22
23. Benlhabib H, Mendelson CR. Epigenetic regulation of surfactant protein A gene (SP-A) expression in fetal lung reveals a critical role for Suv39h methyltransferases during development and hypoxia. Mol Cell Biol. 2011;31:1949-58
24. Gessaman JD, Selker EU. Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa. Proc Natl Acad Sci U S A. 2017;114:E9598-e607
25. Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S. et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691-701
26. Velazquez Camacho O, Galan C, Swist-Rosowska K, Ching R, Gamalinda M, Karabiber F. et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017:6
27. Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593-9
28. Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323-37
29. Carvalho Alves-Silva J, do Amaral Rabello D, Oliveira Bravo M, Lucena-Araujo A, Madureira de Oliveira D, Morato de Oliveira F. et al. Aberrant levels of SUV39H1 and SUV39H2 methyltransferase are associated with genomic instability in chronic lymphocytic leukemia. Environ Mol Mutagen. 2017;58:654-61
30. Mutonga M, Tamura K, Malnassy G, Fulton N, de Albuquerque A, Hamamoto R. et al. Targeting Suppressor of Variegation 3-9 Homologue 2 (SUV39H2) in Acute Lymphoblastic Leukemia (ALL). Transl Oncol. 2015;8:368-75
31. Dang-Nguyen TQ, Haraguchi S, Furusawa T, Somfai T, Kaneda M, Watanabe S. et al. Downregulation of histone methyltransferase genes SUV39H1 and SUV39H2 increases telomere length in embryonic stem-like cells and embryonic fibroblasts in pigs. J Reprod Dev. 2013;59:27-32
32. Allali-Hassani A, Wasney GA, Siarheyeva A, Hajian T, Arrowsmith CH, Vedadi M. Fluorescence-based methods for screening writers and readers of histone methyl marks. J Biomol Screen. 2012;17:71-84
33. Piao L, Nakakido M, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. Automethylation of SUV39H2, an oncogenic histone lysine methyltransferase, regulates its binding affinity to substrate proteins. Oncotarget. 2016;7:22846-56
34. Sone K, Piao L, Nakakido M, Ueda K, Jenuwein T, Nakamura Y. et al. Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nat Commun. 2014;5:5691
35. Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717-24
36. Piao L, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget. 2015;6:16939-50
37. Kochin V, Kanaseki T, Tokita S, Miyamoto S, Shionoya Y, Kikuchi Y. et al. HLA-A24 ligandome analysis of colon and lung cancer cells identifies a novel cancer-testis antigen and a neoantigen that elicits specific and strong CTL responses. Oncoimmunology. 2017;6:e1293214
38. Yu W, Lu W, Chen G, Cheng F, Su H, Chen Y. et al. Inhibition of histone deacetylases sensitizes EGF receptor-TK inhibitor-resistant non-small-cell lung cancer cells to erlotinib in vitro and in vivo. Br J Pharmacol. 2017;174:3608-22
39. Lin PC, Chang WH, Chen YH, Lee CC, Lin YH, Chang JG. Cytotoxic effects produced by arecoline correlated to epigenetic regulation in human K-562 cells. J Toxicol Environ Health A. 2011;74:737-45
40. Cheng CC, Chang J, Huang SC, Lin HC, Ho AS, Lim KH. et al. YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9a-mediated stemness in lung cancer cells. PLoS One. 2017;12:e0182149
41. Franci C, Zhou J, Jiang Z, Modrusan Z, Good Z, Jackson E. et al. Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS One. 2013;8:e58183
42. Shuai W, Wu J, Chen S, Liu R, Ye Z, Kuang C. et al. SUV39H2 promotes colorectal cancer proliferation and metastasis via tri-methylation of the SLIT1 promoter. Cancer Lett. 2018
43. Hung SY, Lin HH, Yeh KT, Chang JG. Histone-modifying genes as biomarkers in hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:2496-507
44. Yamabuki T, Daigo Y, Kato T, Hayama S, Tsunoda T, Miyamoto M. et al. Genome-wide gene expression profile analysis of esophageal squamous cell carcinomas. Int J Oncol. 2006;28:1375-84
45. Vieira FQ, Costa-Pinheiro P, Ramalho-Carvalho J, Pereira A, Menezes FD, Antunes L. et al. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr Relat Cancer. 2014;21:51-61
46. Askew EB, Bai S, Parris AB, Minges JT, Wilson EM. Androgen receptor regulation by histone methyltransferase Suppressor of variegation 3-9 homolog 2 and Melanoma antigen-A11. Mol Cell Endocrinol. 2017;443:42-51
47. Zheng Y, Li B, Wang J, Xiong Y, Wang K, Qi Y. et al. Identification of SUV39H2 as a potential oncogene in lung adenocarcinoma. Clin Epigenetics. 2018;10:129
48. Fan Z, Li L, Li M, Zhang X, Hao C, Yu L. et al. The histone methyltransferase Suv39h2 contributes to nonalcoholic steatohepatitis in mice. Hepatology. 2017;65:1904-19
49. Yang G, Weng X, Zhao Y, Zhang X, Hu Y, Dai X. et al. The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat Commun. 2017;8:14941
50. Shao J, Li L, Xu H, Yang L, Bian Y, Fang M. et al. Suv39h2 deficiency ameliorates diet-induced steatosis in mice. Biochem Biophys Res Commun. 2017;485:658-64
51. Zhou K, Zhang Q, Liu Y, Xiong Y, Wu S, Yang J. et al. Aberrant histone modification in CD19(+) B cells of patients with chronic lymphocytic leukemia. Onco Targets Ther. 2017;10:1173-9
52. Wen H, Li Y, Malek SN, Kim YC, Xu J, Chen P. et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS One. 2012;7:e51203
53. Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ. et al. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217-22
54. Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M. et al. Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature. 2018;560:504-8
55. Kusevic D, Kudithipudi S, Iglesias N, Moazed D, Jeltsch A. Clr4 specificity and catalytic activity beyond H3K9 methylation. Biochimie. 2017;135:83-8
56. Vougiouklakis T, Saloura V, Park JH, Takamatsu N, Miyamoto T, Nakamura Y. et al. Development of novel SUV39H2 inhibitors that exhibit growth suppressive effects in mouse xenograft models and regulate the phosphorylation of H2AX. Oncotarget. 2018;9:31820-31
57. Schuhmacher MK, Kudithipudi S, Jeltsch A. Investigation of H2AX methylation by the SUV39H2 protein lysine methyltransferase. FEBS Lett. 2016;590:1713-9
58. Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H. et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106-20
59. Liu L, Kimball S, Liu H, Holowatyj A, Yang ZQ. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget. 2015;6:2466-82
60. Ozdag H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L. et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90
61. Pena-Llopis S, Wan Y, Martinez ED. Unique epigenetic gene profiles define human breast cancers with poor prognosis. Oncotarget. 2016;7:85819-31
62. Lu W, Katzenellenbogen BS. Estrogen Receptor-beta Modulation of the ERalpha-p53 Loop Regulating Gene Expression, Proliferation, and Apoptosis in Breast Cancer. Horm Cancer. 2017;8:230-42
63. Nacht AS, Pohl A, Zaurin R, Soronellas D, Quilez J, Sharma P. et al. Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J. 2016;35:1822-43
64. Michalak EM, Visvader JE. Dysregulation of histone methyltransferases in breast cancer - Opportunities for new targeted therapies? Mol Oncol. 2016;10:1497-515
65. Paschall AV, Yang D, Lu C, Choi JH, Li X, Liu F. et al. H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance. J Immunol. 2015;195:1868-82
66. Li Q, Wang X, Lu Z, Zhang B, Guan Z, Liu Z. et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS One. 2010;5:e13732
67. Wang DY, Zou LP, Liu XJ, Zhu HG, Zhu R. Hepatitis B virus X protein induces the histone H3 lysine 9 trimethylation on the promoter of p16 gene in hepatocarcinogenesis. Exp Mol Pathol. 2015;99:399-408
68. Sakamoto A, Akiyama Y, Shimada S, Zhu WG, Yuasa Y, Tanaka S. DNA Methylation in the Exon 1 Region and Complex Regulation of Twist1 Expression in Gastric Cancer Cells. PLoS One. 2015;10:e0145630
69. Yang Y, Meng Q, Wang C, Li X, Lu Y, Xin X. et al. MicroRNA 675 cooperates PKM2 to aggravate progression of human liver cancer stem cells induced from embryonic stem cells. J Mol Med (Berl). 2018
70. Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016;143:3037-44
71. Arts RJ, Blok BA, van Crevel R, Joosten LA, Aaby P, Benn CS. et al. Vitamin A induces inhibitory histone methylation modifications and down-regulates trained immunity in human monocytes. J Leukoc Biol. 2015;98:129-36
72. Lu C, Liu K. Epigenetic regulation of PD-L1 expression and pancreatic cancer response to checkpoint immunotherapy. Translational Cancer Research. 2017;6:S652-S4
73. Tong CW, Wang JL, Jiang MS, Hsu CH, Chang WT, Huang AM. Novel genes that mediate nuclear respiratory factor 1-regualted neurite outgrowth in neuroblastoma IMR-32 cells. Gene. 2013;515:62-70
74. Wang JL, Chang WT, Tong CW, Kohno K, Huang AM. Human synapsin I mediates the function of nuclear respiratory factor 1 in neurite outgrowth in neuroblastoma IMR-32 cells. J Neurosci Res. 2009;87:2255-63
75. Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK. et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535-46
76. De Mol E, Fenwick RB, Phang CT, Buzon V, Szulc E, de la Fuente A. et al. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem Biol. 2016;11:2499-505
77. Liao WT, Lu JH, Lee CH, Lan CE, Chang JG, Chai CY. et al. An Interaction between Arsenic-Induced Epigenetic Modification and Inflammatory Promotion in a Skin Equivalent during Arsenic Carcinogenesis. J Invest Dermatol. 2017;137:187-96
78. Chen JH, Yeh KT, Yang YM, Chang JG, Lee HE, Hung SY. High expressions of histone methylation- and phosphorylation-related proteins are associated with prognosis of oral squamous cell carcinoma in male population of Taiwan. Med Oncol. 2013;30:513
79. Wang W, Yi M, Chen S, Li J, Li G, Yang J. et al. Significance of the NOR1-FOXA1/HDAC2-Slug regulatory network in epithelial-mesenchymal transition of tumor cells. Oncotarget. 2016;7:16745-59
Author contact
![]() Corresponding author: Lili Yang Ph.D., Huanhuxi Road, Tiyuanbei, Hexi District, Tianjin 300060, China; Email:yanglilicom, Phone: 086-022-23340123, ex 6003, FAX: 086-022-23537796
Corresponding author: Lili Yang Ph.D., Huanhuxi Road, Tiyuanbei, Hexi District, Tianjin 300060, China; Email:yanglilicom, Phone: 086-022-23340123, ex 6003, FAX: 086-022-23537796

 Global reach, higher impact
Global reach, higher impact