Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(10):2312-2318. doi:10.7150/jca.30376 This issue Cite
Review
On The Role of Myelin and Lymphocyte Protein (MAL) In Cancer: A Puzzle With Two Faces
Department of Research in Biochemistry, Instituto Nacional de Enfermedades Respiratorias “Ismael Cosío Villegas”. Mexico City, 14080. Mexico.
Received 2018-10-2; Accepted 2019-2-7; Published 2019-5-21
Abstract
Myelin and lymphocyte protein (MAL) is an integral membrane protein constituent of lipid rafts, and it is implicated in apical transport of proteins in polarized epithelial cells. However, beyond the involvement of MAL in apical sorting and as its function as a raft stabilizer, it is still not totally clear how MAL participates in cell proliferating processes. More controversial and interesting is the fact that MAL has been implicated in carcinogenesis in two opposite ways. First, this protein is overexpressed in ovarian cancer and some kinds of lymphomas where it seems to favor cancer progression. Conversely, it has been reported that downregulation of the MAL gene by promoter hypermethylation is a hallmark of several adenocarcinomas. So far, there is not enough experimental evidence to help us understand this phenomenon, and no MAL mutations or MAL isoforms have been associated with these opposite functions. This review provides an updated summary of the structure and functions of MAL, and we will discuss the possible mechanisms underlying its roles as a tumor suppressor and a tumor progression factor.
Keywords: Myelin and lymphocyte protein, tumor suppressor, oncogene, lipid rafts
Introduction
Structure and biological roles of MAL. MAL belongs to a family that includes BENE, plasmolipin, and MAL-2 [1-3]. All these proteins share a MARVEL (MAL and related proteins for vesicle trafficking and membrane link) domain, which is highly conserved across species and is present in nearly 20 open reading frames (ORFs) in the human genome, including in occludins, physins, gyrins and the chemokine-like factor superfamily (CKLFSF) [4,5].The architecture of this domain is M-shaped, constituted by 4 helical trans-membrane segments spanning about 130 amino acids (aa) with cytoplasmic N- and C- terminal regions [4,5]. Despite its variable size the C-terminal moiety seems to be important in MARVEL-proteins. For example, occludins display a notably larger C-terminal domain compared with other proteins of the MARVEL family, and it is implicated in proper tight junction assembly, endocytosis, and it might be involved in basolateral transport [6]. In contrast, the C-terminus of MAL is short and consists of only 7 aa, but it includes some motifs required for its retrieval to the endoplasmic reticulum (ER) and endocytosis, particularly the tetra-peptide Leu-Ile-Arg-Trp which has a critical role in distributing and targeting MAL into membrane rafts (MR). In this sense, it was demonstrated that point mutations, deletions, insertions, including GFP fusion at C-terminus were reasons why MAL could be no longer be incorporated into MR resulting in misdistribution of MAL [7,8]. Literature regarding MAL and MARVEL domain structures is scarce; however Arthur and Stowell determined the 3 D structure of synaptophysin I by electron microscopy and single-particle three-dimensional (3 D) reconstitution [9]. This typical MARVEL protein forms hexameric basket-like complexes, resembling other known gap junction channels. In this context it is relevant to emphasize that MAL's MARVEL domain can self-associate yielding multimeric complexes, which in turn could participate in making membrane domains that may conceivably serve as platforms for apical delivery of larger proteins from the trans-Golgi network (TGN) than GPI-anchored proteins [10,11].
Alignment of MAL sequences including predicted amino acids interacting with CPET [29]. Alignment was performed with program Clustal W2 [91]. Residues colored in red are those predicted to establish contact with toxin by hydrophobic interactions, and residues colored in green, form hydrogen bonds with specific residues in toxin molecule. Bold letters indicates identity among analyzed species. Final row indicates: o, outside, m, intramembrane, and i, intracellular topology of MAL. Aligned species are from: Hs, Homo sapiens (Accession number: P21145), Bs, Bos Taurus (Accession number: Q3ZBYO), Rn, Rattus norvegicus (Accession number: Q64349), Cl, Canis lupus familiaris (Accession number: Q28296), Ch, Capra hircus (Accession number: XP_017910309), and Mf, Macaca fascicularis (Accession number: XP_014968319).

MAL is a 17 kDa protein resident of glycosphingolipid and cholesterol-enriched microdomains (GEMs) or MR. It is usually found at the TGN and vesicles derived from the TGN, early endosome (EE) membranes, and the apical region of plasma membrane (PM) [12]. The MAL gene was first identified in human T-lymphocytes [13], and almost ten years later MAL protein was discovered in myelin synthesizing cells [14,15]. MAL is part of the machinery for apical delivery of proteins, and constantly recycles from the TGN to the PM in MDCK and FRT cell lines [12,16-18]. Hepatocytes lack endogenous MAL expression, but they do express MAL-2, which, in contrast to MAL, delivers proteins to the basolateral domain. In fact, apical delivery of proteins in hepatocytes follows an “indirect” pathway in which apical proteins are first transferred to the basolateral membrane and are then internalized and transcytosed to the apical membrane domain [3]. Interestingly, experimental data demonstrated that transgenic expression of MAL in the hepatoma-derived hybrid cell line WIF-B restored trafficking of GPI-anchored apical proteins in to the “direct” pathway [19]. Other findings that emphasize the role of MAL in assembly, stabilization of MR, and protein targeting include the following: 1) Apical sorting and transport of influenza virus hemagglutinin in polarized cells require proper function of MAL [20]. 2) Oligodendrocytes from MAL-deficient mice showed altered trafficking and sorting of neurofascin 155 (NF155), Caspr, and the potassium channel Kv 1.2, resulting in paranodal-axon structural abnormalities [21]. 3) In human T-lymphocytes, MAL is essential for the sorting and transport into the PM of the Src-like tyrosine kinase, Lck (lymphocyte-specific protein tyrosine kinase), and participation of formin 2 (IFN2) is required [22]. In this sense, MAL is also important for T cell receptor-(TCR) mediated signaling, and regulates vesicle docking through microtubule-membrane interplay in T-cells [23,24]. 4) Moreover, it has been shown that interactions between MAL and glycosphingolipids are crucial in the formation of microdomains in myelin-forming cells and in epithelial cells from the kidney and stomach [25]. 5) More recently, it was demonstrated in T-cells that MAL is implicated in regulation of exosome trafficking and secretion. In fact MAL interacts with the HIV-Nef protein, increasing exosome release, and the lack of MAL impairs this process and has negative effects on the sorting of the exosomal marker CD63 [26]. However, in addition to these afore-mentioned functions, functions of MAL go further than being purely structural and sorting roles. Indeed, it was described that in ruminant animals, the Clostridium perfringens ε-toxin (CPET) is responsible for a serious central nervous system (CNS) disease which mostly affects the white matter, and is characterized by focal demyelination. Target cells are exclusively mature oligodendrocytes and MAL was identified as a necessary element for binding of CPET [27,28]. In vitro studies showed that human MAL (hMAL) is capable of binding CPET and the second extracellular loop (SEL, aa 115 to 125) seems to be involved in CPET-MAL interaction because the insertion of a FLAG tag in this domain totally abolished the hMAL-CPET interaction [27]. More recently, Khalili and coworkers [29] reported structural details of the CPET and MAL interactions following an in silico strategy. The results from a 2D-interaction plot revealed that the main interaction between CPET and MAL could involve residues V51, Q52 and W54 from the first extracellular loop (FEL) and aa from the second and third transmembrane helices. In this sense the authors proposed that although MAL is not the primary receptor for CPET, it could be the entry point into cytoplasm whether this toxin independently forms a pore or not. Differences in predicted CPET/MAL contact amino acids among distinct MAL animal sources are shown in Fig. 1. 2D-interaction plot results do not predict aa from the SEL making contact with CPET [29], however, disruption of this loop by introducing a FLAG tag could induce structural changes affecting toxin recognition [27]. Even though there are minimal changes in the predicted contact aa between rat MAL and human MAL, the rodent protein is approximately 100 times more sensitive to binding CPET [27]. When we compare homologous sequences implicated in MAL/CPET interaction from ruminants, rodent, and primates most of aa involved in the interaction are conserved [29] (Fig. 1). Structural myelin damage in rat and mouse brains has been described when purified CPET was administered experimentally, a fact which supports that CPET is also bound by rodent MAL in vivo [30]. No myelin-associated disease related to CPET has been clearly demonstrated in humans or rodents.
The Two Roles of Mal in Cancer
MAL as tumor a suppressor protein. The oldest reports found in Medline about the relationship between MAL and cancer came from studies performed in China in 1999 and 2000. In both cases, they reported that down-regulation of MAL was associated with esophageal cancer [31,32]. Afterwards, Mimori et al. confirmed the absence of MAL-mRNA in practically all esophageal carcinoma cell lines studied and in cells from other human carcinomas [33]. They demonstrated that the loss of expression of the MAL gene in esophageal carcinoma cells is due to promoter hypermethylation. In this context, chemical-demethylation of DNA restored cellular levels of MAL-mRNA, and transgenic expression of MAL in tumor cells and cell lines reduced tumor growth in nude mice, diminished cell motility, blocked G1/S transition, and increased expression of Fas receptor and apoptosis in vitro. All these effects point at MAL functioning as a tumor suppressor [33]. On the other hand, down-regulation of MAL alone [34] and both MAL and BENE was confirmed in cervical squamous cancer [35], similar to the loss of MAL expression in dysplastic esophageal lesions in humans and rats [36]. MAL-promoter hypermethylation and concomitant MAL-down-regulation have been described in other epithelial malignancies such colon cancer [37,38], breast cancer [39,40], stomach cancer [41,42], salivary gland cancer [43], other head and neck carcinomas [44,45], non-small cell lung cancer (NSCLC) [46], and bladder cancer [47]. In mammals, cytosine methylation occurs in highly repeated CpG dinucleotide regions called CpG islands, which are located in promoters and intragenic regions of many genes, mainly cell cycle regulators and tumor suppressors [48,49]. Two CpG islands, M1 and M2 have been described in the MAL gene. In head and neck squamous cell carcinoma, Cao and coworkers recognized the promoter region from -680 to -573, upstream from the +1 site as the M1 CpG-island, and M2 was located within the transcribed region, ranging from +390 to +669. However, in gastric cancer, despite M1 having nearly the same location, M2 was assigned closer to +1, from -92 to -7 [41]. This does not agree with the M2 island reported from head and neck carcinomas [45], and interestingly, even though the authors still consider MAL a tumor suppressor gene they showed that hypermethylation of M2, but not M1, correlates with a higher survival of patients with gastric cancer. Similar results were found in Merkel cell carcinoma, where lower expression of MAL was a significantly favorable overall survival parameter [50]. These controversial findings are a good starting point in order to describe the opposing functions of MAL in cancer.
MAL as a tumor progression factor. Around the same time that MAL expression was found to be decreased in esophageal carcinoma, overexpression of MAL was described as a specific feature of primary mediastinal large B-cell lymphoma (PMBL) [51,52], and high expression of MAL was also related to progression of acute adult T-cell leukemia/lymphoma (ATL) [53]. In both cases, higher expression of MAL was a hallmark that allowed the differentiation of PMBL from diffuse large B-cell lymphoma (DLBL) [54] and acute of from chronic ATL [53]. More recently, Hsi and coworkers established an adverse link between MAL expression and prognosis in patients with classic Hodgkin's lymphoma [55]. It is clear that MAL overexpression is a feature of some kinds of lymphocyte-derived cancers and we may think that it is different for epithelial cancers. However, MAL overexpression in ovarian carcinoma was reported in the early 2000's, and MAL was found to be among the most upregulated genes in clear cell-ovarian carcinoma [56]. Subsequently, it was demonstrated that MAL was a significantly upregulated gene in short-term survivors (less than 3 years) with invasive serous ovarian cancers [57]. Hypomethylation of the region mapped from -462 to -266 in the MAL gene promoter partially correlates with increased levels of MAL-mRNA in ovarian tumor cells as well as in ovarian carcinoma cell lines [58]. The results from these studies indicated that MAL enhances the neoplastic phenotype and drug-resistance by mechanisms that are still not totally clear. In this sense the “bad face” of MAL is observed in ovarian carcinoma and lymphomas; in both cases MAL expression is a poor prognosis factor.
Possible Mechanisms Involved in the Mal Dual Behavior
The mechanisms by which MAL can operate in two opposite directions are still elusive and poorly understood [59]. This is because several mechanisms could be involved in these processes and they may work independently in either one or the other direction, nonetheless we could find some clues to gain a better understanding of this dual behavior. MAL expression in T-lymphocytes was initially found only in late stages of development, it meaning in mature T-cells, and MAL-mRNA was not detected in proliferating T-cells [13]. On the other hand, MAL is not expressed in B-lymphocytes, but it is highly expressed in B-cell lymphomas [13,51,52,54]. MAL is also over-expressed in Jurkat cells (leukemia T-cells), indicating that abnormal expression of MAL in lymphocytes could be associated with cell-proliferation and transformation [23,60]. MAL is normally expressed in some tissues during fetal development. In rat kidneys and immature Schwann cells, MAL plays an important role in epithelial differentiation along with glycosphingolipids [25,61,62] and axonal segregation, respectively. Findings in transgenic mice overexpressing MAL revealed structural changes in the kidneys and the central and peripheral nervous system associated with increased cell proliferation [63]. On the other hand, the upregulation of the MAL gene was observed in a rat model of traumatic spinal cord injury, and interestingly, the increase in MAL expression was seen in neurons but not in oligodendrocytes [64]. As we mentioned before, in lymphocytes, MAL is essential in transport of acylated forms of Src-like kinases Lck and Fyn to the GEMs; in agreement with this, MAL gene silencing impairs kinases transport 22,65]. Interestingly, an increase in MAL expression of up to 30 times has been reported in Src-transformed cells [66], but physio-pathological consequences are not established. Thus, in lymphocytes, the proliferative effect associated with MAL overexpression could at least in part be mediated by the Lck-signaling pathway (Ras-MAPK-mediated cell proliferation) [67]; transformed lymphocytes could have permanently activated this pathway and MAL overexpression could favor cell-transformation through Ras activation [68] (Fig. 2). Therefore, as it has been noted in different studies, very accurate control of MAL expression in normal tissues is crucial in order to avoid deleterious effects caused by MAL overexpression [62-64, 69,70]. In fact, the expression of MAL in adult tissues is very selective and is also cell-type specific within each tissue [71]. So far, no evidence has been reported regarding point mutations in the MAL gene as causative of human cancers [44,61]. However, Beder and coworkers found that both loss of heterozygosity and hypermethylation of the MAL promoter are relevant mechanisms for down regulation of MAL in head and neck squamous carcinomas [44]. Thus, both hyper- and hypo-methylation of the MAL gene are associated with cancer progression, and so far, this dilemma only can be somewhat explained by the distinct expression levels of MAL in different cell types. We do not know why higher expression of this protein in leukemic and ovarian cancer cells is advantageous, but for most carcinomas it is not. Furthermore, to the best of our knowledge, there are no reports indicating MAL regulation by other epigenetic mechanisms such microRNAs. Hence, taking these findings together it is clear that MAL switching from tumor suppressor to pro-oncogenic factor perhaps is not because of an intrinsic property of the MAL protein; rather, it is probably related to the oncogenic activity of proteins such as Lck, which in turn are related to MAL and MR functions [67,72]. Other raft-associated proteins have been implicated in cancer such as caveolin-1, which is an important scaffold protein of caveolae [73,74]. These structures are abundant in the PM of endothelial cells where they can transport molecules from the blood into tissues, and intracellularly participate in signaling pathways in response to mechanical stress [75-78] and other signals [78,79]. Similar to MAL, caveolins have functional roles other than as scaffolds in lipid rafts, but caveolins have a wider distribution in organelle membranes than MAL, and they display different roles depending on the organelle [78]. At this point it is important to note that in polarized MDCK cells MAL and caveolin 1 occupy different membrane microdomains [80]. MAL can self-associate, and it could be possible that in the same way that caveolae allow the transport of molecules from the extracellular space into the cytoplasm, MAL-containing MR may be able to exert similar functions, as it happens for CPET [27-29]. These molecules can modify cellular processes through specific pathways, including cell proliferation, motility and survival. MAL could help modify the microenvironment favoring tumor development. In a mechanism comparable to the Epstein-Barr virus oncoprotein LMP-1, which self-associates in GEMs and activates MEK-ERK and PIk3-Akt pathways [81], MAL could favor cell transformation.
Conclusions and a New Perspective
From our current knowledge of MAL structure and functions, including those typical of GEMs, it is challenging to understand how this proteolipid could participate in oncogenesis or tumor progression in two conflicting ways. Moreover it is too risky try to associate any cancer-related activity of MAL without taking into account interacting proteins and their own functional roles. In this sense several signaling intermediaries in addition to no receptor tyrosine kinases (Src, Lck and Fyn), such as cellular receptors (TCR), cytoplasmic tyrosine kinases (ZAP-70), phospholipases (PLC-γ), small GTPases and others still unknown, could be involved in oncogenic activities of MAL- More experimental data are necessary in order to stablish any relationship between MAL and possible effectors. All these mechanisms correlate with pro-oncogenic properties of MAL; however, beyond RAF-mediated apoptosis (Fig. 2), little is known about the mechanisms that could be mediating its tumor suppressor activity. In this context a very interesting protein which we are interested in is a membrane mucin, MUC1. This mucin is overexpressed in several carcinomas [82-84]. In breast carcinoma cells Fanayan and coworkers demonstrated using the yeast two-hybrid system that MUC1 is a partner for MAL-2 [85]. This interaction could explain abnormal trafficking of MUC1 to the baso-lateral membrane in breast cancer cells [82,85]. In the same report, the authors also showed that MUC1 interacts with MAL; however, they did not discuss the interplay between MAL and MUC1. More recently, it was demonstrated that MUC1 increases hypermethylation and can repress the activity of tumor suppressor genes in cancer cells. This effect it is mediated by increasing the expression of DNA-methyl transferases 1 and 3b [86-88], and it could be very interesting to test if MUC1 turns off MAL gene expression in adenocarcinoma cells. With regard to this matter, in our laboratory we are studying the relationship between MAL and MUC1 using a transgenic model based in HEK293 cells stably transfected with a 22-VNTR-human MUC1 construct [89]. We have found a significant decrease in cellular levels of MUC1 by transiently transfecting with MAL-cDNA-containing plasmids [90]. This initial finding could account for a possible conflicting relationship between MAL and MUC1, and interfering with MUC1-oncogenic activity could be a mechanism by which MAL exerts its tumor suppressor activity (Fig. 2).
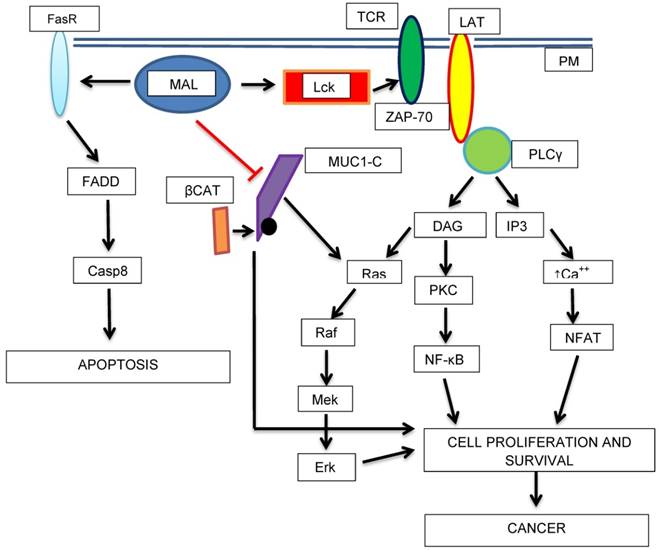
Schematic diagram showing the potential roles of MAL as a tumor progression factor and as a tumor suppressor. The transport of Lck (Lymphocyte-specific tyrosine kinase) to the PM in is mediated by MAL. Lck phosphorylates TCR (T-cell antigen receptor) and ZAP-70 (Z-chain-associated protein kinase 70) which in turn activates LAT (Linker of Activated T-cells). Then, cell-proliferation activating pathways are turned-on. On the other hand, MAL activates apoptosis (higher expression of Fas receptor, FasR or CD95). MAL could acts as a tumor suppressor protein by blocking the proliferative, and in turn, the oncogenic activity of MUC1-C-β-catenin complex.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Perez P, Puertollano R, Alonso MA. Structural and biochemical similarities reveal a family of proteins related to the MAL proteolipid, a component of detergent-insoluble membrane microdomains. Biochem Biophys Res Com. 1997;232:618-21
2. Wilson SH, Bailey AM, Nourse CR. et al. Identification of MAL2, a novel member of the mal proteolipid family, through interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics. 2001;76:81-8
3. de Marco MC, Martín-Belmonte F, Kremer L. et al. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 2002;159:37-44
4. Jin C, Ding P, Wang Y. et al. Regulation of EGF receptor signaling by the MARVEL domain-containing protein CKLFSF8. FEBS Lett. 2005;579:6375-82
5. Sánchez-Pulido L, Martı́n-Belmonte F, Valencia A. et al. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:597-01
6. Feldman GJ, Mullin JM, Ryan MP. Occludin: structure, function and regulation. Adv Drug Deliv Rev. 2005;57:883-17
7. Puertollano R, Alonso MA. A short peptide motif at the carboxyl terminus is required for incorporation of the integral membrane MAL protein to glycolipid-enriched membranes. J Biol Chem. 1998;273:12740-5
8. Caduff J, Sansano S, Bonnet A. et al. Characterization of GFP-MAL expression and incorporation in rafts. Microsc Res Tech. 2001;52:645-55
9. Arthur CP, Stowell MHB. Structure of Synaptophysin: a hexameric MARVEL-domain channel protein. Structure. 2007;15:707-14
10. Magal LG, Yaffe Y, Shepshelovich J. et al. Clustering and lateral concentration of raft lipids by the MAL protein. Mol Biol Cell. 2009;20:3751-62
11. Yaffe Y, Hugger I, Yassaf IN. et al. The myelin proteolipid plasmolipin form oligomers and induces liquid-ordered membranes in the Golgi complex. J Cell Science. 2015;128:2293-02
12. Puertollano R, Alonso M A. MAL, an integral element of the apical sorting machinery, is an itinerant protein that cycles between the trans-Golgi network and the plasma membrane. Mol Biol Cell. 1999;10:3435-47
13. Alonso MA, Weissman SM. cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc Natl Acad Sci USA. 1987;84:1997-01
14. Kim T, Fiedler K, Madison DL. et al. Cloning and characterization of MVP17: a developmentally regulated myelin protein in oligodendrocytes. J Neurosci Res. 1995;42:413-22
15. Schaeren-Wiemers N, Valenzuela DM, Frank M. et al. Characterization of a rat gene, rMAL, encoding a protein with four hydrophobic domains in central and peripheral myelin. J Neurosci. 1995;15:247-52
16. Cheong KA, Zacchetti D, Schneeberger EE. et al. VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc Natl Acad Sci USA. 1999;96:6241-8
17. Martin-Belmonte F, Kremer L, Albar J.P. et al. Expression of the MAL gene in the thyroid: the MAL proteolipid, a component of glycolipid-enriched membranes, is apically distributed in thyroid follicles. Endocrinology. 1998;139:2077-84
18. Martin-Belmonte F, Arvan P, Alonso MA. MAL mediates apical transport of secretory proteins in polarized epithelial Madin-Darby Canine Kidney Cells. J Biol Chem. 2001;276:49337-42
19. Ramnarayanan SP, Cheng ChA, Bastaki M. et al. Exogenous MAL reroutes selected hepatic apical proteins into the direct pathway in WIF-B cells. Mol Biol Cell. 2007;18:2707-15
20. Puertollano R, Martín-Belmonte F, Millán J. et al. The MAL Proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby Canine Kidney Cells. J Cell Biol. 1999;145:141-51
21. Schaeren-Wiemers N, Bonnet A, Erb M. et al. The raft-associated protein MAL is required for maintenance of proper axon-glia interactions in the central nervous system. J Cell Biol. 2004;166:731-42
22. Anton O, Batista A, Millan J. et al. An essential role for the MAL protein in targeting Lck to the plasma membrane of human T lymphocytes. J Exp Med. 2008;205:3201-13
23. Andres-Delgado L, Anton OM, Madrid R. et al. Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood. 2010;116:5919-29
24. Anton OM, Andres-Delgado L, Reglero-Real N. et al. MAL protein controls protein sorting at the supramolecular activation cluster of human T lymphocytes. J Immunol. 2011;186:6345-56
25. Frank M, van der Haar ME, Schaeren-Wiemers N. et al. rMAL is a glycosphingolipid-associated protein of myelin and apical membranes of epithelial cells in kidney and stomach. J Neurosci. 1998;18:4901-13
26. Ventimiglia LN, Fernández-Martín L, Martínez-Alonso E. et al. Cutting edge: Regulation of exosome secretion by the integral MAL protein in T cells. J immunol. 2015;195:810-4
27. Rumah KR, Ma Y, Linden JR. et al. The myelin and lymphocyte protein MAL is required for binding and activity of Clostridium perfringens ε-toxin. PLOS Pathogens. 2015;11:e1004896
28. Linden JR, Ma Y, Zhao B. et al. Clostridium perfringens epsilon toxin causes selective death of mature oligodendrocytes and central nervous system demyelination. mBio. 2015;6:e02513-14
29. Khalili S, Jahangiri A, Sadat Hashemi Z. et al. Structural pierce into molecular mechanism underlying Clostridium perfringens epsilon toxin function. Toxicon. 2016;127:90-9
30. Morris WE, Goldstein J, Redondo LM. et al. Clostridium perfringens epsilon toxin induces permanent neuronal degeneration and behavioral changes. Toxicon. 2017;130:19-28
31. Wang Z, Wang M, Xu X. et al. Studies of MAL gene in human esophageal cancer by RNA in situ hybridization. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2000;5:329-31
32. Xu Z, Wang M, Cai Y. MAL gene is down-regulated substantially in human esophageal cancer. Zhonghua Zhong Liu Za Zhi. 1999;4:250-2
33. Mimori K, Shiraishi T, Mashino K. et al. MAL gene expression in esophageal cancer suppresses motility, invasión and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene. 2003;22:3463-71
34. Overmeer RO, Henken FE, Bierkens M. et al. Repression of MAL tumor suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;219:327-36
35. Hatta M, Nagai H, Okino K. et al. Down-regulation of members of glycolipid-enriched membrane raft gene family, MAL and BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004;30:53-8
36. Mimori K, Nishida K, Nakamura Y. et al. Loss of MAL expression in precancerous lesions of the esophagus. Ann Surg Oncol. 2007;14:1670-7
37. Lind GE, Ahlquist T, Kolberg M. et al. Hypermethylated MAL gene a silent marker of early colon tumorigenesis. JTransl Med. 2008;6:13-23
38. Kalmár A, Péterfia B, Hollósi P. et al. DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1, PTGDR and SFRP1in colorectal adenoma and cancer. BMC Cancer. 2015;15:736-49
39. Horne HN, Lee PS, Murphy SK. et al. Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 2009;7:199-09
40. Guerrero-Preston R, Hadar T, Ostrow KL. et al. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 2014;32:505-12
41. Buffart TE, Overmeer RM, Steenbergen RDM. et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 2008;99:1802-7
42. Choi B, Han T-S, Min J. et al. MAL and TMEM220 are novel DNA methylation markers in human gastric cancer. Biomarkers. 2017;22:35-44
43. Maruya Sh-I, Kim H-W, Weber RS. et al. Gene expression screening of salivary gland neoplasms. J Mol Diag. 2004;6:180-90
44. Beder LB, Gunduz M, Hotomi M. et al. T-lymphocyte maturation-associated protein gene as a candidate metastasis suppressor for head and neck squamous cell carcinomas. Cancer Sci. 2009;100:873-80
45. Hutajulu SH, Indrasari SR, Indrawat LP. et al. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol Cancer. 2011;10:48-56
46. Suzuki M, Shiraishi K, Eguchi A. et al. Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol Rep. 2013;29:1308-14
47. Blaveri E, Simko JP, Korkola JE. et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044-55
48. Pfeifer GP. Defining driver DNA methylation changes in human cancer. Int J Mol Sci. 2018;19:1166-78
49. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010-22
50. Iwasaki T, Matsushita M, Nonaka D. et al. Lower expression of CADM1 and higher expression of MAL in Merkel cell carcinomas are associated with Merkel cell polyomavirus infection and better prognosis. Hum Pathol. 2016;48:1-8
51. Copie-Bergman C, Gaulard P, Maouche-Chretien L. et al. The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood. 1999;94:3567-75
52. Copie-Bergman C, Plonquet A, Alonso MA. et al. MAL Expression in Lymphoid Cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-Cell lymphomas. Mod Pathol. 2002;15:1172-80
53. Kohno T, Moriuchi R, Katamine S. et al. Identification of genes associated with the progression of adult T cell leukemia (ATL). Jpn J Cancer Res. 2000;91:1103-10
54. Chan JKC. Mediastinal large B-cell lymphoma: new evidence in support of its distinctive identity. Adv Anat Pathol. 2000;7:201-9
55. Hsi ED, Sup SJ, Alemany C. et al. MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Hematopathol. 2006;125:776-82
56. Schwartz DR, Kardia SLR, Shedden KA. et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722-9
57. Berchuck A, Iversen ES, Lancaster JM. et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Carcer Res. 2005;11:3686-96
58. Lee PS, Teaberry VS, Bland AE. et al. Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer. 2010;126:1378-89
59. Beder LB, Yamanaka N. MAL (mal, T-cell differentiation protein). Atlas Genet Cytogenet Oncol Haematol. 2011;15:331-5
60. [dataset] Rouillard AD, Gundersen GW, Fernandez NF. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. 2016; Article ID baw100.
61. Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274-81
62. Frank M, Schaeren-Wiemers N, Schneider R. et al. Developmental expression pattern of the myelin proteolipid MAL indicates different functions of MAL for immature Schwann cells and in a late step of CNS myelinogenesis. J Neurochem. 1999;73:587-97
63. Frank M, Atanasoski S, Sancho S. et al. Progressive segregation of unmyelinated axons in peripheral nerves, myelin alterations in the CNS, and cyst formation in the kidneys of myelin and lymphocyte protein-overexpressing mice. J Neurochem. 2000;75:1927-39
64. Zhang L, Cui Z, Shen A. et al. Upregulation of myelin and lymphocyte protein (MAL) after traumatic spinal cord injury in rats. J Mol Histol. 2013;44:125-34
65. Millan J, Alonso MA. MAL, a novel integral membrane protein of human T lymphocytes, associates with glycosylphosphatidylinositol-anchored proteins and Src-like tyrosine kinases. Eur J Immunol. 1998;28:3675-84
66. Shen Y, Chen C-S, Ichikawa H. et al. Src induces podoplanin expression to promote cell migration. J Biol Chem. 2010;285:9646-56
67. Alonso MA, Millán J. The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J Cell Sci. 2001;114:3957-65
68. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459-65
69. Schmid D, Zeis T, Sobrio M. et al. MAL overexpression leads to disturbed expression of genes that influence cytoskeletal organization and differentiation of Schwann Cells. ASN Neuro. 2014 July-September: 1-22
70. Buser AM, Schmid D, Kern F. et al. The myelin protein MAL affects peripheral nerve myelination: a new player influencing p75 neurotrophin receptor expression. Eur J Neuroscience. 2009;29:2276-90
71. Marazuela M, Alonso MA. Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histol Histopathol. 2004;19:925-33
72. Ventimiglia LN, Alonso MA. The role of membrane rafts in Lck transport, regulation and signalling in T-cells. Biochem J. 2013;454:169-79
73. Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494-06
74. Volonte D, Vyas AR, Chen Ch. et al. Caveolin-1 promotes the tumor suppressor properties of oncogene-induced cellular senescence. J Biol Chem. 2018;293:1794-09
75. Massey KA, Schnitzer JE. Caveolae and cancer. In: (ed.) Lierch R, Berdel WE, Kessler T. Angiogenesis Inhibition, Recent Results in Cancer Research. 1st. ed. Berlin Heidelberg: Springer-Verlag. 2010:217-31
76. Boyd NL, Park H, Yi H. et al. Chronic shear induces caveolae formation and alters ERK and Akt responses in endothelial cells. Am J Physiol Heart Circ Physiol. 2003;285:H1113-22
77. Park H, Go YM, Darji R. et al. Caveolin-1 regulates shear stress-dependent activation of extracellular signal-regulated kinase. Am J Physiol Heart Circ Physiol. 2000;278:H1285-93
78. Fridolfsson HN, Roth DM, Insel PA. et al. Regulation of intracellular signaling and function by caveolin. FASEB J. 2014;28:3823-31
79. Patel HH, Murray F, Insel PA. Caveolae as organizers of pharmacologically relevant signal transduction molecules. Annu Rev Pharmacol Toxicol. 2008;48:359-91
80. Millan J, Puertollano R, Fan L. et al. Caveolin and MAL, two protein components of internal detergent-insoluble membranes, are in distinct lipid microenvironments in MDCK cells. Biochem Biophys Res Com. 1997;233:707-12
81. Meckes DGJr, Menaker NF, Raab-Traub N. Epstein-Barr Virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol. 2013;87:1301-11
82. Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874-85
83. Lau SK, Weiss LM, Chu PG. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: an immunohistochemical study. Am J Clin Pathol. 2004;122:61-9
84. Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332-42
85. Fanayan S, Shehata M, Agterof AP. et al. Mucin 1 (MUC1) is a novel partner for MAL2 in breast carcinoma cells. BMC Cell Biol. 2009;10:7-19
86. Rajabi H, Tagde A, Alam M. et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35:6439-5
87. Rajabi H, Tagde A, Kufe D. MUC1-C drives DNA methylation in cancer. Aging. 2016;8:3155-6
88. Tagde A, Rajabi H, Stroopinsky D. et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget. 2016;7:38974-87
89. Kinlough CL, Poland PA, Bruns JB. et al. MUC1 membrane trafficking is modulated by multiple interactions. J Biol Chem. 2004;279:53071-7
90. Lara-Lemus R, Saldaña-Villa AK, Vázquez-Almazán B. La proteína de mielina y linfocitos y la mucina-1 en un modelo de expresión transgénica. Mens Bioquim. 2018;42:48-56
91. McWilliam HLW, Uludag M, Squizzato S. et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597-600
Author contact
![]() Corresponding author: Phone: 52 55 54871705. Email: betony44com
Corresponding author: Phone: 52 55 54871705. Email: betony44com

 Global reach, higher impact
Global reach, higher impact