Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(6):1417-1433. doi:10.7150/jca.28406 This issue Cite
Review
Genetic Polymorphisms of DNA Repair Pathways in Sporadic Colorectal Carcinogenesis
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China
Received 2018-7-9; Accepted 2019-1-12; Published 2019-2-23
Abstract
DNA repair systems play a critical role in maintaining the integrity and stability of the genome, which mainly include base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR) and double-strand break repair (DSBR). The polymorphisms in different DNA repair genes that are mainly represented by single-nucleotide polymorphisms (SNPs) can potentially modulate the individual DNA repair capacity and therefore exert an impact on individual genetic susceptibility to cancer. Sporadic colorectal cancer arises from the colorectum without known contribution from germline causes or significant family history of cancer or inflammatory bowel disease. In recent years, emerging studies have investigated the association between polymorphisms of DNA repair system genes and sporadic CRC. Here, we review recent insights into the polymorphisms of DNA repair pathway genes, not only individual gene polymorphism but also gene-gene and gene-environment interactions, in sporadic colorectal carcinogenesis.
Keywords: DNA repair, polymorphism, colorectal cancer, carcinogenesis
Introduction
DNA repair is an orchestrated system of defenses evolved to protect the genomic integrity and involved in the process preventing carcinogenesis. DNA repair systems play a critical role in maintaining the integrity and stability of the genome, which mainly include base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR) and double-strand break repair (DSBR)[1]. Interindividual differences in DNA repair capacities are important determinants of susceptibility to cancer. Cellular DNA is constantly under damage from endogenous and exogenous stimuli, leading to a dynamic cellular balance between damage and repair[2]. Defects in human DNA repair system would increase the instability of genome, and unrepaired DNA damage may thereby enhance genetic susceptibility to cancer and give rise to carcinogenesis. The polymorphisms in different DNA repair genes that are mainly represented by single-nucleotide polymorphisms (SNPs) can potentially modulate the individual DNA repair capacity and therefore exert an impact on individual genetic susceptibility to cancer.
Colorectal cancer (CRC) is the third most common cancer in men and the second in women worldwide[3]. Among them, Sporadic colorectal cancer is the overwhelming majority, which arises from the colorectum without known contribution from germline causes or significant family history of cancer or inflammatory bowel disease[4]. In recent years, emerging studies have investigated the association between polymorphisms of DNA repair system genes and sporadic CRC. Here, we review recent insights into the polymorphisms of DNA repair pathway genes in sporadic colorectal carcinogenesis by searching different combinations of “DNA repair”, “polymorphism/variant” and “colorectal cancer/ colon cancer/rectal cancer” from Pubmed and web of science.
BER pathway gene polymorphisms and sporadic CRC susceptibility
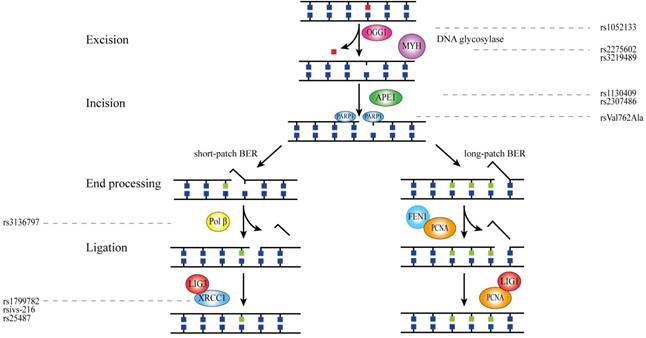
Base excision repair (BER) corrects small base errors which do not significantly alter the DNA helix structure. These damages mainly arise from oxidation, deamination and alkylation[5]. Upon DNA base damage, BER is initiated and four core steps are involved in this process: (1) damaged DNA base removal; (2) incision of the subsequent abasic site; (3) DNA ends processing; (4) ligation of the remaining nick in the DNA backbone[6]. From the beginning of the third step, BER diverges into two sub-pathways of short-patch(only one defective base) and long-patch (more than one defective base ) according to the number of defective bases, and each sub-pathway requires unique functional proteins[7]. OGG1 and MYH are involved in the first step of BER while APE1 and PARP1 participate in the incision of abasic site[8, 9]. Short-patch sub-pathway contains polβ, LIG3 and XRCC1 while FEN1, PCNA and LIG1 contribute to the long-patch sub-pathway[10].
Recognition related BER polymorphisms
OGG1
The OGG1 gene located at chromosome 3p26.2, consisting of seven exons and encodes a glycosylase including 345 amino acids. OGG1 protein repairs 8-hydroxyguanine (8-oxoG), a frequently mutagenic lesion among base modification[11].
As the most common OGG1 polymorphism, the rs1052133 polymorphism results in an amino acid substitution from serine to cysteine in codon 326 at exon 7 [12]. The GG genotype of rs1052133 polymorphism was first linked to increased CRC risk by Moreno, V. et al.'s study in Spanish population[13]. Subsequently, Canbay, E. et al. revealed in Turkish people that G allele was associated with higher risk of CRC compared with C allele[12]. And CG genotype was found to increase susceptibility to CRC according to Przybylowska, K. et al. in Polish population[14]. However, several investigations did not demonstrate similar significance[15-23]. Additionally, one research in Taiwanese found that the CG genotype of rs1052133 polymorphism was related with increased CRC risk but no significant association was demonstrated for 11657A/G polymorphism[24]. It is worth noting that significant interaction was observed between rs1052133 polymorphism and smoking: smokers with variant homozygous GG genotype showed an increased risk of CRC[25].
MYH
MYH, also known as MUTYH, is mapped to chromosome 1p34.1 and encodes a glycosylase. This glycosylase initiates the BER pathway by catalyzing the removal of adenine bases of DNA which is inappropriately paired with guanine, cytosine, or 8-oxo-7,8-dihydroguanine[6].
Altogether three studies detected the role of MYH polymorphisms in colorectal carcinogenesis. Tao, H. et al. investigated four MYH SNPs of IVS1+11C>T(rs2275602), IVS6+35G>A(rs3219487), IVS10-2A>G and 972G>C(rs3219489) for an association with altered CRC risk in Japanese[26]. They suggested that (CT+TT) genotype carriers of rs2275602 polymorphism demonstrated increased risk of CRC compared with individuals carrying CC genotype, while no significant relation was identified in the other three polymorphisms. Kasahara, M. et al. found in Japanese that dominant genetic model of rs3219489 polymorphism was associated with increased CRC risk[20]. Similar significant association was subsequently detected by Przybylowska, K. et al. in a research based on Polish population[14].
Incision related BER polymorphisms
APE1
APE1 consists of five exons and four introns spanning 2.21 kb on chromosome 14q11.2 and encodes a protein of 317 amino acids. APE1 deletes abasic sites formed by OGG1 as well as MUTYH and assembles DNA polymerase β and DNA ligase III in BER[27].
Zhang, S. H. et al. found significant interaction of rs1760944 polymorphism with BMI: a protective effect of the T/G genotype was revealed on the development of CRC among subjects with a BMI < 25 kg/m2, although no significant association was detected between this polymorphism and CRC risk[15]. For APE1 rs2307486 polymorphism in exon 3, carriers of AG genotype demonstrated increased risk of CRC compared with GG genotype in Polish[28]. In addition, several investigations have reported significant association between APE1 rs1130409 G/T polymorphism and altered risk of CRC: four studies found that G allele was the risk allele[12, 15, 20, 29] while Jelonek, K. suggested that T allele significantly increased CRC risk in Polish population[30]. Another study indicated that GG genotype carriers of rs1130409 polymorphism demonstrated significantly lower APE1 mRNA expression than TT genotype carriers, which might be an evidence for the risk role of G allele[31]. Two teams found on significant relation of rs1130409 polymorphism with CRC risk in Chinese[32] and Czech[25], respectively. Ching-Y. et al. studied two APE1 polymorphisms (Asp148Glu and T-656G) in Taiwanese but no significant result was found[24].
PARP1
PARP1 gene is mapped to chromosome 1q41-q42, encoding a chromatin-associated poly (ADP-ribosyl) transferase which can detect single-strand breaks and contribute to BER through its interaction with the XRCC1[33].
One study in Singapore Chinese revealed a positive association between the PARP1 codon 940 Lys/Arg genotype and CRC risk[22]. However, no significant relation was found between Val762Ala polymorphism and CRC risk in this study. Another study by Li, Y. et al. suggested that AlaAla genotype of Val762Ala polymorphism significantly increased CRC risk in both homozygous and recessive model in Chinese [32]. For rs8679 polymorphism in 3'UTR region, Alhadheq, A. M. et al. showed no significant association between the polymorphism and risk of CRC in Saudis population[34].
End processing related BER polymorphisms
POLB
POLB (DNA polymerase beta) gene is located at chromosome 8p11.2, which has 16 exons and 15 introns. Polβ is the major DNA polymerase implicated in the initiation of both short-patch and long-patch BER[35].
Only one POLB SNP, rs3136797 (P242R) polymorphism, has been reported. Moreno, V. et al. investigated 28 SNPs of 15 DNA repair genes including POLB and indicated that POLB P242R polymorphism was significantly associated with a reduced risk of CRC[13]. However, the minor allele is very rare and only a few heterozygous individuals were observed, which still required future investigations to confirm.
FEN1
FEN1 (flap structure-specific endonuclease 1), mapped to chromosome 11q12, is essential in efficient 5' flap removal during long-patch base excision repair and the maturation of Okazaki fragments in DNA replication[36].
Until now, only one study by Liu L. et al. detected -69G>A and 4150G>T polymorphisms of FEN1 in cancers of digestive tract including hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer (126 cases) in Chinese population[37]. However, the results suggested no significant relation of these two variants with CRC risk.
Ligation related BER polymorphisms
XRCC1
XRCC1 gene, located at chromosome 19q13.2, has 17 exons and 16 introns. The protein encoded by this gene works as a scaffolding protein and interacts with PARP1, OGG1 and APE1 to facilitate the processes of BER[15].
A number of studies have suggested that rs1799782 C/T in exon 6 of XRCC1 gene could increase CRC risk: Nissar, S. et al.[38] and Li, Y. et al.[32] found CT genotype was related with increased CRC risk in Kashmiri and Chinese population, respectively; The TT genotype was also found to increase CRC risk by two studies[38, 39]. In addition, rs1799782 polymorphism demonstrated significant interaction with smoking: (CT+TT) genotype smokers had 1.6 folds increased risk of CRC[22]. For XRCC1 IVS2-216G>A polymorphism, only one research by Berndt, S. I. et al. revealed that AA genotype was the protective genotype[29]. Although four studies have investigated XRCC1 rs25489 A/G polymorphism of exon 9 in Japanese[40] , American[18], Norwegian[41] and Korean[42], no significant association was indicated. In addition, significant interaction was observed between GG genotype of rs25489 polymorphism and alcohol drinking to increase the risk of CRC [40]. Another most frequently studied polymorphism was 25487 A/G, which is located at exon 10 and changes amino acid from Arg to Gln. Many studies have found significant relation between this polymorphism and increased CRC risk in AG vs. GG model[42-45] and AA vs. GG model[14, 30, 40, 44-46]. But other two studies observed that AG genotype[47] and AA genotype[48] of rs25487 polymorphism can decrease the risk of CRC. Some other researches failed to find significant relation between rs25487 and CRC risk in multiple population[15, 18-20, 22, 25, 32, 41, 49-56]. It is worth noting that three studies have found significant interactions of rs25487 polymorphism with smoking[15] and drinking[46, 54]: A allele carriers of rs25487 showed interaction with alcohol intake to decrease risk of CRC but AG genotype of rs25487 interacts with smoking to increase the CRC risk.
NER pathway gene polymorphisms and sporadic CRC susceptibility
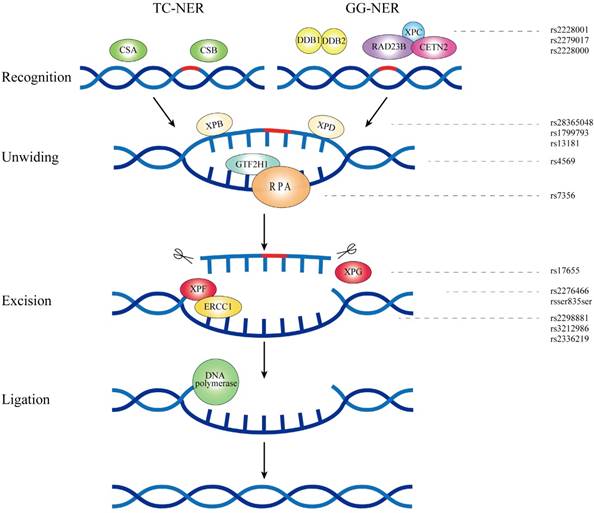
As an essential and versatile system, NER monitors and repairs several types of DNA damage which involves UV-induced cyclobutane pyrimidine dimers, DNA crosslinks and bulky adducts[57]. Transcription coupled NER (TC-NER) and global genome NER (GG-NER) are two NER sub-pathways while the only difference between them is the way of DNA damage recognition[58]. NER consists of four steps: damage recognition, damage demarcation and unwinding, damage incision and new strand ligation. Each step requires indispensable functional proteins, and over 30 factors participate in this precise process[59]. XPA and XPC participate in the first step of NER while XPD together with RPA2 and GTF2H1 play an important role in the damage demarcation and unwinding. Damage incision mainly involves three core proteins of ERCC1, XPF and XPG[60].
DNA damage recognition related NER polymorphisms
XPA
XPA, located at 9q22.33, contains 10 exons and encodes a zinc finger protein which participates in DNA damage recognition of NER. Interacting with DNA and a number of NER proteins, XPA assembles the NER incision complex to the domain where DNA damage occurs[61].
Significant association of BER pathway gene polymorphisms with sporadic CRC susceptibility.
| Variables | Location | Author | Year | Population | Case | Control | Genotypes | OR(95%CI) | Interaction |
|---|---|---|---|---|---|---|---|---|---|
| XRCC1 | 19q13.2 | ||||||||
| rs1799782 | Exon 6 | Dai, Q. | 2015 | Chinese | 438 | 438 | CT vs. CC | 1.19(0.90-1.57) | N.A. |
| TT vs. CC | 1.43(1.20-2.24) | N.A. | |||||||
| Nissar, S. | 2015 | Kashmiri | 100 | 100 | CT vs. CC | 2.01(1.03-3.94) | N.A. | ||
| TT vs. CC | 5.2(1.42-19.5) | ||||||||
| Li, Y. | 2013 | Chinese | 451 | 631 | CT vs. CC | 1.45(1.11-1.89) | N.A. | ||
| TT vs. CC | 1.48(0.91-2.39) | ||||||||
| (CT+TT) vs. CC | 1.45(1.12-1.88) | ||||||||
| Stern, M. C. | 2007 | Chinese | 310 | 1176 | CT vs. CC | 0.9(0.7-1.2) | Interaction with smoking | ||
| TT vs. CC | 0.8(0.5-1.3) | ||||||||
| rs1001581 | Intron 2 | Berndt, S. I. | 2007 | American | 767 | 773 | AA vs. GG | 0.74(0.55-0.99) | None with smoking, alcohol |
| rs25489 | Exon 9 | Yin, G. | 2012 | Japanese | 685 | 778 | AG vs. GG | 0.88(0.66-1.17) | Interaction with alcohol drinking |
| AA vs. GG | 3.07(0.80-11.79) | ||||||||
| rs25487 | Exon 10 | Zhang, S. H. | 2014 | Chinese | 247 | 300 | AG vs. GG | 0.74 (0.52-1.07) | Interaction with smoking |
| AA vs. GG | 1.06 (0.58-1.93) | ||||||||
| Poomphakwaen | 2014 | Thai | 230 | 230 | AG vs. GG | 1.28(0.86-1.90) | Interaction with smoking and drinking | ||
| AA vs. GG | 4.96(1.90-12.95) | ||||||||
| Przybylowska | 2013 | Polish | 182 | 245 | AG vs. GG | 1.31(0.81-2.20) | N.A. | ||
| AA vs. GG | 2.03(1.23-3.97) | ||||||||
| Procopciuc | 2013 | Romanian | 150 | 162 | AG vs. GG | 1.75(1.09-2.82) | N.A. | ||
| AA vs. GG | 3.49(1.55-8.02) | ||||||||
| Yin, G. | 2012 | Japanese | 685 | 778 | AG vs. GG | 1.13(0.91-1.41) | None with alcohol drinking | ||
| AA vs. GG | 1.57(1.01-2.42) | ||||||||
| Zhao, Y. | 2012 | Chinese | 485 | 970 | AG vs. GG | 1.33(1.02-1.68) | N.A. | ||
| AA vs. GG | 2.47(1.63-3.50) | ||||||||
| Wang, J. | 2010 | Indian | 302 | 291 | AG vs. GG | 1.41(0.99-2.03) | Interaction with drinking | ||
| AA vs. GG | 1.20(0.71-2.03) | ||||||||
| Jelonek, K. | 2010 | Polish | 113 | 153 | A allele vs. G allele | 1.51(1.07-2.15) | N.A. | ||
| Stern, M. C. | 2005 | American | 753 | 799 | AG vs. GG | 1.1(0.9-1.3) | N.A. | ||
| AA vs. GG | 0.7(0.5-1.0) | ||||||||
| Hong, Y. C. | 2005 | Korean | 209 | 209 | AG vs. GG | 2.18(1.23-3.88) | N.A. | ||
| AA vs. GG | 1.03(0.31-3.67) | ||||||||
| (AG+AA) vs. GG | 2.00(1.15-3.47) | ||||||||
| Krupa, R. | 2004 | Polish | 51 | 100 | AG vs. GG | 0.73(0.55-0.95) | N.A. | ||
| AA vs. GG | 1.13(0.85-2.34) | ||||||||
| Abdel-Rahman | 2000 | Egyptian | 48 | 48 | AG vs. GG | 3.92(1.40-11.20) | N.A. | ||
| AA vs. GG | 4.20(0.63-34.90) | ||||||||
| OGG1 | 3p26.2 | ||||||||
| rs1052133 | Exon 7 | Zhang, S. H. | 2014 | Chinese | 247 | 300 | CG vs. CC | 0.86(0.53-1.40) | None with smoking, alcohol or BMI |
| GG vs. CC | 0.91(0.56-1.50) | ||||||||
| Przybylowska | 2013 | Polish | 182 | 245 | CG vs. CC | 1.83(1.21-2.70) | N.A. | ||
| GG vs. CC | 1.04(0.23-4.81) | ||||||||
| Canbay, E. | 2011 | Turkish | 79 | 247 | G allele vs. C allele | 2.77(1.40-5.48) | N.A. | ||
| Pardini, B. | 2008 | Czech | 532 | 532 | CG vs. CC | 0.91(0.70-1.18) | Interaction with smoking | ||
| GG vs. CC | 1.43(0.79-2.59) | ||||||||
| (CG+GG) vs. CC | 0.96(0.75-1.23) | ||||||||
| Moreno, V. | 2006 | Spanish | 377 | 329 | GG vs. CC | 2.31(1.05-5.09) | N.A. | ||
| Hansen, R. | 2005 | Norwegian | 166 | 397 | CG vs. CC | 0.56(0.32-0.97) | N.A. | ||
| GG vs. CC | 0.57(0.17-1.83) | ||||||||
| Ching-Yu Lai | 2016 | Taiwanese | 727 | 736 | CG vs. CC | 1.51 (1.11-2.05) | |||
| GG vs. CC | 1.23 (0.90-1.69) | ||||||||
| (CG+GG) vs. CC | 1.38 (1.03-1.85) | ||||||||
| APE1 | 14q11.2 | ||||||||
| rs1130409 | Exon 5 | Zhang, S. H. | 2014 | Chinese | 247 | 300 | GT vs. TT | 0.94(0.64-1.38) | None with smoking, alcohol or BMI |
| GG vs. TT | 2.41(1.50-3.89) | ||||||||
| Li, Y. | 2013 | Chinese | 451 | 631 | GT vs. TT | 1.10(0.83-1.49) | N.A. | ||
| GG vs. TT | 1.13(0.77-1.66) | ||||||||
| Canbay, E. | 2011 | Turkish | 79 | 247 | G allele vs. T allele | 3.43(1.76-6.7) | N.A. | ||
| Jelonek, K. | 2010 | Polish | 113 | 153 | T allele vs. G allele | 2.00(1.39-2.87) | N.A. | ||
| Kasahara, M. | 2008 | Japanese | 68 | 121 | (GT+GG) vs. TT | 2.33(1.21-4.48) | N.A. | ||
| Berndt, S. I. | 2007 | American | 767 | 773 | GT vs. TT | 1.33(1.04-1.69) | None with smoking, alcohol | ||
| (GG+GT) vs. TT | 1.27(1.01-1.60) | ||||||||
| rs2307486 | Exon 3 | Kabzinski, J. | 2015 | Polish | 150 | 150 | AG vs. GG | 2.07(1.21-3.55) | N.A. |
| AA vs. GG | 1.34(0.72-2.51) | ||||||||
| rs1760944 | Promoter | Zhang, S. H. | 2014 | Chinese | 247 | 300 | TG vs. TT | 0.75(0.51-1.10) | Interaction with BMI |
| GG vs. TT | 0.78(0.49-1.25) | ||||||||
| PARP1 | 1q41-q42 | ||||||||
| rs1136410 | Exon 17 | Li, Y. | 2013 | Chinese | 451 | 631 | ValAla vs. ValVal | 1.19 (0.89-1.59) | N.A. |
| AlaAla vs. ValVal | 1.75 (1.20-2.57) | ||||||||
| AlaAla vs. (ValAla+ValVal) | 1.57 (1.12-2.20) | ||||||||
| (CT+CC) vs. TT | 0.584(0.387-0.881) | ||||||||
| rs3219145 | Exon 21 | Stern, M. C. | 2007 | Chinese | 310 | 1176 | LysArg vs. LysLys | 1.7(1.0-3.0) | N.A. |
| ArgArg vs. LysLys | 7.0(0.6-84) | ||||||||
| MUTYH | 1p34.1 | ||||||||
| rs2275602 | Intron 1 | Tao, H. | 2008 | Japanese | 685 | 778 | (CT+TT) vs. CC | 1.46(1.02-2.07) | N.A. |
| rs3219487 | Intron 5 | Tao, H. | 2008 | Japanese | 685 | 778 | AG vs. GG | 1.14(0.88-1.49) | N.A. |
| AA vs. GG | 0.97(0.32-2.93) | ||||||||
| IVS10 -2A/G | Tao, H. | 2008 | Japanese | 685 | 778 | (AG+GG) vs. AA | 0.67(0.39-1.14) | N.A. | |
| rs3219489 | Exon 12 | Przybylowska | 2013 | Polish | 182 | 245 | CG vs. CC | 2.69(1.47-4.94) | N.A. |
| GG vs. CC | 3.35(1.80-6.49) | ||||||||
| Kasahara, M. | 2008 | Japanese | 68 | 121 | (CG+GG) vs. CC | 3.53(1.44-8.70) | N.A. | ||
| Tao, H. | 2008 | Japanese | 685 | 778 | CG vs. GG | 0.96(0.75-1.22) | N.A. | ||
| CC vs. GG | 0.90(0.67-1.22) | ||||||||
| FEN1 | 11q12 | ||||||||
| (-69 A/G) | Promoter | Liu, L. | 2012 | Chinese | 126 | 162 | AG vs. AA | 1.35(0.70-2.72) | N.A. |
| GG vs. AA | 1.60(0.79-3.44) | ||||||||
| rs4246215 | 3'UTR | Liu, L. | 2012 | Chinese | 126 | 162 | GT vs. TT | 1.35(0.65-2.74) | N.A. |
| GG vs. TT | 1.58(0.77-3.44) | ||||||||
| POLB | 8p11.2 | ||||||||
| rs3136797 | Exon 9 | Moreno, V. | 2006 | Spanish | 377 | 329 | (+/-) vs. (+/+) | 0.23(0.05-0.99) | N.A. |
BER pathway gene polymorphisms and sporadic CRC susceptibility.
Only XPA rs1800975 polymorphism in 5'UTR has been investigated by two studies. Joshi, A. D. et al. explored 301 CRC cases and 362 controls of American population but found no significant relation of this polymorphism with CRC risk[62]. Similarly, Hansen, R. D. et al. found no significant association in 397 CRC cases and 800 controls in Denmark[63].
XPC
XPC, mapped to chromosome 3p25.1, consists of 18 exons and is one of the eight core genes in NER system. XPC contributes to damage sensing as well as single-stranded DNA binding during NER process [64].
Polymorphism of rs2228001 (Lys939Gln) in exon 16 has been studied in relation with CRC susceptibility in Malaysian[65], Chinese[66, 67], Turkish[17], Czech[25] and Denmark[63]. Liu, D. et al.'s research in Chinese revealed that AC and (AC+CC) genotype of rs2228001 polymorphism were both related with increased CRC risk compared with wild-type AA genotype[66]. Ahmad Aizat, A. A. et al. found that CC genotype significantly increased the risk of CRC in Malaysian population[65]. Similar correlation was confirmed by Mucha, B. et al.'s study in Polish, which also found significant increased CRC risk of CC genotype[68]. Although no significant relation was found between rs2228001 polymorphism and CRC risk, significant interaction of this polymorphism with red meat was found to increase CRC risk by Hansen, R. D. et al.[63]. For rs2279017 A/C polymorphism at intron 11, Gil, J. et al. suggested increased CRC risk of AC genotype in Polish[69] while another study in American did not find any significant result[62]. The results of rs2228000 C/T polymorphism were still inconclusive: Sun, K. et al.'s study in Chinese[70] and Paszkowska-Szczur, K. et al.'s study in Polish[71] suggested that C allele was the risk allele. However, Steck, S. E. et al. [72] revealed that T allele was the risk allele. In addition, Rui-Xi Hua et al. did not find significant association between rs2228000 polymorphism and CRC risk[67].
DNA damage unwinding related NER polymorphisms
XPD (ERCC2)
XPD, located at 19q13.32, contains 24 exons and encodes a protein which participates in transcription- coupled repair of NER. XPD contributes to the DNA unwinding as well as the damaged DNA fragments excision[61].
Two most frequently studied XPD SNPs are polymorphisms of rs1799793 A/G in exon 10 and rs13181 A/C in exon 22. For rs1799793 polymorphism, Paszkowska-Szczur, K. suggested that both AG genotype and AA genotype were associated with increased risk of CRC compared with wild-type GG genotype in Polish[71]. However, several other investigations did not found similar results in populations of Polish[73], Chinese[22, 74, 75], American[62] or Denmark[63]. Controversies still exist concerning the role of rs13181 polymorphism in relation to CRC susceptibility. Two researches indicated that CC genotype of rs13181 polymorphism was associated with increased risk of CRC compared with the AA genotype in Polish[73] and Romanian[45], respectively. However, Rezaei, H. et al. [76] and Stern, M. C. et al.[77] obtained the opposite conclusion that CC genotype was related with decreased CRC risk in American as well as Iranian. In addition, Stern, M. C. et al. found significant interaction of AC and AA genotype of rs13181 polymorphism with alcohol intake in increasing susceptibility of CRC. In addition, Gil, J. et al. found that the (AC+AA) genotype was associated with decreased CRC susceptibility in polish[69]. Although many other studies investigated the relation between the rs13181 polymorphism and CRC risk in multiple populations[17, 22, 25, 41, 54-56, 62, 63, 74, 75, 78, 79], no significance was found. For rs3810366 polymorphism in promoter, only one team explored the association of this SNP with CRC susceptibility but observed no significance in Chinese [75].
RPA2 and GTF2H1
RPA2 is located at chromosome 1p35.3, encoding a subunit of the heterotrimeric complex RPA which protects single-stranded DNA from nucleases. This heterotrimeric complex binds to single-stranded DNA and contributes to the formation of nucleoprotein complex which plays a key role in DNA unwinding [80]. GTF2H1 is mapped to chromosome 11p15.1, comprising 17 exons and 16 introns. GTF2H1 encodes a member of core-TFIIH basal transcription factor which is involved in transcription initiation and NER pathway[81].
Naccarati, A. et al. found that GG and CG carriers of GTF2H1 rs4596 polymorphism was associated with 0.79 fold decreased CRC risk compared with CC genotype carriers in Czechs [81]. They also observed that the GG genotype of RPA2 rs7356 in 3'UTR region was associated with increased risk of CRC compared with AG and AA genotype. Importantly, RPA2 protein was widely expressed in CRC and miRNA reduced RPA2 expression by preferentially binding to variant G allele of rs7356 polymorphism. These findings partially explained the reason why rs7356 G allele was associated with decreased CRC susceptibility.
DNA damage incision related NER polymorphisms
ERCC1
ERCC1, located at 19q13.32, contains 14 exons and the protein encoded by this gene assembles XPF to form a heterodimer. The heterodimer endonuclease promotes the 5' incision in repairing DNA lesion as well as contributes to DNA recombination repair and inter-strand crosslinks repair[82].
For ERCC1 rs2298881 A/C polymorphism in intron 1, Yang, H. et al.[83] suggested that the CC genotype was related with increased CRC risk compared with AA genotype in Chinese. They found no significant relation of rs11615 C/T polymorphism in exon 4 with CRC susceptibility in Chinese while another team obtained different result. Te-Cheng Yueh. et al.[84] found that the TT genotype of rs11615 C/T polymorphism was associated with 1.86-fold increased CRC risk compared with CC genotype in Chinese. Significant relation between AA genotype of rs3212986 A/C polymorphism in 3'UTR region and increased CRC risk was observed compared with CC genotype[74, 85] in Chinese but no significance was found in American[62]. Importantly, significant interaction was indicated in increasing risk of CRC between TT genotype of rs3212986 polymorphism and cigarette smoking[82]. Additionally, Dai, Q. et al. found that the AA genotype of rs2336219 A/G polymorphism in 3'UTR correlated with increased risk of CRC compared with wild-type GG genotype in Chinese [39]. A number of researches investigated rs11615 C/T polymorphism in CRC susceptibility but indicated no significance in Chinese and Norwegian population[74, 82-84, 86].
XPF (ERCC4)
XPF, located at 16p13.12, contains 13 exons and 12 introns, spanning approximately 28.2 kb. Its encoding protein XPF forms a complex with ERCC1, which is responsible for the 5' incision of DNA damage repair[82].
For polymorphisms of XPF rs2276466 C/G in 3'UTR and rs6498486 A/C in promoter, Hou, R. et al.[82] explored their relationships with CRC risk in Chinese population but indicated no significant association. Another team[83] found no significant association between the rs2276466 C/G polymorphism and risk of CRC. Additionally, no significant association between rs1800067 polymorphism and CRC susceptibility was observed by Joshi, A. D. et al.[62] in American. The synonymous substitution of rs1799801 at exon 13 has been investigated by Kabzinski, J. et al.[87], the result of which indicated that CT genotype correlated with decreased susceptibility of CRC compared with the CC genotype.
XPG (ERCC5)
XPG is mapped to chromosome 13q33, encoding a structure-specific endonuclease XPG which is composed of 1186 amino acids. XPG contributes to the 3' incision of DNA damage and enables DNA repair complex to stabilize to the domain of damage DNA[61].
For polymorphism of XPG rs17655 C/G in exon 15, Du, H. et al.[88] found that the variation from G allele to C allele was associated with increased risk of CRC in Chinese. Additionally, another team observed that CG genotype of rs17655 polymorphism was related with 1.33-fold increased CRC susceptibility in Chinese compared with GG genotype[66]. In 1901 cases and 1976 controls, rs2094258, rs751402, rs2296147, rs1047768 and rs873601 polymorphisms of ERCC1 were studied by Rui-Xi Hua et al.[89] in relation with CRC risk and most of the results demonstrated significance. In this research, they observed that four SNPs (rs2094258C/T in promoter, rs751402C/T in 5' UTR, rs1047768 C/T in exon 2 and rs873601 in 3'UTR) were associated with increased CRC risk, three of which (rs2094258, rs751402 and rs873601) also correlated with XPG mRNA expression. Other three studies suggested no significant association between rs17655 C/G polymorphism and risk of CRC in Chinese[70], American[62] or Czech[25]. For XPG 1558His/Asp polymorphism, Kabzinski, J. et al. failed to show significant association with susceptibility of CRC in Polish[73].
MMR pathway gene polymorphisms and sporadic CRC susceptibility
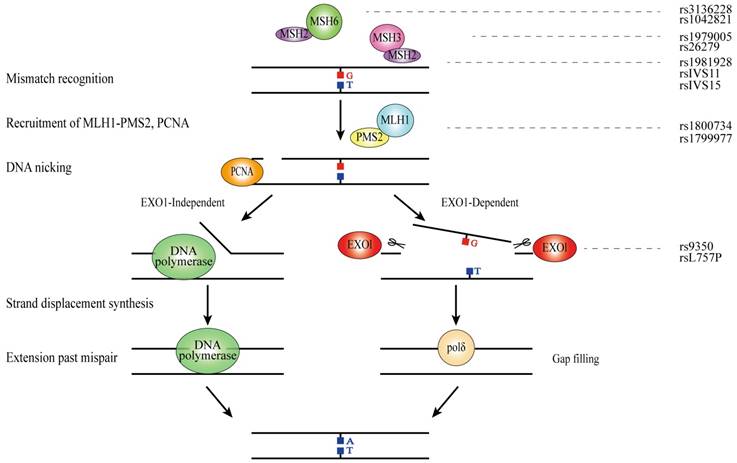
DNA mismatch repair (MMR) is a highly conserved biological pathway that is involved in maintaining genomic stability[90]. MMR recognizes and corrects the biosynthetic errors aroused during DNA replication as well as the mispaired bases which is generated in DNA recombination or caused by oxidative DNA damage[91]. MMR decreases 100-1000 folds DNA errors and protects them from mutations during cellular proliferation[92]. Human MMR process is classified into four steps: (1) the mismatch recognition by MutS homologs (MSH2, MSH3 and MSH6) and recruitment of MutL homologs (MLH1, MLH3, PMS1 and PMS2); (2) strand discrimination to mark the erroneous DNA strand; (3) strand removal by unwinding and excision reactions (EXO1); (4) DNA-re-synthesis and ligation to complete the repair reaction[93].
Significant association of NER pathway gene polymorphisms with sporadic CRC susceptibility.
| Variables | Location | Author | Year | Population | Case | Control | Genotypes | OR(95%CI) | Interaction |
|---|---|---|---|---|---|---|---|---|---|
| XPC | 3p25.1 | ||||||||
| rs2228001 | Exon 16 | Ahmad Aizat | 2013 | Malaysian | 255 | 255 | AC vs. AA | 1.27(0.87-1.84) | N.A. |
| CC vs. AA | 1.88(1.08-3.28) | ||||||||
| Liu, D. | 2012 | Chinese | 1028 | 1085 | AC vs. AA | 1.40(1.16-1.69) | N.A. | ||
| CC vs. AA | 0.98(0.84-1.13) | ||||||||
| (AC+CC) vs. AA | 1.31(1.10-1.56) | ||||||||
| Hansen, R. D. | 2007 | Dane | 397 | 800 | AC vs. AA | 1.08(0.83-1.42) | Interaction with red meat | ||
| CC vs. AA | 1.16(0.77-1.77) | ||||||||
| Mucha, B. | 2018 | Polish | 221 | 270 | AC vs. AA | 1.07(0.65-1.76) | N.A. | ||
| CC vs. AA | 1.82(1.08-3.06) | ||||||||
| rs2279017 | Intron 11 | Gil, J. | 2012 | Polish | 133 | 100 | AC vs. CC | 2.07(1.14-3.78) | N.A. |
| rs2228000 | Exon 9 | Sun, K. | 2015 | Chinese | 890 | 910 | CT vs. TT | 1.06(0.87-1.30) | N.A. |
| CC vs. TT | 2.19(1.60-3.01) | ||||||||
| Paszkowska | 2015 | Polish | 758 | 1841 | CT vs. CC | 0.59(0.49-0.72) | N.A. | ||
| TT vs. CC | 0.29(0.20-0.41) | ||||||||
| Steck, S. E. | 2014 | African American | 244 | 331 | CT vs. CC | 1.7(1.1-2.6) | N.A. | ||
| rs1799793 | Exon 10 | Paszkowska | 2015 | Polish | 758 | 1841 | AG vs. GG | 1.92(1.41-2.62) | N.A. |
| AA vs. GG | 6.92(4.61-10.36) | ||||||||
| rs13181 | Exon 22 | Kabzinski, J. | 2015 | Polish | 235 | 240 | AC vs. AA | 0.60(0.35-1.02) | N.A. |
| CC vs. AA | 14(6.31-31.05) | ||||||||
| Rezaei, H. | 2013 | Iranian | 88 | 88 | AC vs. AA | 1.33(0.68-2.62) | N.A. | ||
| CC vs. AA | 0.10(0.03-0.30) | ||||||||
| Procopciuc | 2013 | Romanian | 150 | 162 | AC vs. AA | 1.49(0.91-2.44) | N.A. | ||
| CC vs. AA | 3.02(1.15-8.25) | ||||||||
| Gil, J. | 2012 | Polish | 133 | 100 | (AC+CC) vs. AA | 0.45(0.22-0.91) | N.A. | ||
| Stern, M. C. | 2006 | American | 753 | 799 | AC vs. AA | 1.0(0.8-1.2) | Interaction with smoking or drinking | ||
| CC vs. AA | 0.7(0.4-1.0) | ||||||||
| XPF(ERCC4) | 16p13.12 | ||||||||
| Ser835Ser | Exon 15 | Kabzinski, J. | 2015 | Polish | 146 | 149 | CT vs. CC | 0.57(0.34-0.98) | N.A. |
| TT vs. CC | 1.12(0.60-2.07) | ||||||||
| XPG(ERCC5) | 13q33 | ||||||||
| rs17655 | Exon 15 | Sun, K. | 2015 | Chinese | 890 | 910 | CG vs. GG | 1.01(0.80-1.26) | N.A. |
| CC vs. GG | 1.12(0.85-1.47) | ||||||||
| Du, H. | 2014 | Chinese | 878 | 884 | CG vs. GG | 1.41(1.15-1.74) | N.A. | ||
| CC vs. GG | 1.34(1.00-1.79) | ||||||||
| (CG+CC) vs.GG | 1.40(1.15-1.70) | ||||||||
| Liu, D. | 2012 | Chinese | 1028 | 1085 | CG vs. GG | 1.33(1.09-1.63) | N.A. | ||
| CC vs. GG | 0.93(0.81-1.06) | ||||||||
| (CG+CC) vs.GG | 1.20(0.99-1.46) | ||||||||
| rs2094258 | Promoter | Rui-Xi Hua | 2016 | Chinese | 1901 | 1976 | CT vs. CC | 1.17(1.01-1.36) | |
| TT vs. CC | 1.49(1.18-1.89) | ||||||||
| rs751402 | 5'UTR | Rui-Xi Hua | 2016 | Chinese | 1901 | 1976 | CT vs. CC | 0.82(0.70-0.96) | |
| TT vs. CC | 0.69(0.55-0.86) | ||||||||
| rs1047768 | Exon 2 | Rui-Xi Hua | 2016 | Chinese | 1901 | 1976 | TC vs. TT | 1.00(0.86-1.16) | |
| CC vs. TT | 1.33(1.01-1.75) | ||||||||
| rs873601 | 3'UTR | Rui-Xi Hua | 2016 | Chinese | 1901 | 1976 | AG vs. GG | 1.18(1.00-1.40) | |
| AA vs. GG | 1.41(1.15-1.72) | ||||||||
| ERCC1 | 19q13.32 | ||||||||
| rs2298881 | Intron 1 | Yang, H. | 2015 | Chinese | 279 | 316 | AC vs. AA | 1.37(0.91-1.92) | N.A. |
| CC vs. AA | 2.68(1.47-4.75) | ||||||||
| Hou, R. | 2014 | Chinese | 204 | 204 | AC vs. AA | 1.08(0.71-1.74) | N.A. | ||
| CC vs. AA | 1.45(0.64-3.46) | ||||||||
| rs11615 | Exon 4 | Te-Cheng Y. | 2017 | Chinese | 362 | 362 | CT vs. CC | 1.06 (0.77-1.46) | |
| TT vs. CC | 1.86 (1.20-2.87) | ||||||||
| rs3212986 | 3'UTR | Ni, M. | 2014 | Chinese | 213 | 240 | AC vs. CC | 1.47(0.99-2.18) | None with smoking or drinking |
| AA vs. CC | 2.50(1.10-5.70) | ||||||||
| Hou, R. | 2014 | Chinese | 204 | 204 | GT vs. GG | 1.26(0.81-2.03) | Interaction with smoking | ||
| TT vs. GG | 1.93(0.96-3.94) | ||||||||
| Zhang, Q. | 2018 | Chinese | 200 | 200 | AC vs. CC | 1.20(0.79-1.81) | N.A. | ||
| AA vs. CC | 2.53(1.14-5.60) | ||||||||
| rs2336219 | 3'UTR | Dai, Q. | 2015 | Chinese | 438 | 438 | AG vs. GG | 1.34(0.88-1.77) | N.A. |
| AA vs. GG | 1.46(1.14-2.43) | ||||||||
| RPA2 | 1p35.3 | ||||||||
| rs7356 | 3'UTR | Naccarati | 2012 | Czech | 1098 | 1469 | GG vs. (AG+AA) | 1.33(1.01-1.75) | N.A. |
| GTF2H1 | 11p15.1 | ||||||||
| rs4596 | 3'UTR | Naccarati | 2012 | Czech | 1098 | 1469 | (CG+GG) vs. CC | 0.79(0.64-0.99) | N.A. |
NER pathway gene polymorphisms and sporadic CRC susceptibility.
MutS homologs related MMR polymorphisms
MSH2
MSH2 is located at chromosome 2p21-p16.3, consisting of 21 exons and 20 introns. MSH2 participates in the formation of two heterodimeric complexes of Mutsα and Mutsβ which are involved in insertion-deletion loops in DSBR[94].
In Chinese population, Li, G. et al.[95] found that CT genotype of MSH2 IVS15-214 polymorphism was associated with decreased risk of CRC compared with TT genotype. They observed that the AG genotype of IVS11+107 polymorphism were related with decreased CRC susceptibility compared with AA genotype. Importantly, significant gene-environment interactions were detected of both C allele of IVS15-214 polymorphism and GG genotype of IVS11+107 polymorphism with cereals intake in decreasing CRC susceptibility. In addition, TT genotype of rs1981928 polymorphism was correlated with 0.78 fold reduced CRC risk in English[96]. For rs4987188 polymorphism, several researches showed no significant association with CRC risk in American[62], Canadian[97] or Polish[98]. No significant relationship was observed of another two SNPs of -118 T/C[99] and IVS12-6 T/C[97] polymorphisms with CRC risk in Canadian population.
MSH3
MSH3, also known as DUP, FAP4 and MRP1, is located at 5q14.1 and consists of 24 exons. MSH3 cooperates with MSH2 to form a heterodimer Mutsα which binds to a mismatch and activates the MMR pathway[93].
Only one study by Koessler, T. et al.[96] explored the association between MSH3 rs1979005 C/T polymorphism and CRC risk and found that the TT genotype was associated with decreased risk of CRC compared with CC genotype in English. They observed that the GG genotype of rs26279 A/G polymorphism in exon 23 correlated with 1.31 folds increased risk of CRC compared with wild-type AA genotype.
MSH6
MSH6 is mapped to chromosome 2p16.3 and encodes a MutS family protein which contributes to the mismatched nucleotides recognition before repair. Together with MSH2, MSH6 forms a mismatch recognition heterodimer complex which adjusts the function of MMR by exchanging ATP and ADP when DNA mismatches are bound and divided[94].
For MSH6 rs1042821 G/A polymorphism in exon 1, significant association was found of the AG genotype with increased CRC risk compared with GG genotype in Polish [100] but another team failed to observed significance in mixed population[101]. However, Tulupova, E. et al. found that GA and AA genotype of the same rs1042821 polymorphism in promoter correlated with decreased CRC susceptibility compared with GG genotype in Czech population, the reason of which might be that rs1042821 played different roles in variant transcripts. They also observed that T-allele carriers of MSH6 rs3136228 polymorphism in promoter were associated with increased risk of CRC in Czechs compared with carriers of GG genotype [102]. For MSH6 -159C/T promoter polymorphism, Mrkonjic, M. et al. showed no significance in Canadians[99].
MutL homologs related MMR polymorphisms
MLH1 and PMS2
MLH1, located at 3p22.2, contains 21 exons and PMS2 is mapped to 7p22.1, consisting of 16 exons and 15 introns. MLH1 and PMS2 form a MutL-alpha heterodimer which manages the activity of endonuclease involved in mismatches recognition and loops insertion or deletion[103]. In addition, MutL-alpha heterodimer also plays a key role in mismatched DNA removal[103].
For MLH1 rs1800734 A/G polymorphism in prompter, A allele was found to significantly reduce the risk of CRC compared with G allele in Polish[98], Spanish[104] and Mexican population[51]. However, Nizam, Z. M. suggested that AG genotype was associated with 3.71 folds increased CRC risk compared with GG genotype in Malaysian[105]. Other two researches also investigated the relation of rs1800734 polymorphism with CRC risk but no significance was shown in American[101] and Canadian[97]. For MLH1 rs1799977 polymorphism in exon 8, Nejda, N. et al. observed that both AG and GG genotype were associated with increased risk of CRC compared with AA genotype in Spanish [106]. But other teams failed to find significance in Mexican[51], American[62, 101] or Canadians[97]. Only Raptis, S. et al. studied MLH1 IVS14-19A>G polymorphism but did not obtain significant result[97]. Although H.X. Peng et al. studied the relation of V384D, R217C and rs1799977 polymorphisms with CRC risk, the samples of each genotypes were insufficient to draw reliable conclusion[107]. For PMS2 rs63750451 polymorphism in exon 9, one team explored its relation with CRC risk but show no significance in Polish[100].
DNA nicking related MMR polymorphisms
EXO1
EXO1, mapped to 1q42-q43, consists of 17 exons and encodes a protein with 5' to 3' exonuclease activity and RNase H activity, which participates in DNA nicking of MMR. Additionally, EXO1 is the only known active nuclease in human cells MMR[93].
For EXO1 rs9350 polymorphism in exon 14, Haghighi, M. M. et al. found that CT genotype was associated with 0.17-fold decreased CRC susceptibility compared with CC genotype in Iranian [108]. Another team observed that C allele of rs9350 significantly increased the risk of CRC compared with T allele in American[109]. Importantly, they showed a significant interaction between C allele of rs9350 polymorphism and cigarette smoking in increasing CRC risk.
DSBR pathway gene polymorphisms and sporadic CRC susceptibility
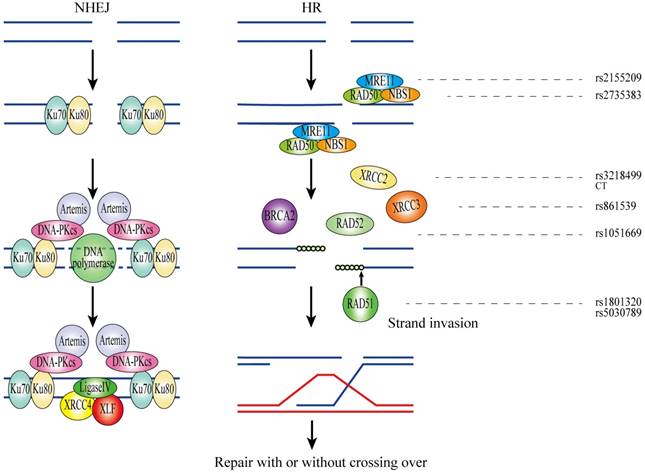
DNA double-strand breaks (DSBs) are highly toxic lesions which result in genetic instability[110]. To preserve genome integrity, a number of DSBR reactions exist in organisms, of which non-homologous end-joining (NHEJ) and homologous recombination (HR) are the two most widely used systems[111]. NHEJ is regarded as an error-prone manner and utilizes limited or no homologous DNA for end joining. Bound to the damaged DNA ends to initiate NHEJ, the Ku70/80 heterodimer recruits and triggers the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) which facilitates the downstream repair processes. Then, scaffold proteins XRCC4 and XLF move to the defect domain and combine with DNA Ligase 4 for repairing the lesions[111, 112]. In contrast, HR is largely error free and requires extensive homology for the repair of DNA DSBs. After the recognition of DSBs in HR, the resection of DSBs is completed by the MRE11/RAD50 /NBS1 complex which then generates a 3' ssDNA overhang. BRCA2, RAD51 as well as RAD51 paralogous (Rad51C, Rad51D, XRCC2, XRCC3) bind to the ssDNA tails and form a presynaptic filament. Subsequently, the formation of D loop in strand invasion is initialized and DSBs were repaired by structure-specific nucleases[113].
Homologous recombination (HR)
End resection related DSBR polymorphisms
MRE11 and NBS1
MRE11 , located at chromosome 11q21, contains 22 exons and encodes a protein with 3' to 5' exonuclease and endonuclease activity. NBS1 is mapped to 8q21.3 and consists of 19 exons and 18 introns. Together with MRE11 and RAD50, NBS1 forms a complex involved in DNA ends resection, which generates 3' single-stranded tails in HR[114].
Significant association of MMR pathway gene polymorphisms with sporadic CRC susceptibility.
| Variables | Location | Author | Year | Population | Case | Control | Genotypes | OR(95%CI) | Interaction |
|---|---|---|---|---|---|---|---|---|---|
| MLH1 | 3p22.2 | ||||||||
| rs1800734 | Promoter | Nizam | 2013 | Malaysian | 52 | 104 | AG vs. GG | 3.71(1.42-9.74) | N.A. |
| AA vs. GG | 2.36(0.88-6.31) | ||||||||
| Michal Mik | 2017 | Polish | 144 | 151 | AG vs.AA | 1.09 (0.58-2.05) | |||
| GG vs AA | 2.07 (1.11-3.83) | ||||||||
| Martinez | 2013 | Spanish | 183 | 236 | AG vs. GG | 0.58(0.39-0.86) | N.A. | ||
| AA vs. GG | 1.16(0.35-3.91) | ||||||||
| Muniz | 2012 | Mexican | 108 | 120 | AG vs. GG | 0.66(0.37-1.17) | N.A. | ||
| AA vs. GG | 0.32(0.13-0.79) | ||||||||
| rs1799977 | Exon 8 | Nejda, N. | 2009 | Spanish | 140 | 125 | AG vs. AA | 2.55(1.48-4.39) | N.A. |
| GG vs. AA | 2.48(1.20-5.11) | ||||||||
| V384D | H.X. Peng | 2016 | Chinese | 156 | 311 | AA | 0.03 (0-0.24) | ||
| AT | 28.18 (3.81-∞) | ||||||||
| TT | ∞ (0-∞) | ||||||||
| MSH2 | 2p21-p16.3 | ||||||||
| IVS15-214T>C | Li, G. | 2015 | Chinese | 451 | 630 | CT vs. TT | 0.62(0.46-0.83) | Interaction for cereals | |
| CC vs. TT | 0.89(0.62-1.26) | ||||||||
| IVS11+107A>G | Li, G. | 2015 | Chinese | 451 | 630 | AG vs. AA | 0.61(0.42-0.88) | Interaction for cereals | |
| GG vs. AA | 0.76(0.52-1.10) | ||||||||
| rs1981928 | Intron 7 | Koessler, T. | 2008 | English | 2299 | 2284 | AT vs. AA | 1.05(0.93-1.18) | N.A. |
| TT vs. AA | 0.78(0.62-0.99) | ||||||||
| MSH3 | 5q14.1 | ||||||||
| rs1979005 | Intron 20 | Koessler, T. | 2008 | English | 2299 | 2284 | CT vs. CC | 0.90(0.76-1.06) | N.A. |
| TT vs. CC | 0.41(0.18-0.94) | ||||||||
| rs26279 | Exon 23 | Koessler, T. | 2008 | English | 2299 | 2284 | AG vs. AA | 1.04(0.92-1.17) | N.A. |
| GG vs. AA | 1.31(1.05-1.62) | ||||||||
| MSH6 | 2p16.3 | ||||||||
| rs1042821 | Exon 1 | Piotr Zelga | 2017 | Polish | 200 | 200 | AG vs.GG | 1.69 (1.1-2.61) | |
| AA vs. GG | 2.08 (0.52-8.42) | ||||||||
| rs3136228 | Promoter | Tulupova | 2008 | Czech | 614 | 614 | (GT+TT) vs. GG | 1.29(1.02-1.62) | N.A. |
| rs1042821 | Promoter | Tulupova | 2008 | Czech | 614 | 614 | (GA+AA) vs. GG | 0.76(0.60-0.98) | N.A. |
| EXO1 | 1q42-q43 | ||||||||
| rs9350 | Exon 14 | Haghighi | 2010 | Iranian | 90 | 98 | CT vs.CC | 0.17(0.03-0.82) | N.A. |
| TT vs.CC | 0.69(0.37-1.28) | ||||||||
| Gao, Y. | 2011 | American | 1338 | 1503 | C allele vs. T allele | 1.30(1.11-1.51) | Interaction with smoking | ||
MMR pathway gene polymorphisms and sporadic CRC susceptibility.
Naccarati, A. et al. found that CC genotype of MRE11 rs2155209 polymorphism was associated with decreased risk of CRC compared with TT genotype in Italian[115]. However, they did not find significant relation between CT genotype of NBS1 rs14448 polymorphism and CRC risk. For NBS1 rs2735383 polymorphism, Li, J. T. et al. observed that CC genotype correlated with increased CRC susceptibility compared with GG genotype in Chinese[116]. In addition, no significant association was found of NBS1 rs1805794 polymorphism in exon 5 with CRC susceptibility in Czech population[25].
Strand invasion and exchange related DSBR polymorphisms
XRCC2
XRCC2 is located at chromosome 7q36.1 and comprises three exons and two introns. XRCC2 protein improves the activity of RAD51 which is involved in strand invasion and exchange reactions in HR[117].
Li, X. B. et al. demonstrated significant association of XRCC2 rs718282 polymorphism with increased CRC risk in Chinese but no significance was found for rs3218384 polymorphism[117]. For XRCC2 rs3218499 polymorphism, Curtin, K. observed that CC genotype correlated with increased CRC risk compared with CG and GG genotypes in the mixed population of English and American[118]. Additionally, two researches failed to find significant relationship between rs3218536 polymorphism in exon 3 and CRC susceptibility in Polish [119] and American[120].
XRCC3
XRCC3, also known as CMM6, is located at chromosome 14q32.3 and contains 10 exons. XRCC3 encodes a member of Rad51-related proteins which function in the maintenance of chromosome stability and initiation of homologous sequence strand invasion[121].
Controversial results were found for the association between XRCC3 rs861539 C/T polymorphism and CRC risk. Zhao, Y. et al. observed that T allele was a risk factor for CRC in Chinese[44] but C allele indicated higher CRC risk according to Mort, R. et al.'s study in English[122]. Other two teams suggested that CT genotype was related with increased CRC risk compared with CC genotype in Kashmirian[123] and Chinese[121], respectively. However, Mucha, B. et al. suggested that CT genotype significantly decreased CRC risk in Polish[124]. Krupa, R. et al. found that CT genotype significantly decreased risk of CRC but TT genotype correlated with increased susceptibility of CRC in Polish[48]. In addition, some other researches failed to indicate significant association of rs861539 polymorphism with CRC risk in Algerian[78], Polish[119], Indian[54], Czech[25], Chinese[55], Norwegian[41] or American [47, 120]. For rs1799794 and rs1799796 polymorphisms of XRCC3, no significant relation was observed in American[120].
RAD51
RAD51, located at chromosome 15q15.1, contains 14 exons and encodes RAD51 which interacts with BRCA1 and BRCA2 in response to the DNA damage in DSBR. RAD51 also cooperates with RAD51 paralogues to handle the strand transfer of DNA in HR[112].
For RAD51 rs1801320 polymorphism, Krupa, R. et al. found that CC genotype was related with decreased CRC risk compared with GG genotype in polish[119]but another team obtained an opposite conclusion in the same population[125]. Nissar, S. et al. suggested that CG genotype was a risk genotype of CRC in Kashmiri[126]. No significant association was found in Yazdanpanahi, N. et al.'s study of RAD51 rs1801320 polymorphism in Iranian[127]. One research investigated the relationship between RAD51 172G/T polymorphism and CRC risk in polish but no significance was found[125]. Mucha, B. et al. indicated that AG genotype of rs5030789 promoter polymorphism was associated with increased CRC susceptibility [128] but no significant association was observed for rs2619679 [128] or rs1801320 polymorphism[129].
RAD52
RAD52 is located at chromosome 12p13.33 and contains 17 exons and 16 introns. RAD52 works as a mediator alone in HR or interacts with RAD51 to participate in the strand invasion and exchange in human cells[112].
Although the relation was studied between several RAD52 SNPs and CRC risk, only Naccarati, A. et al. found that AA genotype of RAD52 rs1051669 polymorphism significantly increased CRC risk compared with GG genotype in Italian [130]. For rs11571378, rs7963551, rs6489769 and rs10774474 polymorphisms, no significance was found in relation with CRC susceptibility[130, 131].
Non-homologous end-joining (NHEJ)
End ligation related DSBR polymorphisms
XRCC4
XRCC4, also known as SSMED, is mapped to chromosome 5q14.2 and consists of 13 exons and 12 introns. Together with XLF, scaffold protein XRCC4 binds DNA ligase IV in order to seal the breaks in NHEJ[112]. Emami, N. studied the relationship of XRCC4 rs6869366 and rs28360071 polymorphisms with CRC risk in Iranian population but demonstrated no significance[132].
DSBR pathway gene polymorphisms and sporadic CRC susceptibility.
Significant association of DSBR pathway gene polymorphisms with sporadic CRC susceptibility.
| Variables | Location | Author | Year | Population | Case | Control | Genotypes | OR(95%CI) | Interaction |
|---|---|---|---|---|---|---|---|---|---|
| XRCC2 | 7q36.1 | ||||||||
| rs718282 | Li, X. B. | 2014 | Chinese | 246 | 262 | (CT+TT) vs. CC | 1.65(1.13-2.40) | N.A. | |
| rs3218384 | Promoter | Li, X. B. | 2014 | Chinese | 246 | 262 | (CG+CC) vs. GG | 1.30(0.89-1.90) | N.A. |
| rs3218499 | Intron 2 | Curtin, K. | 2009 | U.K./U.S. | 1252 | 1422 | CC vs. (CG+GG) | 1.6(1.1-2.2) | N.A. |
| XRCC3 | 14q32.3 | ||||||||
| rs861539 | Exon 7 | Nissar, S. | 2014 | Kashmirian | 120 | 150 | CT vs. CC | 2.53 (1.37-4.66) | N.A. |
| TT vs. CC | 2.29(0.96-5.40) | ||||||||
| Mucha, B. | 2013 | Polish | 194 | 204 | CT vs. CC | 0.57(0.37-0.87) | N.A. | ||
| TT vs. CC | 0.82(0.44-1.55) | ||||||||
| Zhao, Y. | 2012 | Chinese | 485 | 970 | CT vs. CC | 1.82(1.24-2.93) | N.A. | ||
| TT vs. CC | 1.84(1.15-3.12) | ||||||||
| Jin, M. J. | 2005 | Chinese | 140 | 280 | CT vs. CC | 3.25(1.42-7.42) | None with smoking or drinking | ||
| Krupa, R. | 2004 | Polish | 51 | 100 | CT vs. CC | 0.26(0.25-0.27) | N.A. | ||
| TT vs. CC | 9.45(8.77-11.65) | ||||||||
| Tranah, G. J. | 2004 | American | 932 | 1282 | CT vs. CC | 0.95(0.78-1.16) | N.A. | ||
| TT vs. CC | 0.89(0.68-1.17) | ||||||||
| Mort, R. | 2003 | English | 246 | 256 | C allele vs. T allele | 1.52(1.04-2.22) | N.A. | ||
| NBS1 | 8q21.3 | ||||||||
| rs2735383 | 3'UTR | Li, J. T. | 2015 | Chinese | 1076 | 1263 | CG vs. GG | 1.13(0.97-1.41) | N.A. |
| CC vs. GG | 1.68(1.31-2.13) | ||||||||
| CC vs. (CG+GG) | 1.55 (1.27-1.94) | ||||||||
| rs14448 | 3'UTR | Naccarati, A. | 2016 | Italian | 1111 | 1469 | TC vs. TT | 0.78 (0.51-1.19) | |
| RAD51 | 15q15.1 | ||||||||
| rs1801320 | 5' UTR | Nissar, S. | 2014 | Kashmiri | 100 | 120 | CG vs. GG | 3.84(3.84-7.20) | N.A. |
| CC vs. GG | 1.82(0.85-3.88) | ||||||||
| (CG+CC) vs. GG | 3.0(1.6-5.3) | ||||||||
| Romanowicz | 2012 | Polish | 320 | 320 | CG vs. GG | 0.60 (0.38-0.96) | N.A. | ||
| CC vs. GG | 5.84 (3.76-9.09) | ||||||||
| Krupa, R. | 2011 | Polish | 100 | 100 | CG vs. GG | 0.60(0.33-1.12) | N.A. | ||
| CC vs. GG | 0.06(0.02-0.22) | ||||||||
| rs5030789 | Promoter | Mucha, B. | 2015 | Polish | 115 | 118 | AG vs. GG | 1.85(1.06-2.26) | N.A. |
| AA vs. GG | 1.21(0.47-3.12) | ||||||||
| RAD52 | 12p13.33 | ||||||||
| Naccarati, A. | 2016 | Italian | 1111 | 1469 | GA vs. GG | 1.09( 0.86-1.37) | |||
| AA vs.GG | 1.78 (1.13-2.80) | ||||||||
| (GA+AA) vs. GG | 1.17 (0.93-1.46) | ||||||||
| AA vs.(GG+GA) | 1.72( 1.10-2.69) | ||||||||
| MRE11A | 11q21 | ||||||||
| rs2155209 | 3'UTR | Naccarati, A. | 2016 | Italian | 1111 | 1469 | CT vs. TT | 0.94 (0.75-1.19) | N.A. |
| CC vs.TT | 0.66 (0.45-0.96) | ||||||||
| (TC+CC) vs TT | 0.88 (0.70-1.09) | ||||||||
| CC vs.(TT+TC) | 0.68 (0.47-0.97) |
Summary and Future Directions
Genetic polymorphisms in DNA repair genes may modulate DNA repair efficiency thereby influencing the development of sporadic CRC. In recent years, substantial progress has been made towards uncovering the genetic architecture of CRC, which offer great opportunity to benefit the understanding of sporadic CRC development. In this review, we summarized the genetic architecture of DNA repair genes involved in sporadic colorectal carcinogenesis as well as discussed the future directions of how genetic insights improve clinical surveillance, prevention and treatment strategies of sporadic CRC.
Previously, polymorphisms of BER core genes including XRCC1, OGG1, APE1, PARP1, MUTYH and POLB have been linked to altered CRC risk by multiple studies. Important genes involved in NER pathway of XPC, XPD, XPF, XPG and ERCC1 all possess certain polymorphisms which significantly influence CRC susceptibility. For MMR system, key genes of MLH1, MSH2, MSH3, MSH6 and EXO1 demonstrated significant associations with CRC risk. As essential members of DSBR pathway, XRCC2, XRCC3, NBS1, RAD51, RAD52 and MRE11A polymorphisms showed involvement in the determination of CRC susceptibility. The observed significant associations of polymorphisms in BER, NER, MMR and DSBR pathway core genes with sporadic CRC risk suggested an extensive implication of genetic polymorphisms of DNA repair pathways in colorectal carcinogenesis. The promising values of these polymorphisms in CRC prediction and prevention as well as their underlying mechanisms are of great importance. In addition, polymorphisms of DNA repair pathways might be applied in clinical outcomes to guide management of CRC patients. For example, ERCC1 and XRCC1 polymorphisms may influence the clinical outcome of colorectal cancer patients treated with mFOLFOX6 adjuvant chemotherapy[133]. Genetic polymorphisms of MLH3 rs175057 as well as MSH2 rs3771273, rs10188090 and rs10191478 may predict prognosis in patients with locally advanced rectal cancer who received preoperative chemoradiotherapy [134]. XRCC3 Thr241Met polymorphism was associated with time-to-metastasis of CRC[135]. The specific role of the summarized polymorphisms of our review in clinical application and underlying mechanisms required further studies to elucidate.
Acknowledgements
This study is supported by grants from the National Science and Technology Support Program (2015BAI13B07), Public Welfare Foundation of Liaoning Province (No. 2015005002) and Fund for Scientific Research of The First Hospital Of China Medical University (FHCMU- FSR).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nature reviews Cancer. 2016;16(1):20-33
2. Decordier I, Loock KV, Kirsch-Volders M. Phenotyping for DNA repair capacity. Mutat Res. 2010;705(2):107-129
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5-29
4. Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, Kuipers EJ. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637-1649
5. Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst). 2014;19:14-26
6. Krokan HE, Bjoras M. Base Excision Repair. Cold Spring Harbor Perspectives in Biology. 2013;5(4):a012583-a012583
7. Parsons JL, Dianov GL. Co-ordination of base excision repair and genome stability. DNA Repair (Amst). 2013;12(5):326-333
8. Patrono C, Sterpone S, Testa A, Cozzi R. Polymorphisms in base excision repair genes: Breast cancer risk and individual radiosensitivity. World journal of clinical oncology. 2014;5(5):874-882
9. Dianov GL, Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41(6):3483-3490
10. Sharma RA, Dianov GL. Targeting base excision repair to improve cancer therapies. Mol Aspects Med. 2007;28(3-4):345-374
11. Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends in biochemical sciences. 2012;37(4):162-172
12. Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, Balik E, Bulut T, Yamaner S, Bugra D. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27(7):1295-1302
13. Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12(7 Pt 1):2101-2108
14. Przybylowska K, Kabzinski J, Sygut A, Dziki L, Dziki A, Majsterek I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutat Res. 2013;745-746:6-15
15. Zhang SH, Wang LA, Li Z, Peng Y, Cun YP, Dai N, Cheng Y, Xiao H, Xiong YL, Wang D. APE1 polymorphisms are associated with colorectal cancer susceptibility in Chinese Hans. World J Gastroenterol. 2014;20(26):8700-8708
16. Sameer AS, Nissar S, Abdullah S, Chowdri NA, Siddiqi MA. DNA repair gene 8-oxoguanine DNA glycosylase Ser326Cys polymorphism and colorectal cancer risk in a Kashmiri population. DNA Cell Biol. 2012;31(4):541-546
17. Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomarkers. 2010;14(4):559-564
18. Curtin K, Samowitz WS, Wolff RK, Ulrich CM, Caan BJ, Potter JD, Slattery ML. Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(12):3384-3388
19. Sliwinski T, Krupa R, Wisniewska-Jarosinska M, Lech J, Morawiec Z, Chojnacki J, Blasiak J. No association between the Arg194Trp and Arg399Gln polymorphisms of the XRCC1 gene and colorectal cancer risk and progression in a Polish population. Exp Oncol. 2008;30(3):253-254
20. Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, Tsutou A, Tabuchi Y, Tanaka K, Yamamoto M. et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49
21. Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Le Neel T, Le Houerou C, Faroux R, Ollivry J, Lafraise B. et al. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008;8:326
22. Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM, Koh WP, Yu MC. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2363-2372
23. Hansen R, Saebo M, Skjelbred CF, Nexo BA, Hagen PC, Bock G, Bowitz Lothe IM, Johnson E, Aase S, Hansteen IL. et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229(1):85-91
24. Lai CY, Hsieh LL, Tang R, Santella RM, Chang-Chieh CR, Yeh CC. Association between polymorphisms of APE1 and OGG1 and risk of colorectal cancer in Taiwan. World J Gastroenterol. 2016;22(12):3372-3380
25. Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R. et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638(1-2):146-153
26. Tao H, Shinmura K, Suzuki M, Kono S, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K. et al. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 2008;99(2):355-360
27. Laev SS, Salakhutdinov NF, Lavrik OI. Inhibitors of nuclease and redox activity of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1). Bioorganic & medicinal chemistry. 2017;25(9):2531-2544
28. Kabzinski J, Majsterek I, Mik M, Dziki A, Dziki L, Maciejczak L. Impact of APEX Ile64val Gene Polymorphisms of DNA Repair Ber System on Modulation of the Risk of Colorectal Cancer in the Polish Population. Pol Przegl Chir. 2015;87(3):121-123
29. Berndt SI, Huang WY, Fallin MD, Helzlsouer KJ, Platz EA, Weissfeld JL, Church TR, Welch R, Chanock SJ, Hayes RB. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67(3):1395-1404
30. Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, Rzeszowska-Wolny J, Widlak P. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51(3):343-352
31. Santos JC, Funck A, Silva-Fernandes IJ, Rabenhorst SH, Martinez CA, Ribeiro ML. Effect of APE1 T2197G (Asp148Glu) polymorphism on APE1, XRCC1, PARP1 and OGG1 expression in patients with colorectal cancer. Int J Mol Sci. 2014;15(10):17333-17343
32. Li Y, Li S, Wu Z, Hu F, Zhu L, Zhao X, Cui B, Dong X, Tian S, Wang F. et al. Polymorphisms in genes of APE1, PARP1, and XRCC1: risk and prognosis of colorectal cancer in a northeast Chinese population. Med Oncol. 2013;30(2):505
33. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nature reviews Molecular cell biology. 2017;18(10):610-621
34. Alhadheq AM, Purusottapatnam Shaik J, Alamri A, Aljebreen AM, Alharbi O, Almadi MA, Alhadeq F, Azzam NA, Semlali A, Alanazi M. et al. The Effect of Poly(ADP-ribose) Polymerase-1 Gene 3'Untranslated Region Polymorphism in Colorectal Cancer Risk among Saudi Cohort. Dis Markers. 2016;2016:8289293
35. Kaufman BA, Van Houten B. POLB: A new role of DNA polymerase beta in mitochondrial base excision repair. DNA Repair (Amst). 2017;60:A1-A5
36. Dehe PM, Gaillard PH. Control of structure-specific endonucleases to maintain genome stability. Nature reviews Molecular cell biology. 2017;18(5):315-330
37. Liu L, Zhou C, Zhou L, Peng L, Li D, Zhang X, Zhou M, Kuang P, Yuan Q, Song X. et al. Functional FEN1 genetic variants contribute to risk of hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer. Carcinogenesis. 2012;33(1):119-123
38. Nissar S, Sameer AS, Rasool R, Chowdri NA, Rashid F. Polymorphism of the DNA Repair Gene XRCC1 (Arg194Trp) and its role in Colorectal Cancer in Kashmiri Population: a Case Control Study. Asian Pac J Cancer Prev. 2015;16(15):6385-6390
39. Dai Q, Luo H, Li XP, Huang J, Zhou TJ, Yang ZH. XRCC1 and ERCC1 polymorphisms are related to susceptibility and survival of colorectal cancer in the Chinese population. Mutagenesis. 2015;30(3):441-449
40. Yin G, Morita M, Ohnaka K, Toyomura K, Hamajima N, Mizoue T, Ueki T, Tanaka M, Kakeji Y, Maehara Y. et al. Genetic polymorphisms of XRCC1, alcohol consumption, and the risk of colorectal cancer in Japan. J Epidemiol. 2012;22(1):64-71
41. Skjelbred CF, Saebo M, Wallin H, Nexo BA, Hagen PC, Lothe IM, Aase S, Johnson E, Hansteen IL, Vogel U. et al. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study. BMC Cancer. 2006;6:67
42. Hong YC, Lee KH, Kim WC, Choi SK, Woo ZH, Shin SK, Kim H. Polymorphisms of XRCC1 gene, alcohol consumption and colorectal cancer. Int J Cancer. 2005;116(3):428-432
43. Abdel-Rahman SZ, Soliman AS, Bondy ML, Omar S, El-Badawy SA, Khaled HM, Seifeldin IA, Levin B. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159(1):79-86
44. Zhao Y, Deng X, Wang Z, Wang Q, Liu Y. Genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 and risk of colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13(2):665-669
45. Procopciuc LM, Osian G. Lys751Gln XPD and Arg399Gln XRCC1 in Romanians. Association with sporadic colorectal cancer risk and different stages of carcinomas. Chirurgia (Bucur). 2013;108(5):711-718
46. Poomphakwaen K, Promthet S, Suwanrungruang K, Chopjitt P, Songserm N, Wiangnon S. XRCC1 gene polymorphism, diet and risk of colorectal cancer in Thailand. Asian Pac J Cancer Prev. 2014;15(17):7479-7486
47. Stern MC, Siegmund KD, Corral R, Haile RW. XRCC1 and XRCC3 polymorphisms and their role as effect modifiers of unsaturated fatty acids and antioxidant intake on colorectal adenomas risk. Cancer Epidemiol Biomarkers Prev. 2005;14(3):609-615
48. Krupa R, Blasiak J. An association of polymorphism of DNA repair genes XRCC1 and XRCC3 with colorectal cancer. J Exp Clin Cancer Res. 2004;23(2):285-294
49. Gao CM, Ding JH, Li SP, Liu YT, Cao HX, Wu JZ, Tang JH, Tajima K. Polymorphisms in XRCC1 gene, alcohol drinking, and risk of colorectal cancer: a case-control study in Jiangsu Province of China. Asian Pac J Cancer Prev. 2014;14(11):6613-6618
50. Khan NP, Pandith AA, Yousuf A, Khan NS, Khan MS, Bhat IA, Nazir ZW, Wani KA, Hussain MU, Mudassar S. The XRCC1 Arg399Gln gene polymorphism and risk of colorectal cancer: a study in Kashmir. Asian Pac J Cancer Prev. 2013;14(11):6779-6782
51. Muniz-Mendoza R, Ayala-Madrigal ML, Partida-Perez M, Peregrina-Sandoval J, Leal-Ugarte E, Macias-Gomez N, Peralta-Leal V, Meza-Espinoza JP, Moreno-Ortiz JM, Ramirez-Ramirez R. et al. MLH1 and XRCC1 polymorphisms in Mexican patients with colorectal cancer. Genet Mol Res. 2012;11(3):2315-2320
52. Gsur A, Bernhart K, Baierl A, Feik E, Fuhrlinger G, Hofer P, Leeb G, Mach K, Micksche M. No association of XRCC1 polymorphisms Arg194Trp and Arg399Gln with colorectal cancer risk. Cancer Epidemiol. 2011;35(5):e38-41
53. Engin AB, Karahalil B, Karakaya AE, Engin A. Association between XRCC1 ARG399GLN and P53 ARG72PRO polymorphisms and the risk of gastric and colorectal cancer in Turkish population. Arh Hig Rada Toksikol. 2011;62(3):207-214
54. Wang J, Zhao Y, Jiang J, Gajalakshmi V, Kuriki K, Nakamura S, Akasaka S, Ishikawa H, Suzuki S, Nagaya T. et al. Polymorphisms in DNA repair genes XRCC1, XRCC3 and XPD, and colorectal cancer risk: a case-control study in an Indian population. J Cancer Res Clin Oncol. 2010;136(10):1517-1525
55. Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL. Association between polymorphisms of biotransformation and DNA-repair genes and risk of colorectal cancer in Taiwan. J Biomed Sci. 2007;14(2):183-193
56. Karam RA, Al Jiffry BO, Al Saeed M, Abd El Rahman TM, Hatem M, Amer MG. DNA repair genes polymorphisms and risk of colorectal cancer in Saudi patients. Arab journal of gastroenterology: the official publication of the Pan-Arab Association of Gastroenterology. 2016;17(3):117-120
57. Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends in genetics: TIG. 2012;28(11):566-573
58. Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nature reviews Molecular cell biology. 2014;15(7):465-481
59. Liu J, He C, Xing C, Yuan Y. Nucleotide excision repair related gene polymorphisms and genetic susceptibility, chemotherapeutic sensitivity and prognosis of gastric cancer. Mutat Res. 2014;765:11-21
60. Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst). 2015;36:13-18
61. Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nature reviews Cancer. 2005;5(7):564-573
62. Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, Martinez ME, Ahnen DJ, Sandler RS, Lance P, Stern MC. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30(3):472-479
63. Hansen RD, Sorensen M, Tjonneland A, Overvad K, Wallin H, Raaschou-Nielsen O, Vogel U. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res. 2007;619(1-2):68-80
64. Nemzow L, Lubin A, Zhang L, Gong F. XPC: Going where no DNA damage sensor has gone before. DNA Repair (Amst). 2015;36:19-27
65. Ahmad Aizat AA, Siti Nurfatimah MS, Aminudin MM, Ankathil R. XPC Lys939Gln polymorphism, smoking and risk of sporadic colorectal cancer among Malaysians. World J Gastroenterol. 2013;19(23):3623-3628
66. Liu D, Wu HZ, Zhang YN, Kang H, Sun MJ, Wang EH, Yang XL, Lian MQ, Yu ZJ, Zhao L. et al. DNA repair genes XPC, XPG polymorphisms: relation to the risk of colorectal carcinoma and therapeutic outcome with Oxaliplatin-based adjuvant chemotherapy. Mol Carcinog. 2012;51(Suppl 1):E83-93
67. Hua RX, Zhu J, Jiang DH, Zhang SD, Zhang JB, Xue WQ, Li XZ, Zhang PF, He J, Jia WH. Association of XPC Gene Polymorphisms with Colorectal Cancer Risk in a Southern Chinese Population: A Case-Control Study and Meta-Analysis. Genes. 2016;7(10):73
68. Mucha B, Pytel D, Markiewicz L, Cuchra M, Szymczak I, Przybylowska-Sygut K, Dziki A, Majsterek I, Dziki L. Nucleotide Excision Repair Capacity and XPC and XPD Gene Polymorphism Modulate Colorectal Cancer Risk. Clinical colorectal cancer. 2018;17(2):e435-e441
69. Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I, Leszczynski P, Grzebieniak Z, Sasiadek MM. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39(1):527-534
70. Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 2015;36(3):1549-1559
71. Paszkowska-Szczur K, Scott RJ, Gorski B, Cybulski C, Kurzawski G, Dymerska D, Gupta S, van de Wetering T, Masojc B, Kashyap A. et al. Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Mol Biol Rep. 2015;42(3):755-764
72. Steck SE, Butler LM, Keku T, Antwi S, Galanko J, Sandler RS, Hu JJ. Nucleotide excision repair gene polymorphisms, meat intake and colon cancer risk. Mutat Res. 2014;762:24-31
73. Kabzinski J, Przybylowska K, Dziki L, Dziki A, Majsterek I. An association of selected ERCC2 and ERCC5 genes polymorphisms, the level of oxidative DNA damage and its repair efficiency with a risk of colorectal cancer in Polish population. Cancer Biomark. 2015;15(4):413-423
74. Ni M, Zhang WZ, Qiu JR, Liu F, Li M, Zhang YJ, Liu Q, Bai J. Association of ERCC1 and ERCC2 polymorphisms with colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:4112
75. Chang WS, Yueh TC, Tsai CW, Ji HX, Wu CN, Wang SC, Lai YL, Hsu SW, Hsieh MH, Hsiao CL. et al. Contribution of DNA Repair Xeroderma Pigmentosum Group D Genotypes to Colorectal Cancer Risk in Taiwan. Anticancer Res. 2016;36(4):1657-1663
76. Rezaei H, Motovali-Bashi M, Khodadad K, Elahi A, Emami H, Naddaffnia H. Relationship between XPD Lys 751 Gln polymorphism and colorectal cancer risk: a case-control study in a population-based study. Gastroenterol Hepatol Bed Bench. 2013;6(1):18-24
77. Stern MC, Siegmund KD, Conti DV, Corral R, Haile RW. XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev. 2006;15(12):2384-2390
78. Moghtit FZ, Aberkane MS, Le Morvan V, Louhibi L, Bellot R, Bousahba A, Megaiz A, Fodil M, Mediene-Benchekor S, Zemani-Fodil F. et al. No association between XRCC3 Thr241Met and XPD Lys751Gln polymorphisms and the risk of colorectal cancer in West Algerian population: a case-control study. Med Oncol. 2014;31(5):942
79. Sliwinski T, Krupa R, Wisniewska-Jarosinska M, Pawlowska E, Lech J, Chojnacki J, Blasiak J. Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J Exp Med. 2009;218(3):185-191
80. Liu T, Huang J. Replication protein A and more: single-stranded DNA-binding proteins in eukaryotic cells. Acta biochimica et biophysica Sinica. 2016;48(7):665-670
81. Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, Levy M, Polakova V, Lipska L, Vodicka P. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33(7):1346-1351
82. Hou R, Liu Y, Feng Y, Sun L, Shu Z, Zhao J, Yang S. Association of single nucleotide polymorphisms of ERCC1 and XPF with colorectal cancer risk and interaction with tobacco use. Gene. 2014;548(1):1-5
83. Yang H, Li G, Li WF. Association between ERCC1 and XPF polymorphisms and risk of colorectal cancer. Genet Mol Res. 2015;14(1):700-705
84. Yueh TC, Chou AK, Gong CL, Fu CK, Pei JS, Wu MH, Tsai CW, Chang WS, Hsiao CL, Yen ST. et al. The Contribution of Excision Repair Cross-complementing Group 1 Genotypes to Colorectal Cancer Susceptibility in Taiwan. Anticancer Res. 2017;37(5):2307-2313
85. Zhang Q, Zheng X, Li X, Sun D, Xue P, Zhang G, Xiao M, Cai Y, Jin C, Yang J. et al. The polymorphisms of miRNA-binding site in MLH3 and ERCC1 were linked to the risk of colorectal cancer in a case-control study. Cancer medicine. 2018;7(4):1264-1274
86. Skjelbred CF, Saebo M, Nexo BA, Wallin H, Hansteen IL, Vogel U, Kure EH. Effects of polymorphisms in ERCC1, ASE-1 and RAI on the risk of colorectal carcinomas and adenomas: a case control study. BMC Cancer. 2006;6:175
87. Kabzinski J, Majsterek I, Dziki A, Mik M. The Role of the XPF Gene Polymorphism (Xrcc4) Ser835ser in the Risk of Malignant Transformation of Cells in the Colorectal Cancer. Pol Przegl Chir. 2015;87(2):83-85
88. Du H, Zhang X, Du M, Guo N, Chen Z, Shu Y, Zhang Z, Wang M, Zhu L. Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:6700
89. Hua RX, Zhuo ZJ, Zhu J, Zhang SD, Xue WQ, Zhang JB, Xu HM, Li XZ, Zhang PF, He J. et al. XPG Gene Polymorphisms Contribute to Colorectal Cancer Susceptibility: A Two-Stage Case-Control Study. Journal of Cancer. 2016;7(12):1731-1739
90. Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews Cancer. 2012;12(2):104-120
91. Li GM. Mechanisms and functions of DNA mismatch repair. Cell research. 2008;18(1):85-98
92. Friedhoff P, Li P, Gotthardt J. Protein-protein interactions in DNA mismatch repair. DNA Repair (Amst). 2016;38:50-57
93. Goellner EM, Putnam CD, Kolodner RD. Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst). 2015;32:24-32
94. Edelbrock MA, Kaliyaperumal S, Williams KJ. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res. 2013;743-744:53-66
95. Li G, Hu F, Yuan F, Fan J, Yu Z, Wu Z, Zhao X, Li Y, Li S, Rong J. et al. Intronic and promoter polymorphisms of hMLH1/hMSH2 and colorectal cancer risk in Heilongjiang Province of China. J Cancer Res Clin Oncol. 2015;141(8):1393-1404
96. Koessler T, Oestergaard MZ, Song H, Tyrer J, Perkins B, Dunning AM, Easton DF, Pharoah PD. Common variants in mismatch repair genes and risk of colorectal cancer. Gut. 2008;57(8):1097-1101
97. Raptis S, Mrkonjic M, Green RC, Pethe VV, Monga N, Chan YM, Daftary D, Dicks E, Younghusband BH, Parfrey PS. et al. MLH1 -93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J Natl Cancer Inst. 2007;99(6):463-474
98. Mik M, Dziki L, Malinowska K, Trzcinski R, Majsterek I, Dziki A. Polymorphism of MSH2 Gly322Asp and MLH1 -93G>A in non-familial colon cancer - a case-controlled study. Archives of medical science: AMS. 2017;13(6):1295-1302
99. Mrkonjic M, Raptis S, Green RC, Monga N, Daftary D, Dicks E, Younghusband HB, Parfrey PS, Gallinger SS, McLaughlin JR. et al. MSH2 118T>C and MSH6 159C>T promoter polymorphisms and the risk of colorectal cancer. Carcinogenesis. 2007;28(12):2575-2580
100. Zelga P, Przybylowska-Sygut K, Zelga M, Dziki A, Majsterek I. The 116G > A MSH6 and IVS1-1121C > T PMS2 Genes Polymorphisms Modulate the Risk of the Sporadic Colorectal Cancer Development in Polish Population. Pathol Oncol Res. 2017;24(2):231-235
101. Campbell PT, Curtin K, Ulrich CM, Samowitz WS, Bigler J, Velicer CM, Caan B, Potter JD, Slattery ML. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58(5):661-667
102. Tulupova E, Kumar R, Hanova M, Slyskova J, Pardini B, Polakova V, Naccarati A, Vodickova L, Novotny J, Halamkova J. et al. Do polymorphisms and haplotypes of mismatch repair genes modulate risk of sporadic colorectal cancer? Mutat Res. 2008;648(1-2):40-45
103. de Barros AC, Takeda AAS, Dreyer TR, Velazquez-Campoy A, Kobe B, Fontes MRM. DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway. Biochimie. 2018;146:87-96
104. Martinez-Uruena N, Macias L, Perez-Cabornero L, Infante M, Lastra E, Cruz JJ, Miner C, Gonzalez R, Duran M. Incidence of -93 MLH1 promoter polymorphism in familial and sporadic colorectal cancer. Colorectal Dis. 2013;15(3):e118-123
105. Nizam ZM, Abdul Aziz AA, Kaur G, Abu Hassan MR, Mohd Sidek AS, Yeh LY, Mazuwin M, Ankathil R. Contribution of the MLH1 -93G>a promoter polymorphism in modulating susceptibility risk in Malaysian colorectal cancer patients. Asian Pac J Cancer Prev. 2013;14(2):619-624
106. Nejda N, Iglesias D, Moreno Azcoita M, Medina Arana V, Gonzalez-Aguilera JJ, Fernandez-Peralta AM. A MLH1 polymorphism that increases cancer risk is associated with better outcome in sporadic colorectal cancer. Cancer Genet Cytogenet. 2009;193(2):71-77
107. Peng HX, Xu X, Yang R, Chu YM, Yang DM, Xu Y, Zhou FL, Ma WZ, Zhang XJ, Guan M. et al. Molecular analysis of MLH1 variants in Chinese sporadic colorectal cancer patients. Genet Mol Res. 2016;15(2):gmr7689
108. Haghighi MM, Taleghani MY, Mohebbi SR, Vahedi M, Fatemi SR, Zali N, Shemirani AI, Zali MR. Impact of EXO1 polymorphism in susceptibility to colorectal cancer. Genet Test Mol Biomarkers. 2010;14(5):649-652
109. Gao Y, Hayes RB, Huang WY, Caporaso NE, Burdette L, Yeager M, Chanock SJ, Berndt SI. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis. 2011;32(6):882-887
110. Le Guen T, Ragu S, Guirouilh-Barbat J, Lopez BS. Role of the double-strand break repair pathway in the maintenance of genomic stability. Molecular & cellular oncology. 2015;2(1):e968020
111. Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47(4):497-510
112. Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chemistry & biology. 2015;22(1):17-29
113. Tham KC, Kanaar R, Lebbink JH. Mismatch repair and homeologous recombination. DNA Repair (Amst). 2016;38:75-83
114. Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Progress in Biophysics and Molecular Biology. 2015;117(2-3):182-193
115. Naccarati A, Rosa F, Vymetalkova V, Barone E, Jiraskova K, Di Gaetano C, Novotny J, Levy M, Vodickova L, Gemignani F. et al. Double-strand break repair and colorectal cancer: gene variants within 3' UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget. 2016;7(17):23156-23169
116. Li JT, Zhong BY, Xu HH, Qiao SY, Wang G, Huang J, Fan HZ, Zhao HC. Associations between NBS1 Polymorphisms and Colorectal Cancer in Chinese Population. PLoS One. 2015;10(7):e0132332
117. Li XB, Luo H, Huang J, Zhang JD, Yang ZX, Sun XW. XRCC2 gene polymorphisms and its protein are associated with colorectal cancer susceptibility in Chinese Han population. Med Oncol. 2014;31(11):245
118. Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, Smith G, Bishop DT, Cox A, Camp NJ. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2476-2484
119. Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, Wasylecka M, Dziki L, Morawiec J, Blasiak J. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer-a case control study. Mol Biol Rep. 2011;38(4):2849-2854
120. Tranah GJ, Giovannucci E, Ma J, Fuchs C, Hankinson SE, Hunter DJ. XRCC2 and XRCC3 polymorphisms are not associated with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2004;13(6):1090-1091
121. Jin MJ, Chen K, Song L, Fan CH, Chen Q, Zhu YM, Ma XY, Yao KY. The association of the DNA repair gene XRCC3 Thr241Met polymorphism with susceptibility to colorectal cancer in a Chinese population. Cancer Genet Cytogenet. 2005;163(1):38-43
122. Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Br J Cancer. 2003;89(2):333-337
123. Nissar S, Sameer AS, Lone TA, Chowdri NA, Rasool R. XRCC3 Thr241Met gene polymorphism and risk of colorectal cancer in Kashmir: a case control study. Asian Pac J Cancer Prev. 2014;15(22):9621-9625
124. Mucha B, Przybylowska-Sygut K, Dziki AJ, Dziki L, Sygut A, Majsterek I. Association of Thr241Met polymorphism of XRCC3 gene with risk of colorectal cancer in the Polish population. Pol J Pathol. 2013;64(3):185-190
125. Romanowicz-Makowska H, Samulak D, Michalska M, Sporny S, Langner E, Dziki A, Sychowski R, Smolarz B. RAD51 gene polymorphisms and sporadic colorectal cancer risk in Poland. Pol J Pathol. 2012;63(3):193-198
126. Nissar S, Baba SM, Akhtar T, Rasool R, Shah ZA, Sameer AS. RAD51 G135C gene polymorphism and risk of colorectal cancer in Kashmir. Eur J Cancer Prev. 2014;23(4):264-268
127. Yazdanpanahi N, Salehi R, Kamali S. RAD51 135G>C polymorphism and risk of sporadic colorectal cancer in Iranian population. Journal of cancer research and therapeutics. 2018;14(3):614-618
128. Mucha B, Kabzinski J, Dziki A, Przybylowska-Sygut K, Sygut A, Majsterek I, Dziki L. Polymorphism within the distal RAD51 gene promoter is associated with colorectal cancer in a Polish population. Int J Clin Exp Pathol. 2015;8(9):11601-11607
129. Mucha B, Przybylowska-Sygut K, Dziki L, Dziki A, Sygut A, Majsterek I. Lack of association between the 135G/C RAD51 gene polymorphism and the risk of colorectal cancer among Polish population. Pol Przegl Chir. 2012;84(7):358-362
130. Naccarati A, Rosa F, Vymetalkova V, Barone E, Jiraskova K, Di Gaetano C, Novotny J, Levy M, Vodickova L, Gemignani F. et al. Double-strand break repair and colorectal cancer: gene variants within 3' UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget. 2015;7(17):23156-23169
131. Zhang L, Zhang Y, Tang CH, Su CM. RAD52 gene polymorphisms are associated with risk of colorectal cancer in a Chinese Han population. Medicine. 2017;96(49):e8994
132. Emami N, Saadat I, Omidvari S. Susceptibility to Colorectal Cancer and Two Genetic Polymorphisms of XRCC4. Pathol Oncol Res. 2015;21(4):881-885
133. Zhang L, Zhao J, Yu B, Song X, Sun G, Han L, Wang L, Dong S. Correlations between microsatellite instability, ERCC1/XRCC1 polymorphism and clinical characteristics, and FOLFOX adjuvant chemotherapy effect of colorectal cancer patients. Cancer genetics. 2017;218-219:51-57
134. Yang J, Wang X, Zou SM, Li HM, Xiao Q, Feng YR, Huang Y, Feng T, Chen JN, Lin DX. et al. [Genetic variations in MLH3 and MSH2 genes are associated with the sensitivity and prognosis in locally advanced rectal cancer patients receiving preoperative chemoradiotherapy]. Zhonghua Zhong Liu Za Zhi. 2018;40(6):433-440
135. He Y, Penney ME, Negandhi AA, Parfrey PS, Savas S, Yilmaz YE. XRCC3 Thr241Met and TYMS variable number tandem repeat polymorphisms are associated with time-to-metastasis in colorectal cancer. PLoS One. 2018;13(2):e0192316
Author contact
![]() Corresponding authors: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail: yuanyuanedu.cn
Corresponding authors: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact